Abstract
Rod-shaped bacteria elongate by the action of cell-wall synthesis complexes linked to underlying dynamic MreB filaments. To understand how the movements of these filaments relate to cell wall synthesis, we characterized the dynamics of MreB and the cell wall elongation machinery using high-precision particle tracking in Bacillus subtilis. We found that MreB and the elongation machinery moved circumferentially around the cell, perpendicular to its length, with nearby synthesis complexes and MreB filaments moving independently in both directions. Inhibition of cell wall synthesis by various methods blocked the movement of MreB. Thus, bacteria elongate by the uncoordinated, circumferential movements of synthetic complexes that insert radial hoops of new peptidoglycan during their transit, possibly driving the motion of the underlying MreB filaments.
The shape of most bacteria is maintained by the cell wall peptidoglycan (PG), a three-dimensional meshwork composed of glycan strands linked by peptide cross-bridges, but how collections of genes confer defined shapes and widths to this structure is unclear. The PG is a single macromolecule, and the mechanisms that localize the synthesis of this structure dictate the shape of the organism.
The peptidoglycan elongation machinery (PGEM) responsible for rod-shaped growth is composed of synthetic enzymes (called penicillin binding proteins (PBPs)) and conserved membrane proteins (MreC, MreD, RodA, RodZ). These proteins interact with MreB (1–4), a distant actin homolog that assembles into cytoplasmic filaments (5, 6). Depletions of PGEM proteins or disruption of MreB filaments produces round cells (7–9). MreB appears to form helical structures by light microscopy, which have been proposed to organize the PGEM to facilitate the construction of a rod shaped cell (10–12).
MreB is dynamic, and moves directionally in Bacillus subtilis (13, 14) and Caulobacter crescentus (15). However, it is unclear how this motion relates to cell wall synthesis. To explore the origin and function of MreB movement, we characterized and compared the relative dynamics of MreB and the PGEM in Bacillus subtilis.
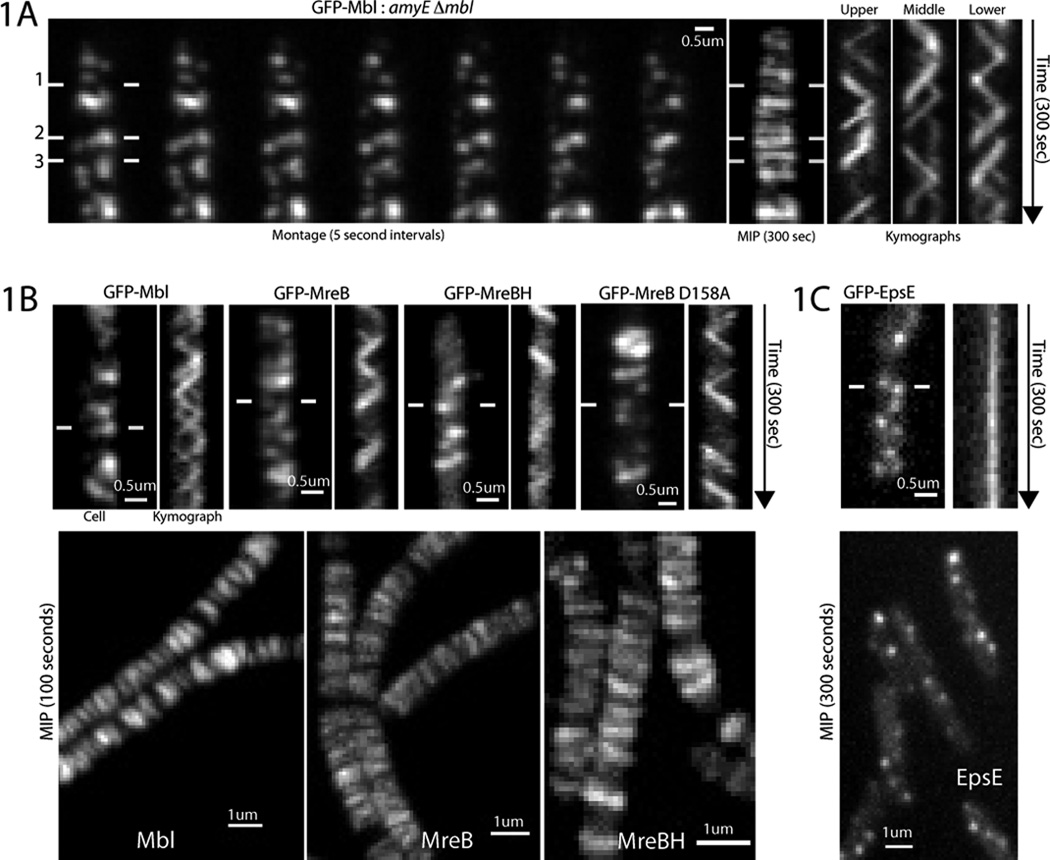
B. subtilis expresses three MreB paralogs (MreB, Mbl and MreBH). We began by imaging their dynamics with confocal microscopy. GFP-Mbl expressed as the sole source of Mbl in the cell displayed well-separated foci that moved linearly across the cell width (Fig. 1A, movies S1A-B). Maximal intensity projections of these movies revealed closely spaced horizontal bands perpendicular to the cell length, suggesting the predominant movement occurred as a rotation around the cell circumference. Kymographs drawn across the cell width generated diagonal lines, indicative of circumferential movement occurring at approximately constant velocity (22 ± 4nm/sec, n=40). Similar circumferential motion was observed with GFP-fusions to all three MreB paralogs regardless of expression context (Fig. 1B, movies S2A-B). The motions of MreB did not arise from rotation of the cell itself: imaging of MreB and EpsE (a protein associated with the flagellar bodies traversing the cell wall) demonstrated that MreB filaments rotate within a static cell (Fig. 1C, movies S3A-B).
Fig. 1. MreB paralogs display circumferential motion independent of the cell body.
(A) Left: Montage of GFP-Mbl motion (BDR2061) from movie S1B. Middle: Maximal Intensity Projection (MIP) of movie S1B. Right: Kymographs drawn between lines in montage. Under our growth conditions Bacillus grows in long septate chains.
(B) Top: Kymographs of GFP-Mbl, GFP-MreB, and GFP-MreBH in merodiploid strains. Far right: Kymograph showing axial motion of GFP-MreB(D158A), a mutation believed to inhibit ATP hydrolysis. Bottom: MIP of movies of GFP-Mbl, GFP-MreB, and GFP-MreBH.
(C) Top: Kymograph of EpsE-GFP. Bottom: MIP of an EpsE-GFP movie.
Previous studies have attributed the motion of MreB to polymerization dynamics or treadmilling (15, 16), models at odds with the observation that purified B. subtilis MreB displays no difference in polymerization in ADP and ATP bound states (5). To investigate the role of polymer dynamics in MreB movement, we imaged GFP-MreB containing two different mutations thought to perturb ATP hydrolysis (7, 16, 17). Consistent with an inhibition of MreB function, expression of these mutants resulted in perturbed cell morphologies (Fig. S1). However, these mutants displayed circumferential movements at speeds similar to those observed above (24±4, 26±3 nm/sec, n=25) (Figs. 1B, S1C, movies S2B, S4), suggesting that a mechanism other than polymerization dynamics drives MreB motion.
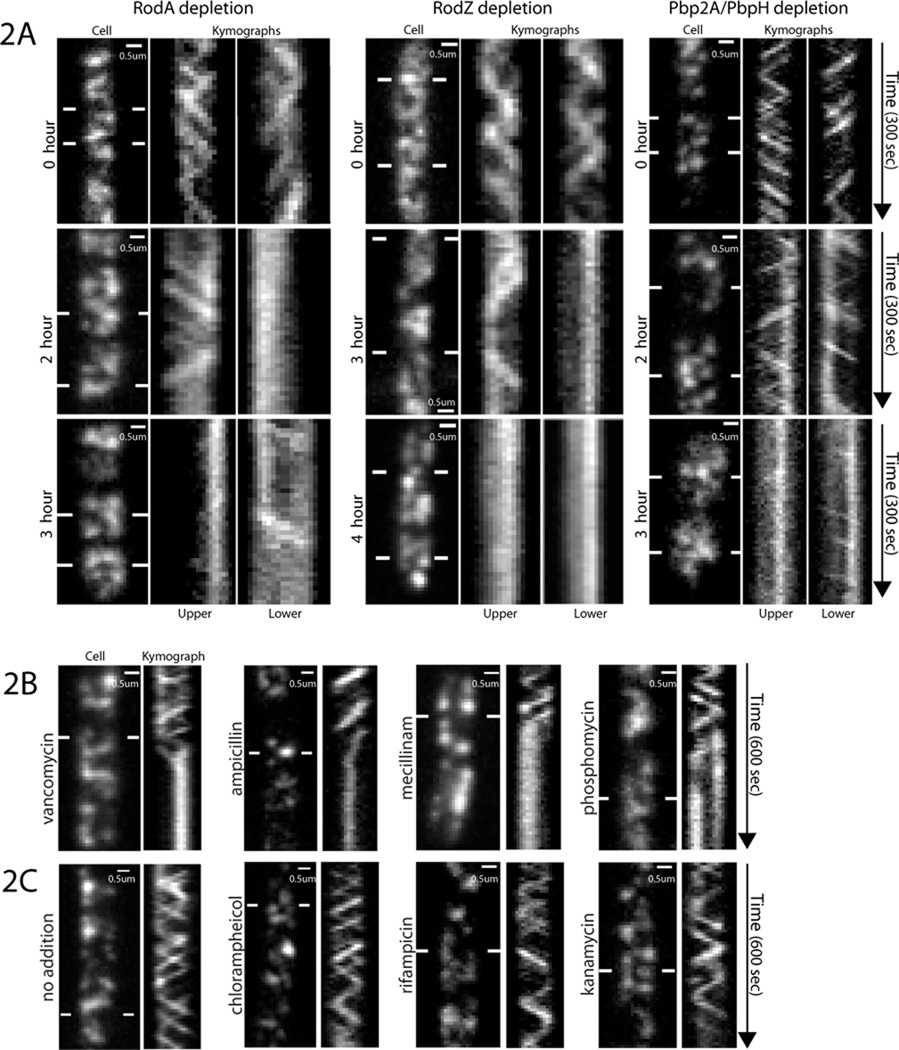
Because MreB interacts with the PGEM (1–4), we hypothesized that MreB movement could be driven by cell wall synthesis. To test this, we monitored GFP-Mbl dynamics while depleting three components of the PGEM: RodA, RodZ, and Pbp2A (Figs. 2A, S2, movies S5A-F). As these proteins depleted over time we observed a gradual cessation of movement. At late stages of depletion, the majority of Mbl was motionless. Notably, these experiments revealed a disconnected structure: at intermediate depletion states (~2 hours), cells displayed immobile filaments while adjacent particles still underwent rotary movement.
Fig. 2. Filament motion requires cell wall synthesis.
(Kymographs are drawn between lines).
(A) Kymographs of GFP-Mbl during depletions of IPTG-inducible genes: 1) RodA- a membrane-spanning component of the PGEM, 2) RodZ- a protein that links MreB to the PGEM and 3) Pbp2A- an elongation-specific transpeptidase, which was depleted in a strain lacking the redundant transpeptidase PbpH. Strains were grown in 2mM IPTG, shifted to media without IPTG, then imaged at the indicated times.
(B) Kymographs showing antibiotics targeting cell wall synthesis freeze GFP-Mbl motion. BDR2061 was imaged following addition of 2µL of antibiotics to a 600µL agar pad. Initial concentrations: 10mg/ml ampicillin (blocks transpeptidation), 5mg/ml mecillinam (blocks transpeptidation), 80µg/ml vancomycin (blocks transglycosylation and transpeptidation), 50mg/ml phosphomycin (blocks PG precursor synthesis, 6uL added).
(C) Kymographs showing off-target antibiotics do not affect GFP-Mbl motion. BDR2061 was incubated with indicated antibiotics for 2 minutes and immediately imaged. Final concentrations: 500µg/ml rifampicin (inhibits transcription), 500µg/ml kanamycin (inhibits translation), 340µg/ml chloramphenicol (inhibits translation).
Similar to genetic depletions, the addition of antibiotics targeting different steps in PG synthesis caused a cessation of filament motion (Fig. 2B, movies S6A-D). This effect was rapid: antibiotic addition to cells under thin agar pads completely stopped filament motion within 10–30 seconds. High concentrations of antibiotics that target other essential processes had no effect on filament movement (Fig. 2C, movies S7A-B), suggesting this effect is specific. Furthermore, the minimal concentrations (MIC) of antibiotics that stopped motion mirrored the minimal concentrations that inhibited cell growth (Fig. S3, Table S1, movies S8A-D), with short treatments near the MIC resulting in partially frozen filaments.
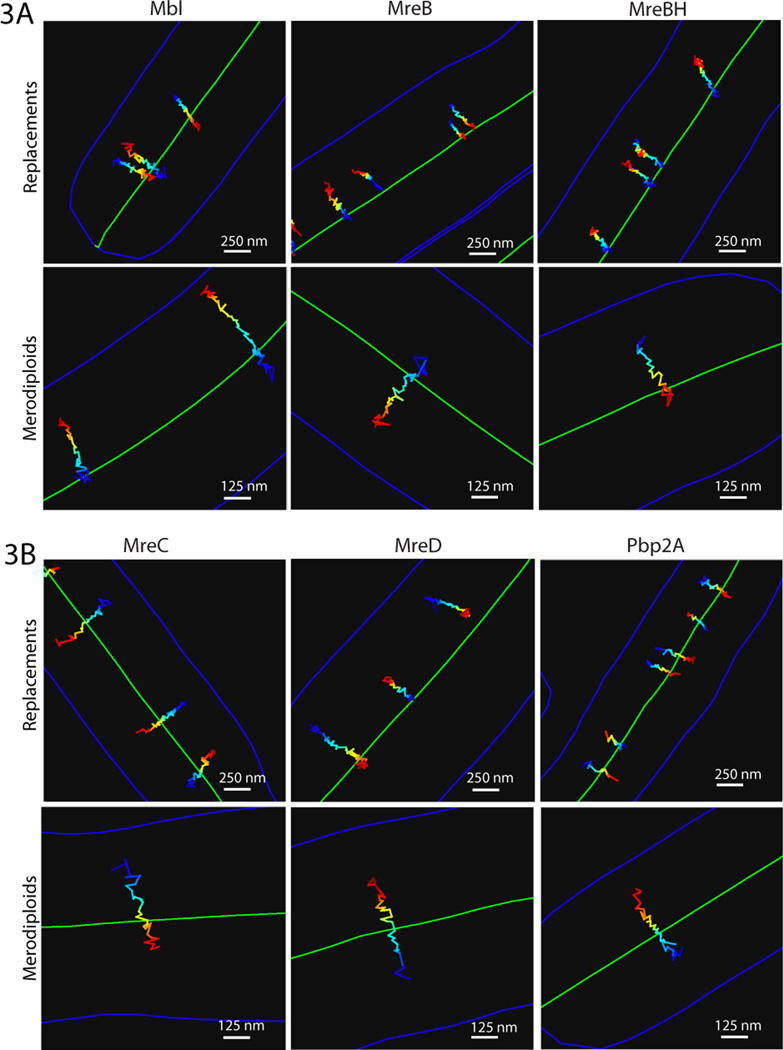
Thus, PG synthesis appears to drive the motion of MreB. For this hypothesis to be correct, both the PGEM and MreB paralogs should move around the cell body in a similar manner. To test this, we characterized the dynamics of the MreB paralogs and three of the PGEM components (MreC, MreD, Pbp2A) using high-precision particle tracking. We titrated the expression of GFP-fusions to low levels to obtain diffraction-limited foci, which we imaged using total internal reflection fluorescence (TIRF) microscopy to examine their dynamics on the bottom half of the bacterium. Low-level expression was obtained in two ways: 1) inducible expression in the background of endogenous protein (merodiploids); and 2) inducible expression as the only source of protein (replacements). The centroids of foci were fit to obtain positional information with sub-pixel precision (18) and their positions tracked over time (movie S9A). These studies demonstrated that all 6 proteins move in linear paths across the cell width (Fig. 3).
Fig. 3. Particle tracking of MreB paralogs and the PGEM shows linear movements across the cell.
Representative traces of (A) MreB paralogs and (B) PGEM components from each expression condition. Trace color encodes time (blue to red) in 300msec steps. Cell outline is blue, midline green. Low-level expression of MreB, Mbl, MreC, and MreD in replacements resulted in wider cells, which we stabilized with magnesium (Figs. S4-5). For the MreB paralogs both expression methods yielded large numbers of foci that moved in linear paths across the cell width (movies S9B-C). Expression of PGEM proteins via both methods revealed that PGEM foci partition into two populations, one moving slowly and directionally and one moving rapidly and non-directionally, which we interpret to be diffusion within the membrane. When expressed at high levels as replacements, a dense mix of both populations was observed. As PGEM expression levels were reduced, the diffusing population effectively disappeared, leaving predominantly directionally moving foci that traversed the cell width (movies S10A-C). When expressed at low levels in merodiploids, both populations of PGEM foci were observed, with the directionally moving population comprising the minority (movie S11). Because we could only accurately track the slow directionally moving particles, all our data refers to this population.
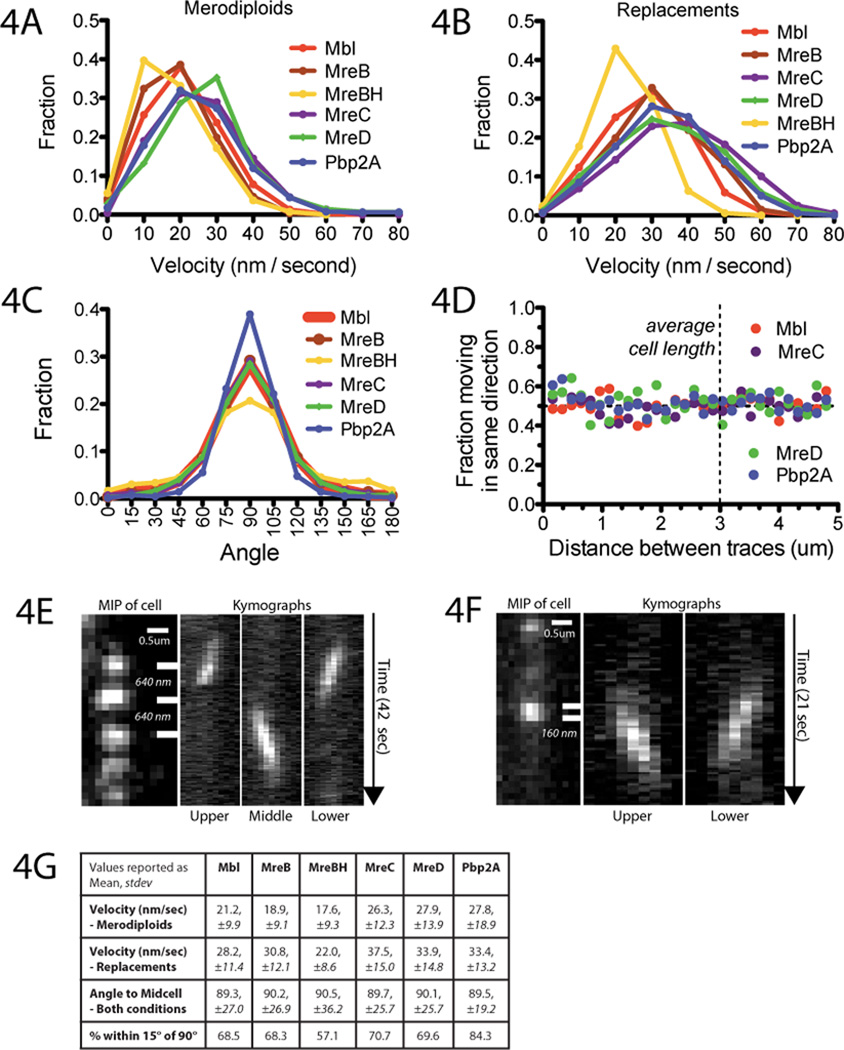
We determined the velocity for all traces over 20 frames in length by two methods. These analyses revealed that all 6 proteins move at similar speeds under each expression condition (Figs. 4A-B,G S6-8, Table S2). When the proteins were expressed at low levels as replacements their mean velocity increased (Fig. 4A vs. 4B), with the exception of the minor paralog MreBH (Fig. S9A). This increase in speed may arise from a cellular response to the reduction in the overall levels of the cell wall synthesis machinery, and we could reproduce this effect in trans by tracking merodiploid GFP-Mbl foci in an MreB deletion strain (Fig. S9B).
Fig. 4. Relative dynamics of the cell wall synthesis machinery and MreB.
Histograms of velocity of GFP-fusions expressed as (A) merodiploids and (B) replacements. Velocity (V) was calculated by fitting MSD vs. t (Fig. S6) to MSD=(Vt)2 + 4Dt, yielding two distinct populations, high (> 5 ×10−4 nm/sec) and low (≤ 5×10−4 nm/sec) (Table S2, Fig. S7). Displayed are high velocity traces that moved in a consistent manner during their lifetime (>0.95 r2-fit to log(MSD) vs. log(t)). Plots of all data without r2-screening are in Fig. S7.
(C) Distributions of the angles that traces cross the cell, determined by combining trajectories with segmented brightfield images (Fig. S10) (17). Shown are traces combined from both expression conditions over 20 frames in length with linear r2-fits > 0.5. Separate plots of each expression condition and different r2-screening criteria are in Fig. S11.
(D) Linear traces (as in 4C) extracted from 100 seconds of imaging were evaluated pairwise to determine their relative directionality. The fraction of traces moving in the same direction is plotted as a function of distance in bins of 160nm.
(E,F) Kymographs of proximal foci in merodiploid GFP-Mbl (E) and replacement GFPPpb2A (F) strains moving in opposing directions across one surface. Distance between kymograph bars is indicated in italics (See also: Fig. S15, movies S13A-D).
(G) Summary table of tracking data.
Next, we determined the angles at which these proteins crossed the cell. The mean angle was 90° for all proteins, with the majority of trajectories falling within 15° of this mean (Figs. 4C, 4G, S11). Analysis of the scaling exponents from the MSD vs. t plots indicated that all 6 proteins move in a directed manner (Fig. S12, Table S2). Thus, all 3 MreB paralogs and 3 PGEM components exhibit directed motions occurring at the same speed and angle to the cell body. Therefore, these proteins move in concert, functioning as a macromolecular unit (1–4). Consistent with this idea, vancomycin stopped the directed movements of all proteins: foci of MreB paralogs became immobile, as did chromosomal replacements of MreC and MreD. In contrast, directionally moving Pbp2A foci transitioned into a rapidly diffusive state (Fig. S13, movie S12A).
Similar to the fragmented structure suggested during PGEM depletions, our single-particle tracking data were not consistent with the existence of a coherent, long-range MreB cytoskeleton. Rather, the directional motions were often discontinuous and independent: foci of both the MreB paralogs and PGEM displayed pauses and reversals in motion, while adjacent foci continued to move unidirectionally (Fig. S14, movies S12B-D). These reversals could not have been due to movement of the proteins around the cell body, as our TIRF imaging only reports movements of foci on the bottom half of the cell.
Having observed discontinuous motions of single foci, we next examined the relative directionality between foci to test if their motions were coordinated along the cell length. We calculated the fraction of traces moving in the same direction over the imaging period as a function of their separation. We found no correlation even at the shortest separations: the relative orientations of all proteins were randomly distributed at all distances (Fig. 4D). This uncoordinated movement was evident in our tracking movies (movies S9A-C, S10A-C), and we have highlighted examples of proximal opposing movements (Figs. 4EF, S15, movies S13A-D). Thus, PGEM and MreB filaments move in both directions around the cell; and we could not resolve any coordination along the cell length.
It thus appears that the coupled motions of the PGEM and MreB reflect the active process of cell wall synthesis: a circumferential motion of disconnected MreB-PGEM complexes moving around the cell in both directions, synthesizing discrete radial bands of PG oriented perpendicular to the cell length (Fig. S16). This model is consistent with the arrangement of B. subtilis PG observed by AFM (19).
MreB filaments are required for elongation-specific PG synthesis (2, 20), suggesting they are integral components of these translocating machines. These filaments may serve as coordinating scaffolds to link the PGEM to the enzymes that synthesize PG precursors (1–4, 10) (Fig. S17). We cannot completely rule out the contribution of polymer dynamics to these motions, because there are no methods to inhibit MreB polymerization without disrupting existing filaments. However, we did not observe any directed motion in the absence of PG synthesis, even with high-precision measurements (Fig S13), suggesting that PG synthesis is the predominant process driving these motions. If these motions are driven by MreB polymerization or another process, this would require induction of equivalent rigor states during depletions of all PGEM components and the antibiotic inhibition of PG crosslinking, polymerization, and precursor synthesis.
Rather than a contiguous helical structure, these observations reveal the mobile, fragmented nature of MreB. Thus, while MreB is required for rod shape maintenance, it cannot function as a cell-spanning structure, much less a coherent “cytoskeleton” in B. subtilis. It remains to be determined how the short-range activities of these independent biosynthetic complexes impart a long-range order to the cell wall.
Supplementary Material
Acknowledgments
We thank P. Graumman, J. Errington, R. Carballido-Lopez, D. Kearns, D. Popham, and D. Scheffers for strains; D. Kahne, S. Walker, T. Bernhardt, and T. Uehara for discussions, Q. Justman for editing, S. Layer for inspiration, T. Emonet for MicrobeTracker, and the HMS Nikon Imaging Center for the use of their facilites. Funding from NIH grants: R01-GM073831 (Rudner), R01-GM096450 (Zhuang), and R01-GM39565 (Mitchison). XZ is a Howard Hughes Medical Investigator. ECG is funded by the Helen Hay Whitney Foundation.
Footnotes
Supporting Online Material
Materials and Methods
SOM Text
Figs. S1 to S17
Tables S1 to S5
Captions for Movies S1 to S13D
References
Supporting movies S1-S13D are availiable at http://mitchison.med.harvard.edu/cellwall/
References and Notes
- 1.Kruse T, et al. Mol Microbiol. 2005 Jan;55:78. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 2.Kawai Y, et al. Mol Microbiol. 2009 Aug;73:719. doi: 10.1111/j.1365-2958.2009.06805.x. [DOI] [PubMed] [Google Scholar]
- 3.Bendezu FO, et al. EMBO J. 2009 Feb 4;28:193. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White CL, et al. Mol Microbiol. 2010 May;76:616. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- 5.Mayer JA, Amann KJ. Cell Motil Cytoskeleton. 2009 Feb;66:109. doi: 10.1002/cm.20332. [DOI] [PubMed] [Google Scholar]
- 6.van den Ent F, et al. Nature. 2001 Sep 6;413:39. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 7.Kruse T, et al. EMBO J. 2003 Oct 1;22:5283. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones LJ, et al. Cell. 2001 Mar 23;104:913. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 9.Gitai Z, et al. Cell. 2005 Feb 11;120:329. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Figge RM, et al. Mol Microbiol. 2004 Mar;51:1321. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 11.Daniel RA, Errington J. Cell. 2003 Jun 13;113:767. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 12.Carballido-Lopez R. Microbiol Mol Biol Rev. 2006 Dec;70:888. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carballido-López R, Errington J. Dev. Cell. 2003 Jan 1;4:19. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- 14.Soufo HJ, Graumann PL. Curr Biol. 2003 Oct 28;13:1916. doi: 10.1016/j.cub.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 15.Kim SY, et al. Proc Natl Acad Sci U S A. 2006 Jul 18;103:10929. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Defeu Soufo HJ, Graumann PL. Mol Microbiol. 2006 Dec;62:1340. doi: 10.1111/j.1365-2958.2006.05457.x. [DOI] [PubMed] [Google Scholar]
- 17.See supporting material on Science Online.
- 18.Thompson RE, et al. Biophys J. 2002 May;82:2775. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayhurst EJ, et al. Proc Natl Acad Sci U S A. 2008 Sep 23;105:14603. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uehara T, Park JT. J Bacteriol. 2008 Jun;190:3914. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.