Abstract
A vast amount of data on the natural resistance of Saccharomyces cerevisiae to a diverse array of chemicals has been generated over the past decade (chemical genetics). We endeavored to use this data to better characterize the “systems” level properties of this phenomenon. By collating data from over 30 different genome-scale studies on growth of gene deletion mutants in presence of diverse chemicals, we assembled the largest currently available gene-chemical network. We also derived a second gene-gene network that links genes with significantly overlapping chemical-genetic profiles. We analyzed properties of these networks and investigated their significance by overlaying various sources of information, such as presence of TATA boxes in their promoters (which typically correlate with transcriptional noise), association with TFIID or SAGA, and propensity to function as phenotypic capacitors. We further combined these networks with ubiquitin and protein kinase-substrate networks to understand chemical tolerance in the context of major post-translational regulatory processes. Hubs in the gene-chemical network (multidrug resistance genes) are notably enriched for phenotypic capacitors (buffers against phenotypic variation), suggesting the generality of these players in buffering mechanistically unrelated deleterious forces impinging on the cell. More strikingly, analysis of the gene-gene network derived from the gene-chemical network uncovered another set of genes that appear to function in providing chemical tolerance in a cooperative manner. These appear to be enriched in lineage-specific and rapidly diverging members that also show a corresponding tendency for SAGA-dependent regulation, evolutionary divergence and noisy expression patterns. This set represents a previously underappreciated component of the chemical response that enables cells to explore alternative survival strategies. Thus, systems robustness and evolvability are simultaneously active as general forces in tolerating environmental variation. We also recover the actual genes involved in the above-discussed network properties and predict the biochemistry of their products. Certain key components of the ubiquitin system (e.g. Rcy1, Wss1 and Ubp16), peroxisome recycling (e.g. Irs4) and phosphorylation cascades (e.g. NPR1, MCK1 and HOG) are major participants and regulators of chemical resistance. We also show that a major subnetwork boosting mitochondrial protein synthesis is important for exploration of alternative survival strategies under chemical stress. Further, we find evidence that cellular exploration of survival strategies under chemical stress and secondary metabolism draw from a common pool of biochemical players (e.g. acetyltransferases and a novel NTN hydrolase).
INTRODUCTION
Elucidating gene functions is one of the most important challenges in genomics and computational biology. Recent technological advances in automation, allied with the genetic tractability of some model organisms (e.g. Saccharomyces cerevisiae), have made the genome-wide application of classical molecular biology and genetics techniques possible. A prime example of this revolution in functional genomics has been generation of homozygous and heterozygous gene deletion collections in budding yeast, representing partial and complete loss-of-function mutants, and their utilization in a variety of genetic studies 1, 2. Earlier studies on such mutant collections in S. cerevisiae indicated that ~17–20% of the yeast genes are essential (or indispensable) for growth under standard laboratory conditions 1, 2. While this straightforward characterization of the “essential gene complement” has a certain importance in understanding the basic organization of biological functions, terms like essentiality and dispensability are frequently misunderstood in the literature. There is a common tendency to associate essentiality identified in such experiments as a condition-independent assessment of the “importance” of a gene. It is in this context that researchers in the past few years are turning their focus to a more natural view of gene-essentiality, i.e. a condition-specific view 1–7. Here yeast mutant libraries have come of great use in assessing the role each gene might have in natural tolerance to varying environmental and chemical conditions 1, 2, 5, 8. In such studies, the fitness (equated with growth) of each mutant is measured in the presence or absence of a given condition (e.g. exposure to a drug in the growth medium). By testing hundreds of chemical and environmental conditions, a recent study reported that more than 95% of the yeast mutants exhibited altered growth in at least one environmental/chemical condition 5. This presents a very different view of gene function from that derived from the previous essentiality studies and offers new openings to decipher gene functions.
In yeast the number of genes with no direct experimental evidence is typically estimated as being over 1000. 9 Although a notable fraction of these proteins contain conserved domains 9 that might help in predicting their biochemistry, these conserved features might not be sufficient to understand their biological role. Furthermore, “uncharacterized” yeast genes are enriched in fungi-specific proteins that might not have conserved features of predictive value. It is here that the chemical genetic experiments might help elucidate functions of such “uncharacterized” genes. Gene expression profiling in S. cerevisiae revealed that genes transiently induced by different environmental stresses are also enriched for uncharacterized genes, suggesting that this subset might be of adaptive value in changing environments 10. Follow-up experiments on chemical profiling studies have indeed begun yielding more precise views of the gene/protein functions – for example ORF Ybr261c was predicted by means of sequence analysis to be a Rossmann fold methyltransferase. However, chemical genetic profiling with additional confirmatory experiments showed that it is likely to methylate components of the translation apparatus, thereby providing greater clarity on its function 11.
Although extremely powerful for uncovering gene functions, the yeast mutant collection has technical limitations that must be considered. For example, detection of growth differentials in naturally slow-growing mutants can be difficult, making assessments of the role of the gene in resistance uncertain. Different researchers might also use different growth conditions and employ different techniques to assess fitness, such as non-competitive growth in arrays and competitive growth in pools, in which barcoded cells are grown together 12. Hence, a certain level of discordance between studies is observed. Integration of data from different studies and obtained using different techniques and conditions is an excellent strategy to complement the limitations and make use of the advantages of each methodology. For example, a study demonstrated that the integration of chemical-genetic data with synthetic genetic interactions can indeed provide useful information on pathways targeted by growth-inhibitory chemicals 13, 14. More recently, Hoon et al described a miniaturized platform comprising three different gene-dosage assays, which was shown to improve the experiments sensitivity and specificity, with potential applications in drug target identification 15.
We have recently shown that the integration of chemical genetics, protein-protein interaction and protein complex data, in conjunction with comprehensive sequence analysis of the protein components of these complexes reveals a previously unexpected role of particular functional systems as natural buffers against chemical stress in yeast 16. We realized that other such computational analysis of the yeast chemical profiling data, which combines sensitive sequence analysis with novel network representations of this information, can provide new insights into the organization of the chemical-stress resistance mechanisms in cells. In the present study we assembled the hitherto largest gene-chemical tolerance dataset (measured using relative fitness) derived from chemical-genetic studies performed over a decade. We represent this data as the chemical phenotype network (CPnet) and used it to uncover some critical principles governing yeast tolerance to bioactive chemical substances. We further overlaid this network with other sources of genome-scale data, such as transcriptional and translational noise, presence/absence of TATA-box, TFIID-/SAGA-dominance status and phenotypic capacitance (see Materials and methods for details) to: (i) elucidate different systems involved in multidrug resistance; (ii) assess roles of two major post-translational modifications, i.e. phosphorylation and ubiquitination, in chemical stress tolerance; (iii) construct a shared chemical phenotype network (SCPnet) that links genes involved in resistance to a significant number of common chemicals. We identified a remarkable absence of overlap between the hubs in the SCPnet and multidrug resistance genes (MDRs), pointing to the presence of a previously unrecognized underlying structure in the yeast response to chemical stress, i.e. deployment of two distinct sets of genes, namely those that by themselves confer tolerance to diverse chemicals (MDRs) and those that functionally partner with a diverse gene set to enable the cell to explore alternative survival strategies when faced with adverse chemicals. Additional analyses were performed to uncover potential biochemical mechanisms by which the SCPnet allows tolerance against chemical stress and provide clues on how evolution molded the yeast genome to keep robustness while maintaining its plasticity.
RESULTS AND DISCUSSION
Systems-level representation of the S. cerevisiae chemical response
Assembly of the chemical phenotype network: multi-drug resistance and its regulation by post-translational modifications
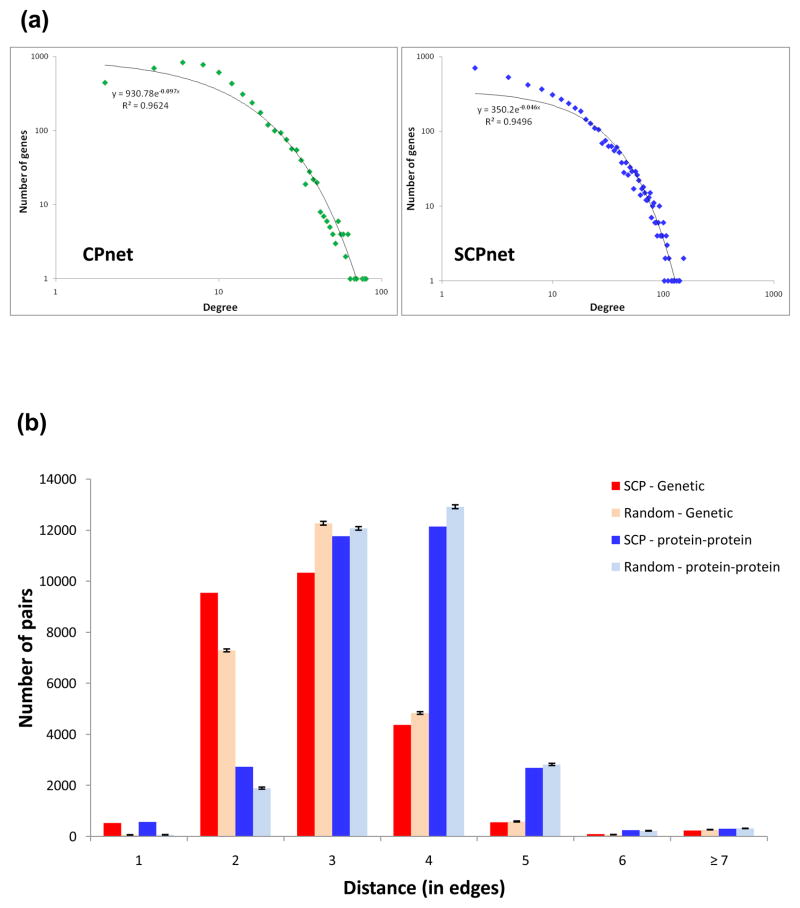
We collated the results from 34 original chemical-genomics (see Materials and methods for references) studies to obtain a comprehensive list of genes whose deletion (either heterozygous or homozygous) causes a growth defect in the respective deletion strains in the presence of a given chemical as opposed to the same strain grown in absence of the chemical. Although the bulk of the data came from large scale studies that simultaneous assayed a diverse list of compounds, we also included as many single-chemical studies as we could, especially given that they provided information on chemicals that were absent from the larger screens. In scoring a gene as being required in natural tolerance to a given chemical, we entirely relied on the assessment of the authors of the primary studies rather than apply our own criteria for this decision. While it is clear that there would be a certain subjectivity and difference in the criteria applied to make this decision in the individual studies, we chose to be inclusive so as to not miss functional effects that might be apparent only under certain conditions. For consistency in the overall rationale employed in this study (see below), we only included those gene deletants that compromised fitness, and not those that conferring increased fitness to chemicals. The final list of genes and chemicals was the non-redundant collation (union) of these 34 datasets and, to our knowledge, represents the largest assembly of such chemical-fitness data. To conceptualize this data we chose a network representation wherein we connected a gene to a chemical against which it conferred tolerance or natural resistance. We term the resultant bimodal network of genes and chemicals the chemical phenotype network (CPnet). The CPnet contains 5,233 ORFs and 425 distinct compounds (nodes) linked by 54,769 edges (Additional file 1) (see Materials and methods for details). In contrast to the ubiquitous power-law degree distributions of biological networks, the degree distribution of the CPnet can be partially approximated by an exponential distribution (y = 930.78e−0.097x, R2 = 0.96 and mode=5; Fig. 1a). We used the transition in slope of the tangent to the degree distribution curve towards the horizontal asymptote (i.e. m > −1) as the threshold to define highly connected genes or hubs in the CPnet. We identified 517 hubs in the CPnet – they are genes required for growth in the presence of multiple chemicals; hence we term them MDR genes (MDRs). Consistent earlier studies (whose datasets were included in our compendium) 3, 5, transporters, transcription factors and proteins involved in endosomal trafficking figure among the MDRs identified in our analysis. For example, transmembrane transporters are significantly over-represented among the MDR genes (Fisher Exact Test (FET); p ≈ 1.2×10−3), suggesting the potential importance of direct efflux as a component of natural resistance. In line with the results of the largest individual dataset and a directed study on heavy metal resistance 5, 17, the endosomal sorting complexes required for transport (ESCRT) and retromer complexes are also major players in the MDR set, suggesting that endosomes-to-vacuole and the retrograde endosome-to-Golgi trafficking of proteins are required for tolerance to a wide range of compounds 5.
Figure 1.
General structural properties of the CP and SCP networks. (a) Degree distribution of the CP (left, green) and SCP (right, blue) networks – both are best fit by exponential distributions. The cut-offs to define hubs are indicated in the figure. (b) We measured the distance between every SCPnet pair of genes (genes forming linked by an edge) in the genetic (red) and protein-protein (blue) interaction networks. Only 2.43% of the SCPnet edges are directly supported by PPIs or GIs. This fraction increases to 30% when one bridging node is allowed in the PPI or GI networks (d = 2). By using degree-preserving rewired replicates of the SCPnet (light-red and light-blue), we no longer observed this trend. These second-degree interactions are more frequent in the GI network, suggesting that SCPnet edges are likely to reflect gene interactions in common pathways, even without close physical interaction between their encoded proteins.
By combining these results with our recent reconstruction of the yeast ubiquitin network 18, we found that the above-defined MDRs are preferentially ubiquitinated relative to the other gene products in the CPnet (FET; p ≈ 3×10−4) (table 1). This is compatible with the well-known role for multiple ubiquitination events in endosomal trafficking and cargo sorting mediated by the ESCRT machinery 19. We also observed that 31 of MDRs are themselves components of the Ub pathway. Of these proteins nearly 50% are specifically involved in regulation of vesicular trafficking or the Golgi complex, and were parts of a distinct densely connected sub-graph that we identified in our previous analysis of the Ub network 18. Like ubiquitination, we also found an enrichment for phosphorylation events (p ≈ 3.2×10−3) among MDRs, suggesting that major players in generic chemical tolerance might be under the regulation of key protein kinase cascades (see below for additional details). Further, the ubiquitinated and phosphorylated MDR proteins present a marginally significant overlap (p ≈ 0.0147) and there might be a certain degree of interplay between the two major post-translational modification events in regulation of the general resistance. However, a more objective assessment of the significance of the above results would require data on condition-specific post-translational modifications (see below).
Table 1.
Genomic and functional features of MDRs and SCP-hubs.
| Feature | MDR | SCP hubs | References |
|---|---|---|---|
| Ubiquitination | ↑ p ≈ 3 × 10−4 | ↓ p ≈1.1 × 10−6 | 18 |
| Phosphorylation | ↑ p ≈ 3.2 × 10−3 | ↓ p ≈ 1.2 × 10−5 | 120–122 |
| TATA box presence a | ↓ p ≈ 0.013 | ↑ p ≈ 10−4 | 30 |
| SAGA dominance a | ↓ p ≈ 2.6 × 10−3 | ↑ p ≈ 0.05 | 31 |
| Phenotypic capacitance | ↑ p ≈ 1.4 × 10−5 | ↓ p ≈ 3.6 × 10−4 | 40 |
| Stress-induced genes | ↓ p ≈ 0.01 | ↑ p ≈ 0.018 | 33 |
| Mediator Srb10 repression | ↓ p ≈ 1.6 × 10−3 | ↑ p ≈ 0.042 | 32 |
| Uncharacterized ORFs | ↓ p ≈ 2.1 × 10−6 | ↑ p ≈ 8.3 × 10−7 | SGD |
| Fast divergence in gene expression | ↓ p ≈ 8.6 × 10−4 | ↑ p ≈ 0.0167 | 34 |
We used a binary classification for SAGA/TFIID regulation and hence the significance levels of the TATA-containing and SAGA-dominated genes enrichment in hSCPs also reflect the under representation of TATA-less and TFIID-dominated genes. The arrows ↑ and ↓ represent over- and under-representation and the p-values were obtained by using the Fisher Exact Test.
Analysis of MDRs reveals novel roles for components of multiple intracellular trafficking systems in chemical tolerance
One of the MDRs that emerged in our analysis is the EH-module-UBA domain protein Ede1, which has been recently shown to be an important player in receptor internalization 20. Based on these observations we propose that a mechanism by which intracellular trafficking system could facilitate chemical tolerance is via receptor down-regulation. Ubiquitination also regulates formation of vesicles from early endosomes that join the trans-Golgi network (TGN), allowing cargos to be recycled to the plasma membrane 21, 22. A key player in this phenomenon is Rcy1, a F-box protein subunit of ubiquitin E3-ligases 23 that emerges as a MDR in our analysis. Rcy1 and its binding partner Skp1 have been previously shown to play a critical role in recycling the v-SNARE protein Snc1 from early endosomes 24. The Rcy1-mediated membrane protein recycling process also involves the phospholipid translocase Cdc50-Drs2 and Ypt31/Ypt32, two closely related Rab GTPases that also affect the half-life and localization of Rcy121. Remarkably, Ypt31 and Ypt32 show striking dissimilarity in their chemical genetics profiles. Ypt31 is required for tolerance to 39 chemicals in our dataset, whereas its paralog is linked to only a single chemical. This strongly suggests a functional divergence between closely related paralogs, with Ypt31 being specifically coupled to Rcy1 in chemical resistance processes. Prior studies show that Rcy1 is part of a canonical Skp1-Cullin-F-box (SCF) complex involving Rcy1, Cdc53 and Hrt1 18, 25. However, Cdc53 and Hrt1 are not highly connected in the CPnet, which is consistent with previous data suggesting their non-involvement in endosomal trafficking 24. Taking these results together, we postulate that Rcy1 is specifically targeted to the trafficking system through the involvement of Ypt31 and mediates distinct ubiquitin-related regulatory events independently of its characterized SCF partners. Thus, Rcy1 could be part of a recycling process of membrane permeases that serve as another mechanism by which endosomal sorting pathways might play a role in generic chemical resistance.
The CPnet also helps to clarify the potential role of another distinct trafficking pathway in general chemical resistance. The EH-module containing calcium-binding protein Irs4p was reported by Hillenmeyer et al 5 as a major drug resistance gene and since this dataset is included in our analyses, we also found Irs4 as a highly connected MDR. The Irs4Δ endocytosis-related phenotypes were previously attributed to the deletion of the regulatory region of a neighboring gene (in the opposite strand), Vps51, which encodes a protein with key roles in recycling several cargoes 26. Irs4 is linked to roughly three times more chemicals than Vps51 (77 and 22, respectively) and the overlap between their chemical profiles is lower than 50%. This suggests that indeed Irs4 has important, Vps51-independent, roles in chemical tolerance. Indeed, recently, Irs4 and its close paralog Tax4 have been shown to lack a endocytotic function but have been implicated in: 1) positively regulating the phosphatase Inp51 to reduce the levels of the second messenger phosphatidylinositol 4,5-bisphosphate 27 and 2) autophagy (formation of a double-membraned vesicle that is subsequently degraded in the vacuole/lysosome) and pexophagy (comparable degradation of the peroxisome) 26. We observed that while the phosphatase Inp51 shared over 60% of the linked chemicals with Irs4, the latter had over three times the connections to chemicals than the former. Further, its paralog Tax4 is only required for resistance to a small number of chemicals. These observations imply that, in contrast to previous suggestions of Irs4 and Tax4 being functionally redundant 26, they are likely to have distinct functions, with Irs4 specializing in chemical resistance processes. Further, the observation that Irs4 is required for tolerance to a far greater set of chemicals than its functional partner Inp51 indicates that the bulk of its tolerance function is most likely via its role in the autophagy pathway. This suggests that in addition to starvation and pathogen response, autophagy and pexophagy might be important general shields against chemical stress effects (see below for further evidence). Thus, our observations indicate that multiple independent intracellular trafficking systems appear to provide distinct generalized survival strategies against environmental chemicals.
The shared chemical phenotype network (SCPnet): uncovering the undergirding of yeast chemical tolerance
Genes with similar chemical profiles reveal novel aspects of the yeast chemical stress response
The CPnet emphasizes the relative importance of individual genes in tolerating a diverse set of substances. It is likely that chemical resistance also requires positive functional interaction between multiple genes to be completely effective, an aspect that is not entirely clear from the CPnet. In this regard we reasoned that genes with significant overlaps in their chemical-genetic profiles might throw light on distinct pathways or potential cooperation between them in tolerance to chemical stresses. Accordingly, we took the CPnet as a starting point, compared the chemical-genetic profiles of every pair of genes, and identified those that shared a higher number of links to chemicals than expected by chance alone. Such genes were represented as nodes of a network and linked by edges, which formally indicate significant similarity in their chemical-genetic profiles. This network was termed the shared chemical phenotype network (SCPnet). The statistical significance of interactions leading to an edge in the SCPnet was computed by a simulation that created 1,000 degree-preserving random CPnets (see Materials and methods for details). The SCPnet has 4,631 genes linked by 40,102 edges (each edge defined by interactions with p ≤ 10−3; Fig 1a, Additional file 2). Interestingly, we observed that only 2.43% of the edges in the SCPnet are mirrored in the protein-protein interaction (PPI) or genetic interaction (GI) networks (i.e. distance (d) = 1, Fig 1b). However, this fraction increases to 30% (or 12,070 edges) when allowing one bridging node in the PPI or GI networks to connect a given pair of genes in the SCPnet (d = 2, Fig 1b). This trend is not observed when the same analysis is performed using degree-preserving rewired replicates of the SCPnet (Fig 1b). Moreover, these second-degree connections are more frequent in the GI than in the PPI network (Fig 1b). Despite not being immediate neighbors, the topological closeness of SCPnet nodes in the PPI and GI networks, indicates that they are likely to participate in common sub-systems even if their gene products do not exhibit close physical interaction. Further, greater representation of topologically close pairs in the GI networks hints that these functional relationships are likely to represent previously unrecovered genetic interactions and other more subtle interactions within a shared functional context.
Like CPnet, the degree distribution of the SCPnet can be partially approximated by an exponential distribution (y = 350.2e− 0.046x and R2 = 0.9496) (Fig 1a). We used this distribution to define the hubs in the SCPnet just as in the CPnet. There were 670 of these, henceforth called SCP-hubs. We investigated the resistance of the SCPnet to random deletion of nodes or preferential removal of hubs (failure and attack, respectively) 28. The SCPnet is considerably more resistant to attack than the GI network and modestly more resistant than the PPI, indicating the possibility of backup among the SCP-hubs (Additional file 3). In most biological networks (e.g. PPI and GI), hubs show a tendency to connect to their low-degree counterparts; hence, these networks are called dissortative 29. In contrast, the SCPnet is highly assortative (assortativity = 0.314), with SCP-hubs forming a strongly connected component. Importantly, the assortativity is completely absent in simulated degree-preserving randomly rewired replicates of the SCPnet (the maximum achieved value was 0.0075). Hence, the observed assortativity is a distinctive property of the real SCPnet and not merely a consequence of its degree distribution. This led us to the question of the biological relevance of this high assortativity in the context of natural chemical tolerance. Since each connection in the SCPnet is based on chemical interactions shared by the two nodes, it is likely that the number of edges a given chemical is involved is proportional to the systems “complexity” required to allow growth in the presence of the chemical. Here “complexity” is defined in an inclusive sense, encompassing all kinds of functional interactions, such as physical inter-molecular interactions, regulatory interactions and compensatory pathways. To explore this idea further, we used the randomized degree-preserving SCPnets to investigate two issues: (i) if the total number of chemicals covered in the randomized networks is the same when compared to the real network; (ii) for each of the covered chemicals, how many edges (gene-gene interactions in the SCPnet) are involved in the tolerance to the given substance. The randomized SCPnets covered approximately 199 chemicals in average, never reaching the 312 chemicals present in the real SCPnet. Furthermore, by comparing the real SCPnet and each randomized network, we noticed that, in addition to the higher coverage of chemicals, the real SCPnet also has a greater number of edges involving the covered chemicals (Additional file Fig 4). Therefore, the assortativity and robustness to attack in the SCPnet arises from a distinctive hidden structure that represents the strong functional interactions between a core set of genes (SCP-hubs) in simultaneously providing tolerance to a wide range of substances.
Hubs in the CP and SCP networks have complementary nature and distinct features
We were interested in understanding the relationship between the SCP-hubs and the hubs in the CPnet (i.e. MDRs). Surprisingly, we discovered that the overlap between the MDRs and SCP-hubs is much lower than expected by chance -- only 29 SCP-hubs are also MDRs; such a low overlap is not recovered in any of 10,000 randomly generated replicates of MDRs and hSCPs. Further investigation showed that MDRs genes and SCP-hubs show highly distinct and often opposing trends across several independent metrics (table 1). For instance, SCP-hubs tend to harbor TATA boxes in their upstream regions, whereas MDRs tend to be TATA-less (table 1). The presence of a TATA box in the promoter regions affects the recruitment of pre-initiation complex: TATA-containing genes preferentially recruit SAGA, whereas TATA-less genes are mainly occupied by TFIID (respectively termed SAGA- and TFIID-dominated genes 30, 31). Keeping with this inference, we also found promoters of SCP-hubs showing a bias towards being SAGA-dominated (table 1), with an exactly opposite trend observed in MDRs, where there is a dominance of TFIID occupancy. SCP-hubs are also enriched in targets of the RNA-polymerase associated mediator complex subunit, the general stress attenuator kinase Srb10 32, suggesting that expression of SCP-hubs are under a general regulatory switch mediated by the opposing actions of Srb10 and SAGA. By analyzing transcriptional profiles, Gasch et al. previously identified a set of genes potentially involved in the adaptation to various environmental stresses 33. Interestingly, the MDRs and SCP-hubs are respectively under- and over-represented among these environmental stress-induced genes (table 1), which correlates with the role of SAGA-mediated transcription in environmental stress responses 31, 33. We also found MDR genes to have a highly constrained gene expression, with virtually no divergence during long-term evolution in poor medium 34, which is consistent with their TATA-less promoters. In contrast, there is an inverse tendency in SCP-hubs (table 1), which is a feature related to the enrichment of TATA-containing genes in their midst 30. TATA-containing promoters were suggested to mediate noisy gene expression, possibly providing adaptive advantages under environmental stress conditions 35–37. Previous studies also showed that there is little correspondence between the genes induced by environmental and radiation stress with those required for sustained growth under the same stress condition 38, 39. Based on this precedence and the above properties of the hubs in the two networks, we propose an explanation for their opposing trends and paradox in terms of their functions: MDRs are likely to be the genes that are typically required for persistent growth in presence of the various chemicals, mounting general responses to them. The SCP-hubs seem to be distinct in providing a cooperative genetic apparatus that prepares the cell for an altered state, exploring different survival strategies in the presence of the chemical. Regulation of SCP-hubs via SAGA binding to TATA-containing promoters potentially allows their noisy expression as a part of the exploration of various biochemical alternatives in search of one that might favor survival. The more stable expression of MDRs, on the other hand, is probably critical for them serving as a sustained defensive mechanism against several deleterious substances.
We found further support for the above concept of the differential roles of MDRs and SCP-hubs in the data generated by recent studies on proteins involved in phenotypic robustness of biological systems in face of constant environmental variation 40. Genes that buffer organisms against such fluctuations/perturbations are typically called phenotypic capacitors 40. Interestingly, MDRs are highly over-represented among phenotypic capacitors, whereas SCP-hubs are largely excluded from this group (table 1). This pattern mirrors the opposing trends discussed above and is consistent with the differential regulation of these genes by promoters differing in the presence of a TATA box. MDRs being constantly expressed via TFIID-dominated promoters are likely to provide a persistent buffer against chemical insults – they are hence critical for robustness of the system in face of such stresses. An exactly opposite role appears to emerge for the SCP-hubs – rather than constraining phenotypic variability they appear to be involved in presenting a diversity of exploratory strategies, some of which might have an adaptive value under the atypical conditions induced by the chemical presence.
Mitochondrial protein synthesis apparatus, rapidly diverging and lineage-specific genes are major players among SCP-hubs
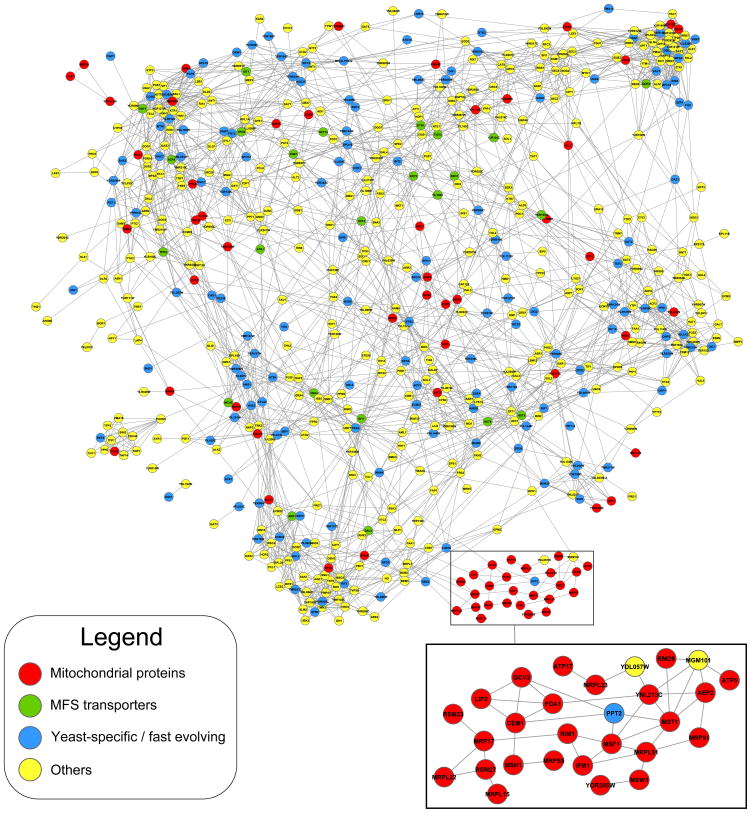
We observed a significant enrichment for genes encoding mitochondrial proteins among SCP-hubs (108 out of 670 SCP-hubs; FET; p ≈ 1.35 × 10−7). Further, the majority of these mitochondrial SCP-hubs form a relatively isolated, densely connected, sub-graph of the SCPnet (Fig. 2). Hence, these mitochondrial functions appear to comprise a functional element distinct from the rest of the SCP-hubs. We noted that ~45% of these mitochondrial SCP-hubs are specifically involved in mRNA maturation and translation (e.g. mitochondrial ribosomal proteins, tRNA synthetases and translation factors; Fig. 2, Additional file 5). Together they might boost mitochondrial productivity at the level of mitochondrial protein synthesis. An important aspect of oxidative metabolism is the more efficient extraction of energy per sugar molecule. In standard sugar-rich environments, yeast grows rapidly using glycolysis to generate ATP. However, it has been observed that stress conditions induce yeast to rapidly change its gene expression program, with slower growth and increased expression of stress-resistance pathways. In this configuration, mitochondrial oxidative phosphorylation is used as the preferred pathway to produce ATP36. Previous studies have shown that increased mitochondrial energy production is required to counter deleterious effects of reactive oxygen stress 41. In light of this, we propose that the mitochondrial SCP-hubs probably have comparable roles in chemical tolerance. Like in the case of classical environmental stresses, a switch to the mitochondrial oxidative metabolism might serve number of purposes: 1) in light of the above observation on the nature of SCP-hubs it is conceivable that the more efficient energy metabolism might help in driving the “noisy” explorative processes elicited by the presence of chemicals. 2) Likewise it might also allow investing energy for driving various detoxification processes such as trafficking, repair and active chemical efflux mechanism in the cells.
Figure 2.
High-confidence interactions between SCP-hubs. We computed cliques 125 in the SCPnet and reasoned that, due to its assortative nature, SCP-hubs should co-occur in several cliques, forming a densely connected sub-network. Thus, we used the point-wise mutual information 18 to identify the most significant edges linking SCP-hubs (p ≤ 0.005). Part of the large mitochondrial component (red) (discussed in the text) is magnified in the bottom of the panel. Although we used a large-scale dataset 119 to assign protein sub-cellular localization in our network, we also found independent evidence supporting the mitochondrial localization for two of the three genes marked as non-mitochondrial in the inset, namely Ppt2 and Mgm101 131, 132. This shows that the SCPnet is able to recover proteins involved in related processes solely based on their chemical-genetic profiles. Yeast-specific/fast evolving proteins (blue) were defined as proteins with close homologs only in the clade saccharomycotina, as identified by a single-pass BLAST search (e-value < 0.001; >30% query/hit coverage). MFS transporters (here colored in green) were found as significantly over-represented among the SCP-hubs. Other classes of genes were not assigned in any special groups and are colored in yellow.
A distinct aspect of the SCP-hubs is revealed by the striking over-representation of experimentally “uncharacterized” genes (as defined by SGD, see Introduction) that we found in this set (p ≈ 8.3 × 10−7). The current lack of functional evidence for the set of ~1,000 “uncharacterized” genes has been attributed to three major (not mutually exclusive) explanations 9: (i) the low expression levels in standard conditions; (ii) the predominance of fungi-specific genes; (iii) functions exerted by their protein products could be important under special conditions, e.g. presence of particular deleterious chemicals. Indeed, we found that SCP-hubs are less transcribed and their protein products are present in lower abundances (WMWT; p ≤ 8 × 10−3). We detected no significant difference in the half-life of the SCP-hubs mRNAs or protein products when compared to those of other genes, indicating that the low protein levels of the SCP-hubs are most probably a result of transcriptional/translational regulation and not a consequence of differential degradation of mRNAs/proteins. We also found an over-representation of genes with close homologs (defined by e-value ≤ 0.001; ≥30% query/hit coverage in a single pass BLAST search) only in yeasts of the saccharomycotina lineage among SCP-hubs (FET; p ≈ 2.5 × 10−3). Consistent with this we also noted a general under-representation of proteins with strong pan-eukaryotic conservation among SCP-hubs (FET; p ≈ 5.6×10−8). The over-representation of these yeast-specific/fast-evolving genes is even more remarkable when mitochondrial proteins are excluded from the SCP-hubs set (FET; p ≈ 1.5 × 10−4). A part of this apparent lineage-specificity arises from accelerated evolution in the yeast lineage – a striking example of this is the yeast mitochondrial translation factor GatF, a glutamate amidotransferase subunit, which while conserved in all cellular organisms is particularly divergent in yeast (it does not detect its orthologs from other eukaryotic lineages in a single-pass BLAST search). The remainder of the lineage-specificity observed among the SCP-hubs emerges from rapidly diverging lineage-specific paralogs and de novo innovations, especially in the form of TM and low complexity proteins (Additional File 5). SCP-hubs tend to have far fewer protein interaction partners than MDRs (WMWT; p < 2.9 × 10−15). In general, SCP-hubs also have less PPIs than non-hubs (FET; p ≈ 7.7 × 10−7), providing a potential explanation for how a large number of new lineage-specific SCP-hubs could emerge relatively rapidly in evolution. Presence of fewer physical interactions probably allows rapid divergence (perhaps via positive selection) due the lack of the constraints arising from the need to conserved interaction interfaces with partners.
Sequence analysis of rapidly diverging and lineage-specific SCP hubs suggests novel chemical survival strategies
We used sensitive sequence analysis methods to understand the possible functions of these lineage-specific or rapidly diverging proteins in protection against deleterious substances of these SCP-hubs. This analysis led to new findings such as a paralogous family of tail-anchored cysteine-rich TM domain proteins (CYSTM proteins), which comprise a novel general stress-resistance superfamily that is likely to be active in several major eukaryotic lineages 42. While transporters are enriched among MDRs we found no significant enrichment for transporters among the SCP-hubs. Only two of the transporters that are MDRs are also represented among the SCP-hubs. Despite the overall lack of enrichment of transporters among SCP-hubs, one of the largest paralogous superfamily of proteins represented among SCP-hubs are transporters of the major facilitator superfamily (MFS) 43. In fact, 1/3rd of the MFS proteins in the network are SCP-hubs, which amounts to a significant enrichment of just this superfamily of transporters (n=24; FET; p ≈ 10−6). Yet, these transporters appear to act in distinct processes – five of them have been previously characterized as players in chemical resistance, while other sets of these transporters are required for uptake of hexoses, iron and in mitochondrial transport of nutrients (Additional file 5). Thus, some of them might augment the energy production either by facilitating nutrient uptake or allowing exploration of alternative nutrient sources in response to the chemical. We also found Yer130c, a predicted transcription factor with C2H2 Zn-fingers, to be a SCP-hub. Orthologous proteins in other fungi such as Candida albicans 44 and Trichoderma atroviride 45 are transcription factors that bind to a specific stress response element to activate programs against acid or osmotic stress. These observations raise the possibility that Yer130c and its orthologs could be a general master regulator of stress response in ascomycete fungi that is required to coordinate natural resistance against a large number of chemicals.
Another stress-induced gene, Ygr127w, was also found to be a SCP-hub in our analysis, with homologous proteins present in animals, plants, fungi, ciliates and various bacterial lineages. It was earlier defined as an uncharacterized gene with a domain of unknown function (PFAM: DUF833). However, through profile-profile comparisons (HHpred program p ≈ 10−8) we were able to show that these proteins contain a catalytic domain of the NTN hydrolase fold with a catalytic cysteine. In particular, they were closest to the peptide/amide-bond hydrolases of the conjugated bile acid hydrolase family 46. We also found that previously uncharacterized proteins among the SCP-hubs include several distinct enzymatic domains such as the ATP-dependent condensation domain (e.g. acyl-CoA synthetase and polyketide synthase-like) (Yor093c), SIS domain aminotransferase (Ymr085w), GCN5-like acetyltransferases (Ygr111w and Yor012w), C-N hydrolase of the nitrilase fold (Yil165c), a distinct member of the gamma-glutamyl cyclotransferase fold (Yer163c), PLP-dependent aminotransferase (Yer152c), Rossmann fold oxidoreductase (Yhr009c) and carbon-sulfur lyase (Yml082w) (Additional file 5). Together, the presence of these catalytic activities among SCP-hubs strongly suggests a major aspect of chemical resistance is the cooperation between diverse enzymatic activities that are likely to modify deleterious substances through adducts such as acetyl groups, redox changes or hydrolysis of key bonds in them to render them less toxic. Many of these enzymatic domains show rapid divergence in the yeast lineage and have emerged through lateral transfer from bacteria or through duplications in certain large multi-gene families. The pattern of lineage-specific innovation seen among the SCP-hubs is also consistent with the above observation on their TFIID dominated promoters: TATA-containing genes tend to have higher expression divergence between closely related species, a trend that is also seen in other distantly related eukaryotes 47. Therefore the lineage-specific innovation at the protein sequence level goes hand in hand with their expression noise at the regulatory level. We propose that this observation, together with the earlier noted stark disparity between CPnet and SCPnet hubs, are the consequences of a unified underlying evolutionary strategy adopted by the organism. SCP-hubs with their multilevel lineage-specific diversity are contributors to the evolvability of the system (i.e. raw material for adaptive evolution), whereas the MDRs are primarily responsible for the system’s robustness.
Effects of major post translational modifications and chemical resistance
To understand the impact of regulatory cascades on the CPnet and the SCPnet we endeavored to place them in the context of key signaling processes. There are several post-transcription modifications ranging from simple chemical groups like phosphate, acetyl or methyl moieties to large molecular adducts like lipids and ubiquitin-like proteins. Previous studies have shown a major role for ubiquitination and phosphorylation in many aspects of signaling environmental changes 48, 49. Additionally, various large-and small-scale studies have been published for these two modifications, which allowed us to systematically assemble genome-scale data and derive networks from them (see Materials and methods for details). To study their intersection with chemical tolerance mechanisms we used the rank plot, a representation that we have recently successfully employed in the comparison of differential behavior of genes in two distinct networks 18. Briefly, the rank plot is a visualization technique that is useful to evaluate the connectedness (proxy for importance) of a given gene in two different functional contexts (networks). The genes are sorted by degrees and these absolute values are then converted to ranks in each context which provide a coordinate system for genes in the plot 18. In comparing the CPnet with the ubiquitin and phosphorylation networks, we considered the fact that nearly all of the phosphorylation/UBL modification events in our reference datasets were detected under standard growth conditions. Hence, if a kinase or Ub pathway protein scores as hub in the CPnet but has low connectivity in the phosphorylation or ubiquitination network then it probably exerts most of its functions under chemical stress. If the converse were true, its signaling roles are likely to be largely relevant in standard growth conditions in mechanisms unrelated to chemical stress. This would allow a means of specifically identifying post-translational modification pathways with key roles in to chemical tolerance.
Chemical tolerance involves multiple regulatory events mediated by the ubiquitin system
Regulation of vesicular trafficking by the ubiquitin-system in natural chemical tolerance
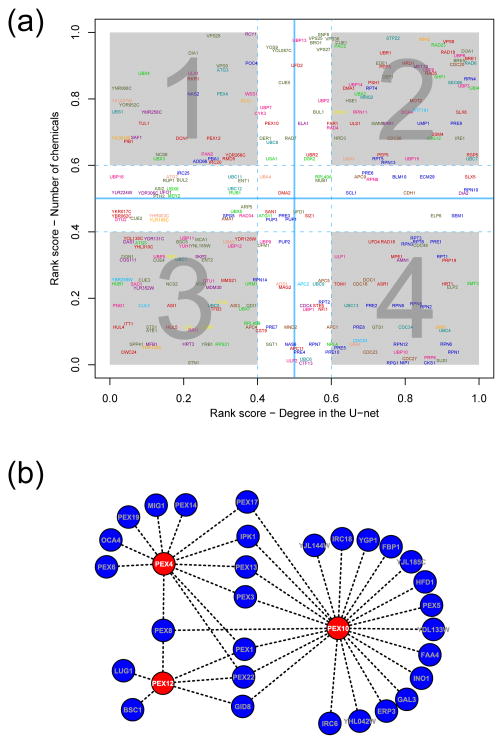
We described above the over-representation of ubiquitination targets among the MDRs and its connection to the role of ubiquitin in vesicular trafficking which is key system in chemical tolerance. The rank plot approach, along with the above-outlined logic provided a means to identify Ub-network genes that have specific influence in chemical tolerance but only a limited role under standard growth conditions. For instance, we found support for a specific role in chemical tolerance for the MDR gene Rcy1 (see above) by virtue of its position in quadrant-1 (contains genes with a specific chemical tolerance role; Fig. 3a) of the rank plot. Also of interest in the same context is the membrane-associated E3 protein Tul1, found in quadrant-1 (Fig. 3a), which has been implicated in ubiquitinating membrane proteins in course of vesicular trafficking 50. We found two other F-box proteins, Ymr258c and Saf1 and another SCF component, the POZ-ankyrin domain protein Yil001w, in this quadrant. Of these Ymr258c, was recently implicated in resistance to methylmercury 51, which is completely consistent with our prediction of a more general role for it in chemical resistance – it remains to be seen if it might function similar to Rcy1 in vesicular trafficking. Yil001w too shows hardly any interactions under standard conditions but is a hub in the SCPnet, suggesting that it might have a specific ubiquitination function in chemical tolerance. Saf1 has been characterized as involved in the SCF-mediated ubiquitination of adenine deaminase Aah1 and its subsequent degradation when yeast cells enter in quiescence 52. It is likely that this F-box E3 subunit might have a role in degradation of enzymes associated with nucleotide salvage that are potential targets of deleterious chemicals and thereby foster natural resistance to them. Likewise, we also found the poorly characterized E1-enzyme of the Ub system with a Trs4-C domain, Ykl027w 53, to be in quadrant-1 of this plot (the only E1 in this quadrant, Fig. 3a). Though this protein has very few functional links under standard conditions, we had shown that it has clear links to the F-box dependent-ubiquitination complexes in the ubiquitin network 18. Its presence in quadrant-1 strongly suggests that its primary role might be in chemical tolerance – it would be of interest for future experimental studies investigate if it might functionally cooperate with any of the other F-box proteins, such as Rcy1, Ymr258c or Saf1 and the SCF component Yil001w, which were also recovered in the same quadrant of the plot.
Figure 3.
Ubiquitin pathway and chemical stress. (a) Rank plot 18 of the ubiquitin pathway components and their relative importance, measured using degree-based ranks, in the non-stressed cellular proteome and CPnet. We reasoned that if a particular Ub pathway component is required for the tolerance against several chemicals and has low degree under largely non-stress conditions, it could have specialized functionalities to operate in chemical stress environments (quadrant 1). On the other hand, components that are mainly active under standard conditions can also be identified (quadrant 4); Color codes for the Ub pathway proteins: proteasome (blue), UBLs (green), Fboxes (purple), E1s (salmon), E2s (dark cyan), E3s (red), DUB (pink), cullins (light blue), POZ (orange), others (dark green), APC (brown), signalosome (yellow). (b) Large peroxisomal component that were recovered from the SCPnet using the Ub pathway proteins Pex4, Pex10 and Pex12 as baits (colored in red). Interestingly, 10 other peroxisomal proteins were identified solely based on shared chemical genetics profiles (colored in blue along with other 19 genes), again showing the biological significance of the SCPnet interactions.
Ubiquitination-mediated peroxisome biogenesis and degradation in chemical resistance
As mentioned above analysis of the CPnet had revealed a potential role for another trafficking system, i.e. autophagy, probably via pexophagy, as a comprehensive tolerance mechanism. Our current analysis of the Ub-network-CPnet rank plot threw further light on the peroxisomal participation in chemical tolerance. Three functionally linked peroxisomal ubiquitination machinery proteins Pex12, Pex10 and Pex4 18 scored high in terms of their ranks in chemical connections (Fig. 3a and are major players in peroxisomal protein import and biogenesis 54. To further delineate this peroxisomal aspect of pervasive chemical tolerance, we used these three proteins as baits to query the SCPnet. As a result we recovered a large peroxisomal sub-network comprising other 13 peroxins, among other genes (Fig. 3b). Importantly, we found that while genes required for de novo synthesis of peroxisome are in this sub-network (i.e. Pex3 and Pex19) 55 those crucial for peroxisomal fission (Inp2, Dnm1 and Vps1) 55 are not present (Fig. 3b). It has been proposed that de novo peroxisome biogenesis is the preferred route to generate new peroxisomes when the cells are under high oxidative stress, perhaps due to heavy damage to the peroxisomes, while peroxisome fission apparently dominates under standard conditions 55. Together, with the above observations of the involvement of pexophagy, it appears that degradation and de novo regeneration of peroxisomes might be a critical aspect of the general chemical tolerance. Perhaps, the combination of pexophagy and regeneration helps eliminate chemically damaged peroxisomes and produce new ones for active detoxification. The recovery of the Ub-pathway components Pex12, Pex10 and Pex4 suggests that this sub-network is probably critical in the process of keeping the peroxisomes functional by importing the requisite proteins.
Identification of deubiquitinating peptidases and proteasomal chaperones specific to chemical resistance
We also uncovered previously unknown roles in chemical tolerance for several other Ub network proteins by means of the rank plot analysis. Of considerable interest in this regard is the deubiquitinating enzyme (DUB) Ubp16, which is conserved across fungi, animals and plants and contains an N-terminal hydrophobic signal peptide. This signal peptide was shown to localize it to the mitochondrial outer membrane with the catalytic domain being lodged in the cytoplasm 56. Surprisingly, we noticed that in the ubiquitin network, which primarily consists of interactions under standard conditions, Ubp16 shows no connections to other proteins 18 and is also not essential for general mitochondrial functions and inheritance 56. However, Ubp16 is required for growth in the presence of 10 chemical compounds (especially metals chlorides; Additional file 1). These observations indicate that this DUB is likely to be specifically involved in the defense against chemicals, probably downstream of a stimulating signal associated with the mitochondrion, by preventing proteasomal degradation of key targets. We also noted two proteasomal chaperone heterodimers in quadrant-1, namely Poc3 (Irc25)-Poc4 and Poc1-Poc2 (Pba1-Add66) (Fig. 3a) 57–59. One possibility is that these chaperones act as buffers to allow efficient proteasomal assembly when the cell is facing the deleterious effects of a chemical in the environment. In support of this proposal we note that poc3Δ and poc4Δ mutants have also been reported to show growth defects under certain stress conditions 57, 58, 60. We also found a conspicuous difference between ubiquitin and SUMO in chemical stress processes (Ubi4 and Smt3, respectively; Fig. 3a). While ubiquitin is very relevant for growth in the presence of several chemicals, SUMO seems to be vital for growth in the presence of a relatively limited set of substances (quadrant- 2 and 4, respectively; Fig. 3a). Yet, we found that a poorly characterized SUMO-network protein, Wss1, which we had formerly predicted to be a metal-dependent desumoylating enzyme 61, to be a major player in chemical tolerance (quadrant-1, Fig. 3a). Wss1 appears to be required for the tolerance of a number of mechanistically distinct DNA-damage agents such as psoralen, berberine chloride, hydroxyurea, cisplatin and mitomycin C. Under standard conditions, this protein shows few interactions and is limited in its expression to a single spot in the cell close to the nuclear membrane 62. Previous evidence has also been presented for Wss1’s involvement in DNA-damage checkpoints 63, 64. Its prominence in the chemical response suggests that it might catalyze specific nuclear desumoylation events that probably regulate DNA-repair or replication pathways that counter the effects of DNA-damaging agents.
Roles for specific phosphorylation cascades
A specific role for the HOG kinase cascade in chemical stress tolerance
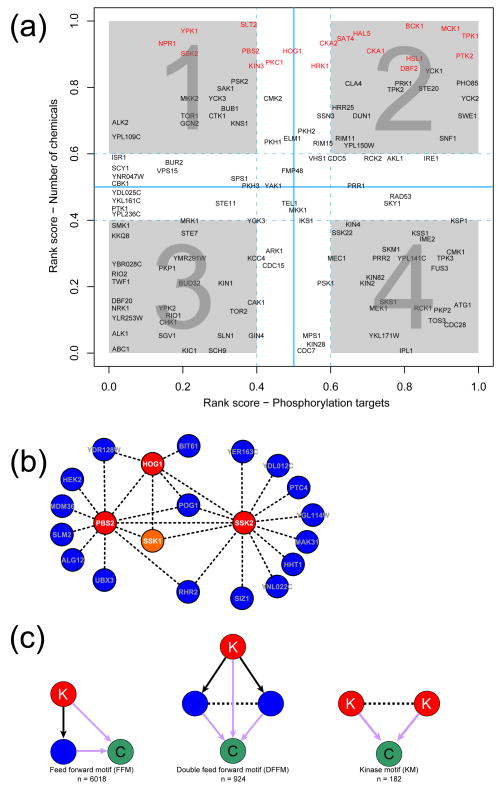
Strikingly, we found that three successive kinases in the HOG signaling cascade Hog1, Ssk2 and Pbs2 scored as MDRs. Furthermore, the upstream receiver domain regulator of the HOG cascade, Ssk1 and the downstream target of the cascade involved in glycerol biosynthesis Rhr2 (Gpp1; a glycerol 3-phosphatase) are also MDRs 65. To investigate if they might actually interact in chemical tolerance we used the three kinases Hog1, Ssk2 and Pbs2 as baits to extract the linked sub-network from the SCPnet. Interestingly, not only did these 3 kinases emerge as a connected clique, but also they were part of a distinct sub-network in which they showed linkages to Rhr2 and Ssk1 (Fig. 4). These linkages strongly suggested that the complete HOG cascade that leads to the glycerol synthesis is involved in involved in natural chemical resistance. Previously, glycerol synthesis has been implicated in a protective response to osmotic, anoxic and oxidative stresses 65, 66. While in part the role of glycerol production in chemical tolerance might arise from its action in countering the osmotic and redox effects of chemicals (e.g. NaCl, hydrogen peroxide and paraquat), all these genes are connected to a chemically diverse set of substances (Additional file 1), indicating a more generalized function. In this light it might be of interest to further experimentally study the role of glycerol production as a general chemo-protective mechanism.
Figure 4.
Phosphorylation and chemical stress. (a) Rank plot 18 of kinases and their relative importance, measured using degree-based ranks, to the non-stressed cellular proteome and CPnet. As in the Ub-network analyses, we reasoned that if a kinase with low number of targets in the kinase-substrate network is required for the tolerance against several chemicals, it should be involved in signaling cascades mediating cellular response/tolerance to stress (quadrant 1). Conversely, kinases specialized in “housekeeping” roles can also be identified using this approach (quadrant 4). Kinases that are MDRs are colored in red and other kinases colored in black; (b) By using three HOG pathway kinases (i.e. Ssk2, Pbs2 and Hog1, colored in red) as baits to query the SCPnet, we were able to recover other components of the signaling cascade (e.g. Ptc4). Potentially novel interaction partners were also recovered, e.g. the CYSTM protein Ydl012c 42. The genes interacting with the kinase triad Ssk2-Pbs2-Hog1 are colored in blue, with the exception of the upstream HOG pathway response regulator, which is represented in orange; (c) Structural motifs we used to analyze the involvement of kinases in chemical response. Chemical-genetic interactions are represented in purple, phosphorylation events are represented by black solid arrows, SCPnet links are represented by dashed black lines. Kinase, substrates and chemical substances are colored in red, blue and green, respectively.
To assess the specific roles of particular protein phosphorylation pathways in chemical tolerance we used the rank plot to compare the importance of a protein kinase for resistance to multiple chemicals relative to its phosphorylation targets inferred under standard conditions along with the logic described above (Fig. 4a). As a result we found that Ssk2 and, to a lesser degree, Pbs2 are more connected in the context of chemical responses than under standard conditions (quadrant-1, Fig. 4a). The HOG pathway has other two MEKK kinases, Ste11 and Ssk22 (a partially redundant paralog of Ssk2), which, unlike Ssk2, are not particularly prominent in terms of their links to chemicals. In fact Ssk22 appears to have fairly high number of targets, but these phosphorylation events seem to be relevant only under standard growth conditions (quadrant-4, Fig. 4a). This suggests that instead of exclusive backup there is a functional divergence between the MEKK kinases of the HOG cascade, with Ssk2 probably being mainly a transducer of chemical stress. The above recovered sub-network of the SCPnet which contains the 3 HOG pathway kinases also links a number of other proteins that might functionally collaborate. One such is the PP2C family phosphatase Ptc4, which might counter the phosphorylation catalyzed by the Hog1 kinase and has been previously shown to be required for lithium tolerance 67. Hence, a particular phosphorylation pattern of substrates generated by the opposing action of the HOG cascade and the Ptc4 phosphatase might be in part required for chemical tolerance. Other linked proteins in this sub-network include the SUMO E3 Siz1, the CYSTM protein Ydl012c 42 and two components of the TorC2 complex, Bit61 and Slm2 protein 68–70. Of special interest is the TorC2 complex, whose protein kinase subunit Tor1 is also positioned in quadrant-1 (Fig. 4a). The PH domain protein Slm2, a TorC2 complex protein (connected to the HOG sub-network in the SCPnet), just as Ssk2, is implicated in actin cytoskeleton reorganization 70, 71. Slm2 is also required for endocytosis of permeases under other stress conditions 68. These connections raise the possibility that the interaction between the HOG cascade and the TorC2 complex might play a role in chemical stress via regulation of cytoskeletal organization or receptor endocytosis.
Resistance mechanisms via regulation of transport and cell-cycle progression by kinases
Another kinase that lies in quadrant-1 is the Npr1 kinase, which is well-known for its role in the stabilization of the high-affinity general amino acid permease Gap1, by antagonizing its down-regulation via Rsp5-mediated ubiquitination 72, 73. Although Npr1 is required for growth in the presence of 30 chemicals, Gap1Δ cells present altered growth in the presence of only 5 compounds, suggesting that Npr1 is likely to phosphorylate other proteins required to the response against chemical stress. Npr1-mediated phosphorylation has also been shown to induce protein degradation of another permease Tat2 (tryptophan permease) 74, 75. Therefore, Npr1 phosphorylation of targets probably has a bi-directional action in both stabilizing and destabilizing specific permeases on the membrane. Thus, it is conceivable that Npr1 might differentially regulate routes for influx or efflux of particular substances and thereby enable chemical resistance. We also found evidence for functional divergence of the paralogs Alk1 and Alk2 (haspin-like kinases) through the rank plot analysis (Fig 4a) 76. Alk1 and Alk2 are highly regulated during the cell cycle 76. Alk2 is required for growth in the presence of 15 chemical compounds, whereas Alk1 is connected to a single substance. Interestingly, overexpression of Alk2 results in mitotic arrest 76, indicating a potential role of this kinase as a cell cycle checkpoint in the presence of potentially deleterious substances.
Structural analysis of the protein kinase network recovers additional roles for phosphorylation in chemical tolerance
To further dissect the role of protein phosphorylation in regulating chemical tolerance we combined the kinase network with the CPnet and SCPnet to derive three kinds of structural motifs that encompass both these networks (Fig 4c): (1) Feed forward motif (FFM)- In this case a kinase phosphorylates a substrate and both of them are required for growth in the presence of a chemical; (2) Double feed forward motif (DFFM)- Here a kinase phosphorylates a pair of proteins connected in the SCPnet and all three are required for the tolerance of a common chemical. DFFMs constitute a stronger version of the FFMs, since in this motif the two kinase substrates are required to survive a statistically significant overlapping set of chemicals; (3) Kinase motif (KM)- here two kinases are important for the response against a common chemical and are also connected in the SCPnet. Overall we detected 6,018 FFMs, 924 DFFMs and 182 KMs (Additional file 6, Fig 4c). In the case of the FFM, we reasoned that if both the kinase and its substrate are required for the tolerance of several chemicals then the phosphorylation event might be a required regulatory step in this context.
The maximum number of FFMs in which a kinase-substrate pair participates is 23, namely the Pbs2-Hog1 pair. We individually examined all the top-ranked kinase-substrate pairs (i.e. most represented across all FFMs) all the way from the above-mentioned first rank pair to those involved in at least approximately half that number of FFMs (i.e. 12 FFMs; Additional file 6). This included 22 distinct kinase-substrates pairs. We found three HOG cascade kinases (i.e. Ssk2, Pbs2, Hog1) among them. Notably, the configuration of these FFMs follows the HOG phosphorylation cascade (Ssk2->Pbs2->Hog1) supporting our above proposal regarding the importance of the HOG pathway in surviving diverse chemical insults. Among the 22 top-ranked kinase-substrate pairs we found five of them to include the kinase Mck1. Of these one combines Mck1 with the two-component system protein Ssk1 that is upstream of the HOG cascade kinases. This suggests the glycogen synthase kinase-3/shaggy pathway initiated by Mck1 might feed into the HOG cascade. Previously, a link was reported between Mck1 and osmotic stress-signaling 77 – it remains to be seen if the HOG-Mck1 interaction might have more general significance in stress response. Of the 5 most frequent kinase pairs found in KMs we found the HOG pathway kinase pairs (Ssk2-Pbs2, Ssk2-Hog1 and Pbs2-Hog1), again reinforcing the involvement of this cascade in the phospho-regulation of chemical tolerance. Other than HOG cascade kinases, the most prevalent kinase pair among the KMs is the Bck1-Slt2 pair (required for growth under the presence of 43 chemicals). This pair has earlier been shown to be an important regulator of cell wall integrity pathway along with the HOG cascade 78, 79. It is possible that their cooperative action in regulating the cell wall stability facilitates tolerance by restricting permeability. Another very prevalent kinase pair among the KMs is Sat4 and Hal5, which have been shown to stabilize transporters on the cell membrane by preventing their endocytosis and vacuolar degradation 80. Hence, as in the case of Npr1 recovered in the rank plot analysis (Fig. 4a), we propose that these kinases might be regulators of chemical resistance by stabilizing key efflux transporters.
Among the DFFMs, one kinase stood out as most frequently represented, namely Tpk1 (i.e. a protein Kinase A isoform). Among these DFFMs we noticed that Tpk1-mediated phosphorylation of the Boi1-Opi1 and the Boi1-Osh3 protein pairs from the SCPnet were required for growth in the presence of 7 and 6 chemicals, respectively. Of these, Boi1 binds acidic phospholipids via its PH domain and is required by the “NoCut” checkpoint pathway, which delays the completion of cytokinesis in response to anaphase defects 81, 82. On the other hand Opi1 is a transcriptional repressor of phospholipid biosynthetic genes 83 and is also activated by Tpk1-catalyzed phosphorylation 84. Osh3 is also a lipid-binding protein with a complex domain architecture that include a GOLD, PH and oxysterol-binding domain 85. In particular, Osh3 is postulated to be involved in non-vesicular transport of sterol, maintaining its cellular distribution and homeostasis 86. Thus it is conceivable that the action of protein kinase A in chemical tolerance is through regulation of membrane structure or composition by regulating lipid metabolism and distribution and multiple levels.
GENERAL CONCLUSIONS
By integrating large- and small- scale datasets in the yeast model we assembled what to the best of our knowledge is the largest currently available chemical-phenotype network derived from gene deletions. The size of this network together with the diversity of the substances covered allowed us to address several general and specific questions concerning the phenomenon of chemical tolerance. While the previous large-scale studies showed that certain key genes and pathways were major players in chemical resistance, the “systems” view of this phenomenon has been poorly explored. Likewise, though it has been clear that systems properties such as robustness have an important role in chemical resistance, the actual components that contribute to such properties and their biochemical mechanisms have not been objectively defined. By means of different network representations (the CPnet and SCPnet) and comparisons of the positions of genes/proteins in different networks (rank plots) we were able to characterize the structure of the cellular systems underlying chemical resistance. In a previous analysis we showed that natural resistance disproportionately depends upon certain chemical complexes, especially those linked to chromatin structure and vesicle tethering, which appear to function as innate buffers against chemical insults 16. We presented evidence that proliferation of certain functionally partially redundant protein complexes in eukaryotic evolution might be related to the advantages of possessing multiple buffers to cover for a diverse range of substances. In this work we used the data to specifically identify individual players that allow the cell to tolerate chemical stresses. Interestingly, these genes, the MDRs, showed a notable enrichment for previously identified phenotypic capacitors – buffers against phenotypic variation 40. Furthermore, several of the protein complexes that we had earlier identified as buffers against chemical effects were also recovered as phenotypic capacitors 16, 40. These observations point to the generality of these players in buffering mechanistically unrelated forces impinging on the cell – i.e. there is a common sub-network of genes that simultaneously provide defenses against diverse potentially deleterious influences and intrinsic fragility. While this article was submitted for review, two publications using different approaches on large-scale chemical genetic data appeared that support the above picture of role of MDRs as capacitors 87, 88. This sub-network appears to represent that part of the system that has primarily evolved for conferring general robustness, which is supplemented in different contexts by different sets of specific players. More strikingly, our analysis of the SCPnet uncovered another set of functionally interacting players that play a role in chemical tolerance. This set of genes appears to be enriched in lineage-specific and rapidly diverging members. In addition to sequence divergence they also show a corresponding tendency for evolutionary divergence in expression patterns and also an increased tendency for noisiness. Further, they appear to be part of a specific expression program that depends on SAGA. We present evidence that this set represents a previously underappreciated component of the chemical response that is involved in probing the environment and exploring alternative survival strategies, representing a systems property known as evolvability, which is frequently (mis)understood as opposite to robustness. However, some studies have proposed that both these concepts can be simultaneously accommodated in biological systems 89–91. Given the role of SAGA-dominated genes in stresses other than chemical, we suspect that such a set of genes might have a more general role in coping with diverse harsh environments. Thus, two distinct systems properties, robustness and evolvability are simultaneously active as forces required to survive environmental variation, including deleterious chemicals.
A critical aspect of this work has been the recovery of the actual genes and associated biochemistry involved in the above-discussed network properties. We were able to identify certain key components of the ubiquitin system, and certain phosphorylation cascades as major participants and regulators of chemical tolerance. In particular we to show that a major sub-network in the SCP are mitochondrial protein synthesis genes, suggesting that boosted mitochondrial function is important for the exploration of alternative survival strategies under chemical stress. We also show that several genes in the SCPnet encode enzymes resembling those commonly found in bacterial and fungal secondary metabolism systems that are active in the stationary phase (e.g. acetylases, peptidases, redox enzymes, aminotransferases and α/β-hydrolases 92). This suggests that the exploration of alternative survival strategies under chemical stress and stationary phase strategies draw from a common pool of biochemical players. Another potential theme in this context could be lineage-specific POZ and F-box containing SCF complexes that might allow targeting of novel sets of proteins for degradation.
In conclusion, the ability of our analysis to extract previously unknown regulatory and functional components of sub-networks, as well as identify novel roles for formerly well-studied pathways, shows the power of such systems approaches in generating new biological knowledge. We hope that this would inspire further experimental studies to extend the findings presented here, and also add impetus for new data generating efforts.
MATERIALS AND METHODS
Datasets
We used data from 34 chemical-genetics publications 1, 3–8, 11, 14, 17, 93–116 to assemble the meta-dataset used here. Chemical compound names were manually inspected to correct typos and redundancy. Protein-protein and genetic interactions were retrieved from the BioGrid database version 2.0.47 117. Other datasets used: ubiquitin network 18, mRNA abundance 32, protein abundance/noise 118, and sub-cellular localization 119, phenotypic capacitance 40, TATA box presence/absence 30, SAGA/TFIID-dominance 31, gene expression evolution in poor medium 34, environmental stress response genes 33. We extensively collected phosphorylation data, from literature searches and large-scale publications to create the kinase-substrate network 120, 121. Information on phosphoproteins was also obtained from a recently published study 122 and used in the enrichment analysis. ORF and gene annotations were obtained from the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/).
Data analyses, network assembly and simulations
The chemical-genetics datasets were processed in order to obtain simple ORF to compound relations. Differences in compound concentration were disregarded. The list of chemical compound names was manually inspected to minimize the occurrence synonyms and correct errors in chemical names, which otherwise would result in redundant ORF to chemical connections. The total, non-redundant set of ORF to chemical interactions constitutes the chemical phenotype (CP) network. After filtering out dubious ORFs and redundant interactions (the same interaction was obtained multiple times in the same or different datasets), our final assembly was composed by 54,572 interactions between 5,233 ORFs and 425 chemical compounds. We then generated 1,000 randomized datasets in a way that each ORF is connected to the same number of chemicals observed in the real data. The overlap strength was computed using the Jaccard index:
Here, Iij represents the number of chemicals linked to both the ORFs i and j, whereas Di and Dj are the total number of chemicals connected to i and j, respectively. The overlap scores were obtained for each ORF pair in the randomized CP networks to obtain an empirical assessment of significance. The set of significant interactions (p ≤ 10−3) represents the shared chemical phenotype (SCP) network, used in this the present work to analyze the co-participation of genes in the response to chemicals. All the nodes in the SCPnet are ORFs, connected by edges that are completely based on chemical-genetics data. The SCPnet was also rewired (degree-preserving) for statistical analysis. Custom Perl scripts (www.perl.org) were used for basic processing and statistical analyses performed using R Statistical language (http://www.r-project.org/). Network properties were computed using custom scripts and the iGraph package for R 123. Robustness to attack/failure and clique detection were performed as previously described 28, 124, 125. High-confidence interactions involving SCP-hubs were identified by point-wise mutual information, computed using the co-occurrence of genes in SCPnet cliques, as we have previously described 18. Graphs were rendered using the Cytoscape software 126.
Sequence searches to define phyletic patterns were performed using single-pass BLAST searches 127, with 30% minimal query and hit coverage and e-value threshold of 10−3 Protein domains were analyzed using HMMer3 128, PSI-BLAST and the PFAM database 129 (e-value ≤ 0.01). Sensitive profile-profile searches were also performed using HHPred 130.
Supplementary Material
Acknowledgments
We acknowledge the Intramural Research Program of the National Institutes of Health, USA for funding our research. We also would like to acknowledge all the authors who have made their genome-scale datasets publicly available. We tried to make the most extensive collection possible and express our regret if we accidentally missed some study in the data gathering process.
ABBREVIATIONS
- CPnet
Chemical phenotype network
- SCPnet
shared chemical phenotype network
- MDRs
multidrug resistance genes
- FET
Fisher Exact Test
- WMWT
Wilcoxon-Mann-Whitney
- PPI
protein-protein interaction
- GI
genetic interaction
- DUB
deubiquitinating enzyme
- UBL
Ubiquitin-like
Footnotes
AUTHORS’ CONTRIBUTIONS
TMV and LA conceived the study, analyzed the results and wrote the paper. TMV implemented the computational methods and integrated the public datasets. SB and SG contributed with high-quality data, ideas and general discussions. The manuscript was read and approved by all the authors.
Contributor Information
Thiago M. Venancio, Email: thiago.venancio@gmail.com.
S. Balaji, Email: balaji.ncbi@gmail.com.
S. Geetha, Email: hemeetha@gmail.com.
L. Aravind, Email: aravind@mail.nih.gov.
References
- 1.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 2.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 3.Ericson E, Gebbia M, Heisler LE, Wildenhain J, Tyers M, Giaever G, Nislow C. PLoS Genet. 2008;4:e1000151. doi: 10.1371/journal.pgen.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Proc Natl Acad Sci U S A. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, et al. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 7.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, et al. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 8.Hoon S, St Onge RP, Giaever G, Nislow C. Trends Pharmacol Sci. 2008;29:499–504. doi: 10.1016/j.tips.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Pena-Castillo L, Hughes TR. Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasch AP. Yeast. 2007;24:961–976. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- 11.Alamgir M, Eroukova V, Jessulat M, Xu J, Golshani A. BMC Genomics. 2008;9:583. doi: 10.1186/1471-2164-9-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuster A, Madan Babu M. Trends Biotechnol. 2008;26:252–258. doi: 10.1016/j.tibtech.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 14.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 15.Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, Giaever G, St Onge RP, Nislow C. Nat Chem Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 16.Venancio TM, Balaji S, Aravind L. Mol Biosyst. 6:165–171. doi: 10.1039/b911821g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruotolo R, Marchini G, Ottonello S. Genome Biol. 2008;9:R67. doi: 10.1186/gb-2008-9-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venancio TM, Balaji S, Iyer LM, Aravind L. Genome Biol. 2009;10:R33. doi: 10.1186/gb-2009-10-3-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raiborg C, Stenmark H. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 20.Dores MR, Schnell JD, Maldonado-Baez L, Wendland B, Hicke L. Traffic. 11:151–160. doi: 10.1111/j.1600-0854.2009.01003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N. Mol Biol Cell. 2005;16:178–192. doi: 10.1091/mbc.E04-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. Mol Biol Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiederkehr A, Avaro S, Prescianotto-Baschong C, Haguenauer-Tsapis R, Riezman H. J Cell Biol. 2000;149:397–410. doi: 10.1083/jcb.149.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M. Mol Cell Biol. 2001;21:3105–3117. doi: 10.1128/MCB.21.9.3105-3117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kus BM, Caldon CE, Andorn-Broza R, Edwards AM. Proteins. 2004;54:455–467. doi: 10.1002/prot.10620. [DOI] [PubMed] [Google Scholar]
- 26.Bugnicourt A, Mari M, Reggiori F, Haguenauer-Tsapis R, Galan JM. Traffic. 2008;9:755–769. doi: 10.1111/j.1600-0854.2008.00715.x. [DOI] [PubMed] [Google Scholar]
- 27.Morales-Johansson H, Jenoe P, Cooke FT, Hall MN. J Biol Chem. 2004;279:39604–39610. doi: 10.1074/jbc.M405589200. [DOI] [PubMed] [Google Scholar]
- 28.Albert R, Jeong H, Barabasi AL. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 29.Newman ME. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67:026126. doi: 10.1103/PhysRevE.67.026126. [DOI] [PubMed] [Google Scholar]
- 30.Basehoar AD, Zanton SJ, Pugh BF. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 31.Huisinga KL, Pugh BF. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 32.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 33.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Proc Natl Acad Sci U S A. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Maury L, Marguerat S, Bahler J. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 37.Murphy KF, Balazsi G, Collins JJ. Proc Natl Acad Sci U S A. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davisand JM, Brown RW. Proc Natl Acad Sci U S A. 2002;99:8778–8783. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tai SL, Snoek I, Luttik MA, Almering MJ, Walsh MC, Pronk JT, Daran JM. Microbiology. 2007;153:877–886. doi: 10.1099/mic.0.2006/002873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy SF, Siegal ML. PLoS Biol. 2008;6:e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant CM, MacIver FH, Dawes IW. FEBS Lett. 1997;410:219–222. doi: 10.1016/s0014-5793(97)00592-9. [DOI] [PubMed] [Google Scholar]
- 42.Venancio TM, Aravind L. Bioinformatics. 26:149–152. doi: 10.1093/bioinformatics/btp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng TT, Virk PS. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 44.Ramsdale M, Selway L, Stead D, Walker J, Yin Z, Nicholls SM, Crowe J, Sheils EM, Brown AJ. Mol Biol Cell. 2008;19:4393–4403. doi: 10.1091/mbc.E07-09-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seidl V, Seiboth B, Karaffa L, Kubicek CP. Fungal Genet Biol. 2004;41:1132–1140. doi: 10.1016/j.fgb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Rossocha M, Schultz-Heienbrok R, von Moeller H, Coleman JP, Saenger W. Biochemistry. 2005;44:5739–5748. doi: 10.1021/bi0473206. [DOI] [PubMed] [Google Scholar]
- 47.Tirosh I, Weinberger A, Carmi M, Barkai N. Nat Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 48.Laney JD, Hochstrasser M. Curr Opin Microbiol. 2004;7:647–654. doi: 10.1016/j.mib.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Schenk PW, Snaar-Jagalska BE. Biochim Biophys Acta. 1999;1449:1–24. doi: 10.1016/s0167-4889(98)00178-5. [DOI] [PubMed] [Google Scholar]
- 50.Reggiori F, Pelham HR. Nat Cell Biol. 2002;4:117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- 51.Hwang GW, Wada N, Kuge S, Naganuma A. J Toxicol Sci. 2009;34:413–416. doi: 10.2131/jts.34.413. [DOI] [PubMed] [Google Scholar]
- 52.Escusa S, Laporte D, Massoni A, Boucherie H, Dautant A, Daignan-Fornier B. J Biol Chem. 2007;282:20097–20103. doi: 10.1074/jbc.M702425200. [DOI] [PubMed] [Google Scholar]
- 53.Burroughs AM, Iyer LM, Aravind L. Proteins. 2008;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eckert JH, Johnsson N. J Cell Sci. 2003;116:3623–3634. doi: 10.1242/jcs.00678. [DOI] [PubMed] [Google Scholar]
- 55.Smith JJ, Aitchison JD. Curr Opin Cell Biol. 2009;21:119–126. doi: 10.1016/j.ceb.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinner A, Kolling R. FEBS Lett. 2003;549:135–140. doi: 10.1016/s0014-5793(03)00801-9. [DOI] [PubMed] [Google Scholar]
- 57.Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 58.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 59.Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. Embo J. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arendt CS, Hochstrasser M. Proc Natl Acad Sci U S A. 1997;94:7156–7161. doi: 10.1073/pnas.94.14.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iyer LM, Koonin EV, Aravind L. Cell Cycle. 2004;3:1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 62.van Heusden GP, Steensma HY. FEMS Microbiol Lett. 2008;282:100–104. doi: 10.1111/j.1574-6968.2008.01113.x. [DOI] [PubMed] [Google Scholar]
- 63.Mohanty BK, Bairwa NK, Bastia D. Proc Natl Acad Sci U S A. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Neill BM, Hanway D, Winzeler EA, Romesberg FE. Nucleic Acids Res. 2004;32:6519–6530. doi: 10.1093/nar/gkh994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pahlman AK, Granath K, Ansell R, Hohmann S, Adler L. J Biol Chem. 2001;276:3555–3563. doi: 10.1074/jbc.M007164200. [DOI] [PubMed] [Google Scholar]
- 66.Bettinger BT, Amberg DC. J Cell Biochem. 2007;101:34–43. doi: 10.1002/jcb.21289. [DOI] [PubMed] [Google Scholar]
- 67.Ruiz A, Gonzalez A, Garcia-Salcedo R, Ramos J, Arino J. Mol Microbiol. 2006;62:263–277. doi: 10.1111/j.1365-2958.2006.05370.x. [DOI] [PubMed] [Google Scholar]
- 68.Bultynck G, Heath VL, Majeed AP, Galan JM, Haguenauer-Tsapis R, Cyert MS. Mol Cell Biol. 2006;26:4729–4745. doi: 10.1128/MCB.01973-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daquinag A, Fadri M, Jung SY, Qin J, Kunz J. Mol Cell Biol. 2007;27:633–650. doi: 10.1128/MCB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wullschleger S, Loewith R, Oppliger W, Hall MN. J Biol Chem. 2005;280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 71.Fadri M, Daquinag A, Wang S, Xue T, Kunz J. Mol Biol Cell. 2005;16:1883–1900. doi: 10.1091/mbc.E04-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Craene JO, Soetens O, Andre B. J Biol Chem. 2001;276:43939–43948. doi: 10.1074/jbc.M102944200. [DOI] [PubMed] [Google Scholar]
- 73.Soetens O, De Craene JO, Andre B. J Biol Chem. 2001;276:43949–43957. doi: 10.1074/jbc.M102945200. [DOI] [PubMed] [Google Scholar]
- 74.Beck T, Schmidt A, Hall MN. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmidt A, Beck T, Koller A, Kunz J, Hall MN. Embo J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nespoli A, Vercillo R, di Nola L, Diani L, Giannattasio M, Plevani P, Muzi-Falconi M. Cell Cycle. 2006;5:1464–1471. doi: 10.4161/cc.5.13.2914. [DOI] [PubMed] [Google Scholar]
- 77.Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I. Plant Physiol. 1999;119:1527–1534. doi: 10.1104/pp.119.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levin DE. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bermejo C, Rodriguez E, Garcia R, Rodriguez-Pena JM, Rodriguez de la Concepcion ML, Rivas C, Arias P, Nombela C, Posas F, Arroyo J. Mol Biol Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perez-Valle J, Jenkins H, Merchan S, Montiel V, Ramos J, Sharma S, Serrano R, Yenush L. Mol Cell Biol. 2007;27:5725–5736. doi: 10.1128/MCB.01375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hallett MA, Lo HS, Bender A. BMC Cell Biol. 2002;3:16. doi: 10.1186/1471-2121-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S, Barral Y. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 83.Santiago TC, Mamoun CB. J Biol Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- 84.Sreenivas A, Carman GM. J Biol Chem. 2003;278:20673–20680. doi: 10.1074/jbc.M300132200. [DOI] [PubMed] [Google Scholar]
- 85.Anantharaman V, Aravind L. Genome Biol. 2002;3:research0023. doi: 10.1186/gb-2002-3-5-research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schulz TA, Prinz WA. Biochim Biophys Acta. 2007;1771:769–780. doi: 10.1016/j.bbalip.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, et al. Science. 327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lehner B. PLoS One. 5:e9035. doi: 10.1371/journal.pone.0009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wagner A. Bioessays. 2008;30:367–373. doi: 10.1002/bies.20728. [DOI] [PubMed] [Google Scholar]
- 90.Wagner A. Proc Biol Sci. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Draghi JA, Parsons TL, Wagner GP, Plotkin JB. Nature. 463:353–355. doi: 10.1038/nature08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Mol Biosyst. 2009;5:1636–1660. doi: 10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blackburn AS, Avery SV. Antimicrob Agents Chemother. 2003;47:676–681. doi: 10.1128/AAC.47.2.676-681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan TF, Carvalho J, Riles L, Zheng XF. Proc Natl Acad Sci U S A. 2000;97:13227–13232. doi: 10.1073/pnas.240444197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang M, Bellaoui M, Boone C, Brown GW. Proc Natl Acad Sci U S A. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Desmoucelles C, Pinson B, Saint-Marc C, Daignan-Fornier B. J Biol Chem. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- 97.Gupta SS, Ton VK, Beaudry V, Rulli S, Cunningham K, Rao R. J Biol Chem. 2003;278:28831–28839. doi: 10.1074/jbc.M303300200. [DOI] [PubMed] [Google Scholar]
- 98.Hanway D, Chin JK, Xia G, Oshiro G, Winzeler EA, Romesberg FE. Proc Natl Acad Sci U S A. 2002;99:10605–10610. doi: 10.1073/pnas.152264899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zewail A, Xie MW, Xing Y, Lin L, Zhang PF, Zou W, Saxe JP, Huang J. Proc Natl Acad Sci U S A. 2003;100:3345–3350. doi: 10.1073/pnas.0530118100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baetz K, McHardy L, Gable K, Tarling T, Reberioux D, Bryan J, Andersen RJ, Dunn T, Hieter P, Roberge M. Proc Natl Acad Sci U S A. 2004;101:4525–4530. doi: 10.1073/pnas.0307122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown JA, Sherlock G, Myers CL, Burrows NM, Deng C, Wu HI, McCann KE, Troyanskaya OG, Brown JM. Mol Syst Biol. 2006;2:20060001. doi: 10.1038/msb4100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. Mol Syst Biol. 2005;1:2005 0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Van Houten B. Genome Biol. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liao C, Hu B, Arno MJ, Panaretou B. Mol Pharmacol. 2007;71:416–425. doi: 10.1124/mol.106.030494. [DOI] [PubMed] [Google Scholar]
- 106.Luesch H, Wu TY, Ren P, Gray NS, Schultz PG, Supek F. Chem Biol. 2005;12:55–63. doi: 10.1016/j.chembiol.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 107.Xia L, Jaafar L, Cashikar A, Flores-Rozas H. Cancer Res. 2007;67:11411–11418. doi: 10.1158/0008-5472.CAN-07-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chamilos G, Lewis RE, Lamaris GA, Albert ND, Kontoyiannis DP. Antimicrob Agents Chemother. 2008;52:1325–1329. doi: 10.1128/AAC.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dilda PJ, Perrone GG, Philp A, Lock RB, Dawes IW, Hogg PJ. Int J Biochem Cell Biol. 2008;40:1016–1029. doi: 10.1016/j.biocel.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 110.Gustavsson M, Barmark G, Larsson J, Muren E, Ronne H. Mol Genet Genomics. 2008;280:233–248. doi: 10.1007/s00438-008-0359-9. [DOI] [PubMed] [Google Scholar]
- 111.Holland S, Lodwig E, Sideri T, Reader T, Clarke I, Gkargkas K, Hoyle DC, Delneri D, Oliver SG, Avery SV. Genome Biol. 2007;8:R268. doi: 10.1186/gb-2007-8-12-r268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jin YH, Dunlap PE, McBride SJ, Al-Refai H, Bushel PR, Freedman JH. PLoS Genet. 2008;4:e1000053. doi: 10.1371/journal.pgen.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Serero A, Lopes J, Nicolas A, Boiteux S. DNA Repair (Amst) 2008;7:1262–1275. doi: 10.1016/j.dnarep.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 114.Tucker CL, Fields S. Comp Funct Genomics. 2004;5:216–224. doi: 10.1002/cfg.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu L, Lopez A, Anaflous A, El Bali B, Hamal A, Ericson E, Heisler LE, McQuibban A, Giaever G, Nislow C, Boone C, Brown GW, Bellaoui M. PLoS Genet. 2008;4:e1000284. doi: 10.1371/journal.pgen.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jo WJ, Loguinov A, Chang M, Wintz H, Nislow C, Arkin AP, Giaever G, Vulpe CD. Toxicol Sci. 2008;101:140–151. doi: 10.1093/toxsci/kfm226. [DOI] [PubMed] [Google Scholar]
- 117.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 119.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 120.Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, Kemmeren P, Holstege FC, Weissman JS, Keogh MC, Koller D, et al. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, et al. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 122.Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Csardi G, Nepusz T. InterJournal. Complex Systems; 2006. p. 1695. [Google Scholar]
- 124.Barabasi AL, Oltvai ZN. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 125.Yu H, Paccanaro A, Trifonov V, Gerstein M. Bioinformatics. 2006;22:823–829. doi: 10.1093/bioinformatics/btl014. [DOI] [PubMed] [Google Scholar]
- 126.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eddy SR. PLoS Comput Biol. 2008;4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Soding J, Biegert A, Lupas AN. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen XJ, Guan MX, Clark-Walker GD. Nucleic Acids Res. 1993;21:3473–3477. doi: 10.1093/nar/21.15.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stuible HP, Meier S, Wagner C, Hannappel E, Schweizer E. J Biol Chem. 1998;273:22334–22339. doi: 10.1074/jbc.273.35.22334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.