Abstract
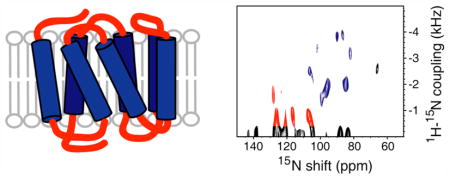
Solid-state NMR spectra with single-site resolution of CXCR1, a G protein-coupled receptor (GPCR), were obtained in magnetically-aligned phospholipid bicelles. These results demonstrate that GPCRs in phospholipid bilayers are suitable samples for structure determination by solid-state NMR. The spectra also enable studies of drug-receptor interactions.
G protein-coupled receptors (GPCRs) are prized targets for structure determination; however, with seven transmembrane helices and more than 300 residues, they are also among the most challenging. Strategically located in the membrane, these proteins regulate the physiological functions of cells in response to external chemical signals. The information is transmitted through the membrane by a change in conformation, and the resulting activation of a cognate G protein triggers myriad signaling pathways in the cytoplasm. About 1000 GPCRs have been identified in the human genome. Although most are for sensory functions, several hundred are potential drug receptors.
The structure of only one GPCR has been determined, and that is of rhodopsin1, which responds to photons and not chemical ligands. Although efforts are being made to model GPCRs on the structure of rhodopsin and with other computational methods2, it is essential to determine their individual three-dimensional structures in order to understand their mechanisms of action and for structure-based drug design. A number of NMR studies of rhodopsin have been reported that illustrate the complexities encountered in applying both solution NMR to micelle3 and magic angle spinning solid-state NMR to bilayer3a, 4 samples of polytopic membrane proteins. NMR studies of other GPCRs have been limited to polypeptides corresponding to portions of the proteins5, isolated following low temperature expression of a GPCR gene in bacteria6, or bound ligands7.
Our approach is to refold GPCRs expressed in bacteria as fusion proteins that form inclusion bodies8. By growing the bacteria in an optimized media formulation9, it is possible to obtain tens of milligrams of protein from each liter of culture without scrambling or loss of the isotopic labels. The purified GPCR is reconstituted into DMPC phospholipid vesicles, followed by the addition of short chain phospholipids (DHPC). The resulting bicelles provide a fully hydrated bilayer environment that both immobilizes and aligns the proteins for solid-state NMR experiments10. The line narrowing essential for high resolution results from the averaging effects of radiofrequency irradiations rather than molecular motions. In aligned samples, the protein structure is mapped directly onto the spectra, and the resonance frequencies provide orientation constraints as input for structure calculations.
Chemokines are small cytokines that control many biological and pathological processes, from immunosurveillance to inflammation, and from viral infection to cancer. The first chemokine receptors to be cloned were CXCR111 (Figure 1) and CXCR212, the two receptors for interleukin 8 (IL-8), following the finding of specific high-affinity binding of IL-8 to polymorphonuclear neutrophils (PMN) 13. The chemokine CXC ligand IL-8 and related agonists (GROa, neutrophil activating peptide 2, NAP-2) recruit and activate PMN by binding CXCR1 and CXCR2. As a result, IL-8 and related molecules have been implicated in a wide range of inflammatory disease states characterized by PMN infiltration in organs. Antichemokine strategies include antibodies, N-terminal modified chemokines, and small molecule antagonists. No small molecule inhibitors of CXCR1 are in an advanced clinical stage with the exception of repertaxin, an allosteric inhibitor of CXCR1 and CXCR214. Thus, any structural information that sheds light on either IL-8 antagonists or small molecule ligands of chemokine receptors that do not bind to the ‘classical’ GPCR sites would be extremely valuable.
Figure 1.
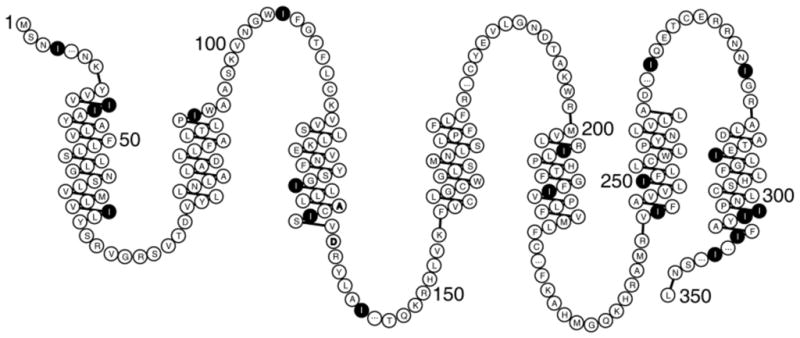
The amino acid sequence of CXCR1 arranged to show the transmembrane helices, loops, and terminal regions predicted by a hydropathy plot23. The locations of the isoleucine residues are marked.
The solid-state NMR spectrum of uniformly 15N labeled CXCR1 in magnetically-aligned bicelles is presented in Figure 2A. Significantly, there is only a small amount of signal intensity discernable at the isotropic frequency (~120 ppm), which indicates that nearly all residues are structured and immobilized along with the rest of the protein in the phospholipid bilayers. Even with carefully optimized cross-polarization conditions, it is possible that signals from some sites are missing due to the effects of motional averaging. Also, there is no evidence of residual “powder pattern” intensity, which demonstrates that this 350-residue GPCR undergoes rotational diffusion rapidly enough (D|| ≥ 105 s−1) to perform the averaging necessary to obtain single line resonances from bilayer samples with their normals perpendicular to the magnetic field10, 15.
Figure 2.
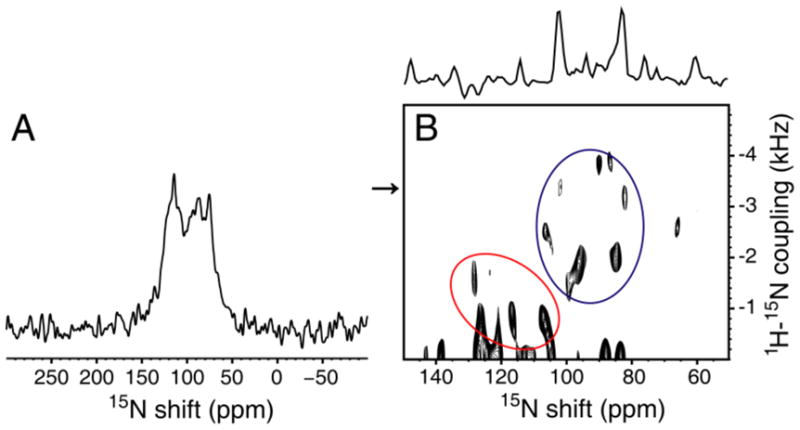
A. Solid-state NMR spectrum of uniformly 15N labeled CXCR1 in magnetically-aligned bicelles. B. Two-dimensional PISEMA spectrum of a 15N Ile labeled sample. The spectral slice through 3.4 kHz is aligned along the top of the spectrum. The spectra were obtained at 750 MHz on 150 μl samples with protein concentrations of 0.6 mM. The mix time was 1 msec, the acquisition time was 5 msec during which SPINAL modulated 1H decoupling was applied24, and the recycle delay was 6 sec. The data were processed with 100 Hz of exponential line broadening applied in t2 followed by Fourier transformation; Maximum Entropy reconstruction was applied to the full data set in t1 with no other apodization25.
PISEMA (Polarization Inversion Spin Exchange at the Magic Angle)16 yields high-resolution separated local field spectra where each resonance from a 15N-labeled amide site in the protein backbone is characterized by orientation-dependent heteronuclear 1H-15N dipolar coupling and 15N chemical shift frequencies. Helices result in characteristic PISA (Polarity Index Slant Angle)17 wheel patterns of resonances that reflect their tilt and polarity in the bilayers. The magnitudes of the chemical shift and dipolar coupling frequencies enable atomic-resolution structures of the proteins to be calculated18.
Figure 2B is a PISEMA spectrum of selectively 15N Ile labeled CXCR1 in bicelles. As indicated in Figure 1, approximately half of the Ile residues are in trans-membrane helices with the others in inter-helical loop or terminal regions. A blue circle denotes the PISA wheel portion (70 ppm–110 ppm) of the spectrum that contains the resonances from residues in the trans-membrane helices; there are nine signals in this region, two of which appear to be more intense than the others. It is reassuring to observe the wheel-like pattern from helical residues; however, because these signals are from residues in six different helices, it is not possible to determine helix tilt or polarity by inspection. Resonances from residues in loop and terminal regions that are likely to have irregular structures have chemical shifts in the 100 ppm–140 ppm range (outlined in red); there are about five signals that we would regard as reliable in this region. Signals with apparent dipolar couplings less than about 500 Hz may be artifacts due to mismatch of the 1H and 15N radiofrequency fields. Consistent with the one-dimensional spectrum in Figure 2, there is little or no intensity near the isotropic frequency that could be ascribed to mobile Ile residues. The spectral slice in Figure 2B includes the weakest signal (102 ppm, 3.4 kHz) in the trans-membrane helical region of the spectrum.
The spectrum in Figure 2B is an early result, and can be improved upon through further optimization of the sample conditions. Also, many aspects of the instrumentation and methods for experiments on aligned samples of proteins are under development and will contribute to higher resolution and sensitivity in the future. In particular, the implementation of triple-resonance and three-dimensional experiments should enable the resolution and assignment of individual resonances from uniformly labeled samples19. The spectrum in Figure 2B demonstrates the complementary use of selective isotopic labeling, which is particularly valuable in this context because the mapping of secondary structure onto the experimental spectra assists the assignment and structure determination processes.20 The ability to obtain spectra with single-site resolution, like those in Figure 2B, means that GPCRs in phospholipid bilayers are suitable samples for structure determination by solid-state NMR. Moreover, it is possible to utilize these spectra for screening of small molecule libraries and the application of SAR by solid-state NMR21, 22 for the development of drugs that target GPCRs.
Supplementary Material
Acknowledgments
This research was supported by Amgen, Inc., and the N.I.H. through grant RO1GM066978 and a postdoctoral fellowship to A.A.D. F32GM065833. It utilized the Biomedical Technology Resource for NMR Molecular Imaging of Proteins supported by grant P41EB002031.
Footnotes
Supporting Information Available: Sample information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 2.(a) Becker OM, Shacham S, Marantz Y, Noiman S. Curr Opin Drug Discov Devel. 2003;6:353–361. [PubMed] [Google Scholar]; (b) Trabanino RJ, Hall SE, Vaidehi N, Floriano WB, Kam VW, Goddard WA., 3rd Biophys J. 2004;86:1904–1921. doi: 10.1016/S0006-3495(04)74256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Getmanova E, Patel AB, Klein-Seetharaman J, Loewen MC, Reeves PJ, Friedman N, Sheves M, Smith SO, Khorana HG. Biochemistry. 2004;43:1126–1133. doi: 10.1021/bi030120u. [DOI] [PubMed] [Google Scholar]; (b) Klein-Seetharaman J, Getmanova EV, Loewen MC, Reeves PJ, Khorana HG. Proc Natl Acad Sci U S A. 1999;96:13744–13749. doi: 10.1073/pnas.96.24.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Klein-Seetharaman J, Reeves PJ, Loewen MC, Getmanova EV, Chung J, Schwalbe H, Wright PE, Khorana HG. Proc Natl Acad Sci U S A. 2002;99:3452–3457. doi: 10.1073/pnas.052713999. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Klein-Seetharaman J, Yanamala NV, Javeed F, Reeves PJ, Getmanova EV, Loewen MC, Schwalbe H, Khorana HG. Proc Natl Acad Sci U S A. 2004;101:3409–3413. doi: 10.1073/pnas.0308713101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Loewen MC, Klein-Seetharaman J, Getmanova EV, Reeves PJ, Schwalbe H, Khorana HG. Proc Natl Acad Sci U S A. 2001;98:4888–4892. doi: 10.1073/pnas.051633098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Crocker E, Patel AB, Eilers M, Jayaraman S, Getmanova E, Reeves PJ, Ziliox M, Khorana HG, Sheves M, Smith SO. J Biomol NMR. 2004;29:11–20. doi: 10.1023/B:JNMR.0000019521.79321.3c. [DOI] [PubMed] [Google Scholar]; (b) Eilers M, Reeves PJ, Ying W, Khorana HG, Smith SO. Proc Natl Acad Sci U S A. 1999;96:487–492. doi: 10.1073/pnas.96.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Patel AB, Crocker E, Reeves PJ, Getmanova EV, Eilers M, Khorana HG, Smith SO. J Mol Biol. 2005;347:803–812. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]; (d) Patel AB, Crocker E, Eilers M, Hirshfeld A, Sheves M, Smith SO. Proc Natl Acad Sci U S A. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Anderson LL, Marshall GR, Crocker E, Smith SO, Baranski TJ. J Biol Chem. 2005;280:31019–31026. doi: 10.1074/jbc.M503690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Zheng H, Zhao J, Wang S, Lin CM, Chen T, Jones DH, Ma C, Opella S, Xie XQ. J Pept Res. 2005;65:450–458. doi: 10.1111/j.1399-3011.2005.00239.x. [DOI] [PubMed] [Google Scholar]; (b) Katragadda M, Maciejewski MW, Yeagle PL. Biochim Biophys Acta. 2004;1663:74–81. doi: 10.1016/j.bbamem.2004.01.012. [DOI] [PubMed] [Google Scholar]; (c) Geng L, Wu J, So SP, Huang G, Ruan KH. Arch Biochem Biophys. 2004;423:253–265. doi: 10.1016/j.abb.2004.01.001. [DOI] [PubMed] [Google Scholar]; (d) Estephan R, Englander J, Arshava B, Samples KL, Becker JM, Naider F. Biochemistry. 2005;44:11795–11810. doi: 10.1021/bi0507231. [DOI] [PubMed] [Google Scholar]; (e) Choi G, Guo J, Makriyannis A. Biochim Biophys Acta. 2005;1668:1–9. doi: 10.1016/j.bbamem.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Tian C, Breyer RM, Kim HJ, Karra MD, Friedman DB, Karpay A, Sanders CR. J Am Chem Soc. 2005;127:8010–1. doi: 10.1021/ja051161b. (also 2006, 128, 5300) [DOI] [PubMed] [Google Scholar]
- 7.Luca S, White JF, Sohal AK, Filippov DV, von Boom JH, Grisshamer R, Baldus M. Proc Natl Acad Sci U S A. 2003;100:10706–10711. doi: 10.1073/pnas.1834523100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Koenig BW, Kontaxis G, Mitchell DC, Louis JM, Litman BJ, Bax A. J Mol Biol. 2002;323:441–461. doi: 10.1016/s0022-2836(02)00745-3. [DOI] [PubMed] [Google Scholar]
- 8.(a) Kiefer H, Maier K, Vogel R. Biochem Soc Trans. 1999;27:908–912. doi: 10.1042/bst0270908. [DOI] [PubMed] [Google Scholar]; (b) Kiefer H, Vogel R, Maier K. Receptors Channels. 2000;7:109–119. [PubMed] [Google Scholar]; (c) 01/14407. PCT Patent WO. 2000
- 9.Giesen AW, Bae LC, Barrett CL, Chyba JA, Chaykovsky MM, Cheng MC, Murray JH, Oliver EJ, Sullivan SM, Brown JM, Dahlquist FW, Homans SW. J Biomol NMR. 2001;19:255–260. doi: 10.1023/a:1011298531256. [DOI] [PubMed] [Google Scholar]
- 10.De Angelis AA, Nevzorov AA, Park SH, Howell SC, Mrse AA, Opella SJ. J Am Chem Soc. 2004;126:15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- 11.Holmes WE, Lee J, Kuang WJ, Rice GC, Wood WI. Science. 1991;253:1278–1280. [PubMed] [Google Scholar]
- 12.Murphy PM, Tiffany HL. Science. 1991;253:1280–1283. [PubMed] [Google Scholar]
- 13.(a) Grob PM, David E, Warren TC, DeLeon RP, Farina PR, Homon CA. J Biol Chem. 1990;265:8311–8316. [PubMed] [Google Scholar]; (b) Samanta AK, Oppenheim JJ, Matsushima K. J Exp Med. 1989;169:1185–1189. doi: 10.1084/jem.169.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertini R, et al. Proc Natl Acad Sci U S A. 2004;101:11791–11796. doi: 10.1073/pnas.0402090101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nevzorov AA, Opella SJ. Handbook of Modern Magnetic Resonance. Kluwer Academic; 2005. pp. 1–10. [Google Scholar]
- 16.Wu CH, Ramamoorthy A, Opella SJ. J Magn Reson. 1994;A 109:270–272. [Google Scholar]
- 17.(a) Marassi FM, Opella SJ. J Magn Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. J Magn Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 18.Nevzorov AA, Opella SJ. J Magn Reson. 2003;160:33–39. doi: 10.1016/s1090-7807(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 19.Vosegaard T, Nielsen NC. J Biomol NMR. 2002:225–247. doi: 10.1023/a:1014987227285. [DOI] [PubMed] [Google Scholar]
- 20.Kamihira M, Vosegaard T, Mason AJ, Straus SK, Nielsen NC, Watts A. J Struct Biol. 2005;149:7–16. doi: 10.1016/j.jsb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 22.Watts A. Nature Rev Drug Discovery. 2005;4:555–568. doi: 10.1038/nrd1773. [DOI] [PubMed] [Google Scholar]
- 23.Skranbanek L, Campagne F, Weinstein H. Nucleic Acids Res. 2003;31:3856–3858. doi: 10.1093/nar/gkg552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha N, Grant CV, Wu CH, De Angelis AA, Howell SC, Opella SJ. J Magn Reson. 2005;177:197–202. doi: 10.1016/j.jmr.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Jones DH, Opella SJ. J Magn Reson. 2006;178:91–99. doi: 10.1016/j.jmr.2005.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


