Abstract
In this work we demonstrate the existence in Vibrio anguillarum 775 (pJM1) of two chromosomal genes encoding outer membrane proteins that operate in the transport of ferric enterobactin. One of them is a novel receptor that we named FetA and the other is the already characterized FvtA that functions in the uptake of iron complexes of both enterobactin and vanchrobactin. Ferric enterobactin transport proficiency was resumed in double mutants for these two genes when they were complemented with either fetA or fvtA, whereas only the cloned fvtA could complement for ferric vanchrobactin transport. Quantitative RT-PCR assays demonstrated that transcription of the fetA gene is regulated by FetR, that is encoded upstream and in reverse orientation from fetA. This gene as well as fetA, are up-regulated in iron limiting condition in a Fur-dependent manner. The two divergent promoters are located in the intergenic region between fetR and fetA that has a putative Fur binding site and an IrgB binding site in the overlapping promoters of fetR and fetA. FetA and FetR show high homology to V. cholerae IrgA and IrgB respectively and the intergenic regions fetA–fetR and irgA–irgB are also highly related suggesting a vertical transmission of the fetA–fetR cluster from V. cholerae to V. anguillarum.
Keywords: Iron, Siderophore, Enterobactin, Outer membrane receptor
Introduction
Vibrio anguillarum 775 (pJM1) serotype O1 is a fish pathogen that causes terminal hemorrhagic septicemia in marine and fresh water fish. Iron uptake systems mediated by siderophores and heme have been reported to be important in the survival of this bacterium under iron limiting conditions as those found in the vertebrate host and in the environment (Actis et al. 2011; Crosa 1980; Naka and Crosa 2011).
Many O1 strains of V. anguillarum carry the pJM1-type virulence plasmid that harbors genes for the biosynthesis of the siderophore anguibactin and its cognate transport system (Actis et al. 2011; Crosa 1980; Crosa and Walsh 2002; Di Lorenzo et al. 2003). Some of the genes responsible for anguibactin biosynthesis are located in the chromosome and are functional homologues of the plasmid harboring the anguibactin biosynthesis genes (Alice et al. 2005; Naka et al. 2008). It is of interest that these very same homologues are part of a biosynthesis cluster coding for the siderophore vanchrobactin (Naka et al. 2008). However, the 775 (pJM1) strain of V. anguillarum does not produce this siderophore. We suggested that V. anguillarum 775 (pJM1) originally had the ability to produce vanchrobactin, but that this strain lost that capability after acquiring the pJM1 plasmid. At that time a transposon from pJM1 might have inserted into the vanchrobactin biosynthesis gene vabF (Naka et al. 2008) possibly because vanchrobactin biosynthesis was no longer necessary, since anguibactin is a stronger iron chelator (Naka et al. 2008). However, although not producing vanchrobactin, the 775 strain can still take up this siderophore using the outer membrane ferric vanchrobactin receptor FvtA, first identified by our laboratory in the 775 (pJM1) strain (Naka et al. 2008), and the cognate cytoplasmic membrane transport genes. Recently it was demonstrated in the V. anguillarum strain RV22, a naturally-occurring pJM1-less strain and a vanchrobactin producer, that the FvtA protein might as well contribute to the transport of ferric enterobactin because a fvtA mutant shows a reduced uptake of ferric enterobactin (Balado et al. 2009). The fact that albeit reduced, transport still occurs, indicates that there must be another receptor to transport enterobactin in the absence of fvtA. In this work we report that a novel receptor, FetA, a homologue of the V. cholerae enterobactin receptor IrgA, functions as a ferric-enterobactin outer membrane receptor in V. anguillarum 775 (pJM1), but not for ferric vanchrobactin unless it is overexpressed. We also demonstrate that expression of the fetA gene in strain 775 is positively controlled by FetR, a LysR type regulator, and that fetA and fetR are iron regulated in a Fur dependent manner.
Materials and methods
Strains, plasmids and growth conditions
Strains and plasmids used in this study are listed in Table 1. V. anguillarum strains were cultured at 25°C in Trypticase soy broth supplemented with 1% NaCl (TSBS) or on agar (TSAS). As iron limiting growth conditions V. anguillarum strains were incubated in M9 minimal medium (Crosa 1980) supplemented with 0.2% Casamino Acids, and 5% NaCl (CM9) and in some experiments the iron chelator ethylenediamine-di-(o-hydroxyphenyl acetic acid) (EDDA). Ferric ammonium citrate was added into CM9 medium for iron rich growth conditions. E. coli strains were cultured at 37°C in LB broth or on LB agar. When necessary, antibiotics were used at following concentrations: for V. anguillarum, chloramphenicol 10 µg/ml and rifampicin 100 µg/ml; for E. coli, ampicillin 100 µg/ml and chloramphenicol 30 µg/ml.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Phenotype | Source or reference |
|---|---|---|
| V. anguillarum strains | ||
| 775 (pJM1) | Wild-type, Washington (serotype O1, pJM1) | Crosa (1980) |
| 775 (pJM1)-pMMB | 775 (pJM1) harboring pMMB208 | Naka et al. (2008) |
| HNVA-10 | 775 (pJM1)ΔfetR | This study |
| RV22 | Wild type, Spain (serotype O2, plasmidless) | Lemos et al. (1988) |
| RV22-pMMB | RV22 harboring pMMB208 | This study |
| CC9-16 | 775 (pJM1) derivative of anguibactin deficient, anguibactin transport system deficient | Walter et al. (1983) |
| CC9-16ΔfvtA | CC9-16ΔfvtA | Naka et al. (2008) |
| HNVA-6 | CC9-16ΔtonB2 | Naka et al. (2010) |
| HNVA-7 | CC9-16ΔfetA | This study |
| HNVA-8 | CC9-16ΔfvtAΔfetA | This study |
| H775-3 | Plasmidless derivative of 775 (pJM1) | Crosa (1980) |
| HNVA-9 | H775-3ΔfetAΔfvtA | This study |
| 775MET11 | 775 fur point mutant isolated in the presence of 10 mM MnCl2 | Tolmasky et al. (1994) |
| E. coli strains | ||
| Top10 | F-mcrA Δ(mrr-hsdRMS-mcrBC) | Invitrogen |
| φ80lacZΔM15 ΔlacX74 recA1 | ||
| araD139Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | ||
| S17-λkpir | λ-pir lysogen; thi pro hsdR hsdM+ recA RP4-2 Tc:Mu-Km:Tn7(Tpr Smr) | Simon et al. (1983) |
| Plasmids | ||
| pCR2.1 | Ampr, Kmr, PCR cloning vector | Invitrogen |
| pMMB208 | A broad-host-range expression vector; Cmr IncQ lacIq Ptac; polylinker from M13mp19 | Morales et al. (1991) |
| pHN5 | pDM4 harboring ΔfvtA | This study |
| pHN6 | pDM4 harboring ΔfetA | This study |
| pHN7 | pDM4 harboring ΔfetR | This study |
| pHN8 | pMMB208 harboring fvtA | This study |
| pHN9 | pMMB208 harboring fetA | This study |
| pHN10 | pMMB208 harboring fetR | This study |
Construction of mutants and their complementation
DNA cloning and PCR were carried out by standard protocols (Sambrook and Russell 2001). Upstream and downstream regions of the genes to be mutated were combined by Splicing by Overlapping Extension (SOE) PCR using primers listed in Table 2 (Senanayake and Brian 1995). First PCR reactions were performed using -F and -1, and -R and -2 primers in separate tubes, while -F and -R primers were used for the second PCR (Table 2). The generated fragments were cloned into the T-vector, and subcloned into suicide vector pDM4. Conjugation of plasmids into V. anguillarum as well as screening of mutants was followed using procedures we previously described (Naka et al. 2008). To complement the mutations, the wild type genes containing the Shine-Dalgarno sequence were PCR amplified, cloned into pMMB208 and conjugated into V. anguillarum strains.
Table 2.
Primers used in this study
| Primers name | Nucleotide sequences |
|---|---|
| Construction of mutants | |
| fvtA mut-F | TTTTGAGCCAGTGGATCAGC |
| fvtA mut-R | CGTGTTTGTCAACATGGATCAG |
| fvtA mut-1 | CTAGAACTGGTAATCAGCGACATCCATGGGCTTCATGACCT |
| fvtA mut-2 | AGGTCATGAAGCCCATGGATGTCGCTGATTACCAGTTCTAG |
| fetA mut-F | TCAAGATGCGACTCTTCTGC |
| fetA mut-R | CTCAATGTGCTCCAAACCATC |
| fetA mut-1 | CCTAACCAATAACGACGTCCATTCAGCGGATTGAGTCTGG |
| fetA mut-2 | CCAGACTCAATCCGCTGAATGGACGTCGTTATTGGTTAGG |
| fetR mut-F | TTCAATGGCTTGCAGTGGTG |
| fetR mut-R | ACCAAGTGGAATCAGCCGTC |
| fetR mut Cla1-1 | GGTGCTTTCCATTCCTCAGGATCGATGGAAAGCTTTGATCGCACTG (underline, ClaI site) |
| fetR mut Cla1-2 | CAGTGCGATCAAAGCTTTCCATCGATCCTGAGGAATGGAAAGCACC (underline, ClaI site) |
| Complementation | |
| fvtA comp SphI F | GCATGCCGATAATGGTTGCAGCAACATC (underline, SphI site) |
| fvtA comp XbaI R | TCTAGACTAGAACTGGTAATCAGCGAC (underline, XbaI site) |
| fetA com SphI F | GCATGCCAAAATTATGAATAATCCACTCCTG (underline, SphI site) |
| fetA com EcoRI R | GAATTCTTAAAAATTAACGTCAATACCTAACC (underline, EcoRI site) |
| fetR com SphI F | GCATGCACAGATGGAATGGTCGATACCG (underline, SphI site) |
| fetR com XbaI R | TCTAGATTATGGTGCTTTCCATTCCTCAG (underline, XbaI site) |
| qRT-PCR | |
| fetA-QRT-F | TCACCTCATCACACGTGTTC |
| fetA-QRT-R | ATTTGGTCGAGTTTGACGCG |
| fetR-QRT-F | AAGGCAACCGTCTAACTTTGAC |
| fetR-QRT-R | TCCAGCCTTGTTGAGCAAAC |
| aroC-QRT-F | GTCGAATCCACCTTATGTGG |
| aroC-QRT-R | ACGGAATGTGGTCGTATTGG |
Bioassays
Bioassays were used to detect ferric-siderophore transport as described previously (Tolmasky et al. 1988). Briefly, 50 µl of overnight cultures of the strains to be tested were mixed with 20 ml CM9 melted agar (1.5% agar and ~40°C) supplemented with Cm (10 µg/ml), IPTG (500 µM) and EDDA (20 µM). After solidification, V. anguillarum 775 (pJM1) as an anguibactin source, V. anguillarum RV22 as a vanchrobactin source, purified enterobactin (1 mg/ml) (EMC microcollections GmbH, Tubingen, Germany) and ferric ammonium citrate (10 mg/ml) were streaked and/or spotted on the agar, and incubated 48 h at 25°C. The existence of growth halos around streaked or spotted iron sources were checked at the 24 and 48 h time points.
Expression analysis of V. anguillarum genes
Total RNAs were extracted using RiboPure-Bacteria™ kit (Ambion, Austin, TX) from V. anguillarum strains grown in different conditions, and treated with TURBO DNA-free™ kit (Ambion, Austin, TX) to remove contaminated residual DNA. The reverse transcription (RT) reaction was performed with SuperScript™ II Reverse Transcriptase (Invitrogen, Carlsbad, CA) using the Random primer (Invitrogen, Carlsbad, CA). Quantitative PCR experiments were carried out using Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) following the procedure described in the manual.
Outer membrane extraction and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
Outer membrane fractions of V. anguillarum were extracted using methods previously described with minor modifications (Crosa and Hodges 1981). Briefly, V. anguillarum was cultured in CM9 medium; late-exponential phase bacteria were harvested and pellets were resuspended into 600 µl of Tris–HCl (pH 7.6) buffer. After sonication of the suspension, cell debris was removed by centrifugation. The supernatants were treated with 1.5% sodium lauroyl sarcosinate for 2 h at 4°C, and centrifuged 1 h at 15,000 rpm at 4°C, then pellets were re-suspended in distilled water. Samples were subjected to SDS-PAGE using Criterion™ XT Precast Gel (10% Bis–Tris) (Bio-Rad, Hercules, CA, USA) for 5 h at 80 V and the gel was stained with Bio-Safe™ Commassie G-250 stain (Bio-Rad).
Results
FetA is an enterobactin receptor in V. anguillarum 775 (pJM1)
The serotype O1 strain 775 (pJM1) carrying the 65 kb pJM1 plasmid can take up the endogenous siderophore anguibactin as well as exogenous siderophores vanchrobactin, enterobactin and ferrichrome (Lemos et al. 1988; Naka et al. 2008). Strain H775-3, from which the pJM1 plasmid had been cured, is still able to use enterobactin as well as vanchrobactin as iron sources indicating that the specific receptor(s) for the ferric complexes of those two siderophores must be encoded on the chromosome. We recently identified in the V. anguillarum 775 (pJM1) strain the fvtA gene that shows 30% identity with the Bordetella pertussis enterobactin receptor gene, bfeA (Naka et al. 2008). We also showed that an fvtA mutant in an anguibactin production negative 775 (pJM1) derivative, does not transport vanchrobactin (Naka et al. 2008). Furthermore, Balado et al. (2009) demonstrated that FvtA in V. anguillarum serotype O2 strain RV22 could be involved in vanchrobactin as well as enterobactin transport. However the fvtA mutant in RV22 is still able to transport enterobactin indicating that this strain must have another enterobactin receptor. We first performed bioassays to determine whether the 775 (pJM1) fvtA mutant can still transport ferric enterobactin. As shown in Table 3, with this mutant we can only detect uptake of ferric enterobactin but not of ferric vanchrobactin, as compared to the control tonB2 mutant of this strain that cannot use any those two ferric siderophores as iron sources. Thus, there must indeed be another receptor for ferric-enterobactin in the chromosome of 775 (pJM1). Based on our whole genome sequencing data of V. anguillarum 775 (pJM1) (Naka et al. 2011) we identified the putative receptor gene named fetA that shares high similarity to the V. cholerae irgA gene (68% identity and 81% similarity in protein level) encoding a ferric enterobactin receptor.
Table 3.
Bioassay to test ferric-siderophore utilization
| Indicator strains | Iron sources | |||
|---|---|---|---|---|
| Anguibactin | Vanchrobactin | Enterobactin | FAC | |
| Wt (pMMB208) | + | + | + | + |
| ΔtonB2 (pMMB208) | − | − | − | + |
| ΔfvtA (pMMB208) | + | − | + | + |
| ΔfetA (pMMB208) | + | + | + | + |
| ΔfvtAΔfetA (pMMB208) | + | − | − | + |
| ΔfvtAΔfetA (pMMB208-fvtA) | + | + | + | + |
| ΔfvtAΔfetA (pMMB208-fetA) | + | +w | + | + |
50 µl of overnight cultures of indicator strains in CM9 broth was mixed with 20 ml of melted CM9 agar (adjusted to ~40°C) supplemented with 1 mM IPTG and 20 µM EDDA. After solidification iron sources such as anguibactin from V. anguillarum 775 (pJM1)-pMMB, vanchrobactin from V. anguillarum RV22-pMMB and 1 µM of purified enterobactin from EMC microcollections GmbH was spotted on the plates, and growth halos around spots were checked after 24 or 48 h incubation at 25°C. Ferric ammonium citrate (FAC) was used as a positive control. +, growth at 24 and 48 h; +w, weakly growth only at 48 h; −, no growth at 24 and 48 h. Wt, CC9-16; ΔtonB2, HNVA-6; ΔfetA, HNVA-7; ΔfvtAΔfetA, HNVA-8
To assess whether the fetA gene product is functional as a receptor for enterobactin we mutated this gene in an fvtA mutant background. Table 3 shows that the double fvtA and fetA mutant thus constructed, is unable to use vanchrobactin and enterobactin as iron sources even when the strain was incubated for long times. When the mutation is complemented with the fetA gene in trans we observed the recovery of ferric enterobactin transport and a very weak ferric vanchrobactin transport was also detected when it is over-expressed. These results indicate that the fetA protein main function must be as a ferric enterobactin receptor.
fetA is an iron regulated outer membrane protein
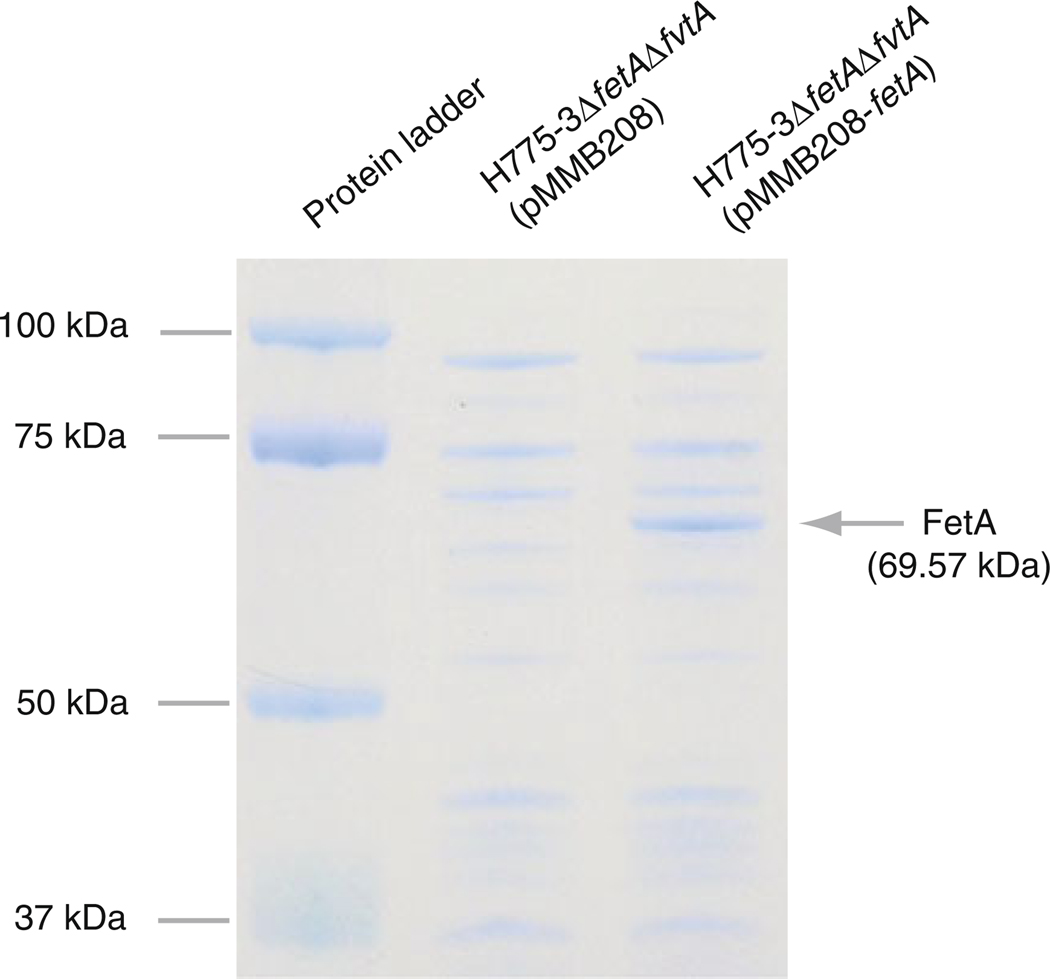
It was difficult to detect the fetA protein in the wild type strain membranes, likely due to its low expression level. Thus we needed to use a fetA overexpressing strain to test whether FetA is located in the outer membrane fractions. The predicted size of FetA is similar to FatA and FvtA, thus to detect FetA we constructed the H775-3ΔfetAΔfvtA strain that does not express fetA, fatA nor fvtA. The fetA gene was overexpressed from pMMB208-fetA in this strain. The H775-3ΔfetAΔfvtA strain carrying empty pMMB208 was used as a negative control. The SDS-PAGE of the outer membrane preparations shown in Fig. 1 provides the correlation between the presence of an outer membrane protein band of 69.57 kDa that corresponds to the mature FetA protein in the H775-3ΔfetAΔfvtA (pMMB208-fetA) while the negative control did not show this band indicating that FetA actually localize in the outer membrane of V. anguillarum.
Fig. 1.
Localization of FetA in the outer membrane fractions. H775-3ΔfetAΔfvtA(pMMB208), HNVA-9 carrying pMMB208 and H775-3ΔfetAΔfvtA(pMMB208-fetA), HNVA-9 carrying pHN9 were grown in CM9 broth supplemented with 5 µg/ml Cm and 500 µM IPTG and outer membrane fractions were extracted by using sarkosyl. Samples were subjected to SDS-PAGE using Criterion™ XT Precast Gel (10% Bis–Tris) for 5 h at 80 V. Protein ladder, Precision Plus Protein Standards (Bio-Rad)
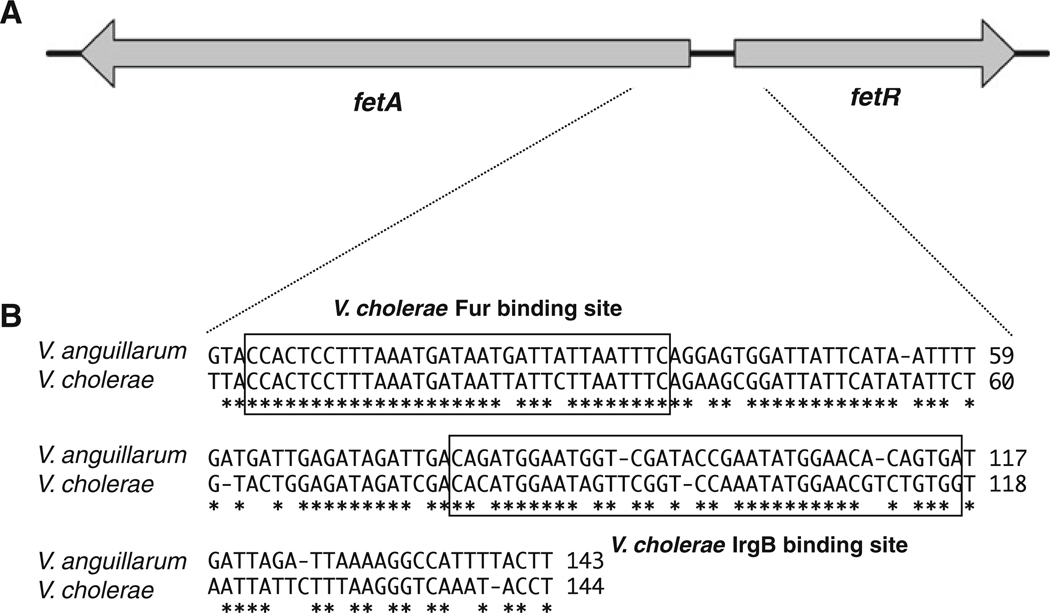
A regulator of fetA expression
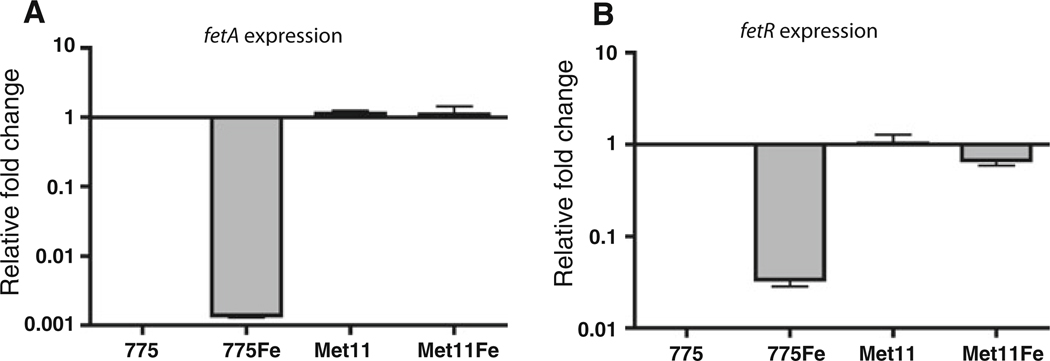
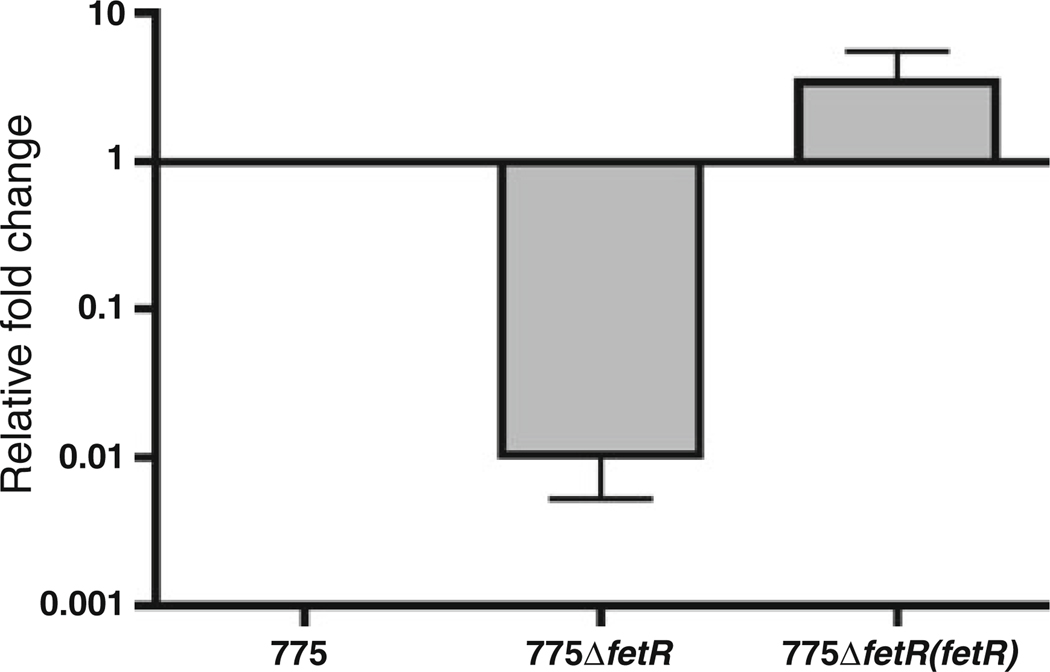
Analysis of the upstream region of fetA unveiled the presence of a gene that we named fetR that is a homologue (72% identity and 83% similarity in protein level) of the V. cholerae irgB gene belonging to the LysR family of transcriptional activators (Fig. 2). In V. cholerae IrgB is a positive regulator of the irgA gene, and irgA and irgB were both iron regulated in a Fur-dependent manner (Goldberg et al. 1990a, b, 1991; Watnick et al. 1998). The comparison of the putative promoter region of fetA and fetR with the corresponding region of V. cholerae revealed that they are highly conserved including a ferric uptake regulator (Fur) and IrgB binding sites identified between irgA and irgB in V. cholerae (Watnick et al. 1998) (Fig. 2). The similarity of both genes as well as the promoter region motivated us to test whether they have similar regulation mechanisms in both bacteria. First, we determined whether fetR and/or fetA are regulated by iron concentration and/or Fur using quantitative RT-PCR (qRT-PCR). As shown in Fig. 3, the expression of both fetR and fetA is clearly down regulated under iron rich condition while expression patterns in a fur mutant were similar in both iron-rich and iron-limiting conditions. These results indicate that fetR as well as fetA are iron regulated in a Fur-dependent manner. Furthermore, we examined the expression of fetA in the fetR wild type and mutant backgrounds using qRT-PCR (Fig. 4). Transcription of fetA was dramatically decreased in the fetR deletion mutant demonstrating that fetR is required for the expression of fetA. Complementation of the fetR mutant in trans restored the fetA expression similar to the wild type level. These results clearly demonstrate that fetR encoded upstream and in reverse orientation from fetA, is required for fetA expression at the level of transcription.
Fig. 2.
Schematic map of V. anguillarum fetA and fetR (a), and the comparison of its intergenic region with that between V. cholerae irgB and irgA (b). a The orientation of the fetA and fetR was shown as arrows. b The nucleotide sequence of the intergenic region of V. anguillarum fetA–fetR as compared with that between V. cholerae irgA and irgB. Asterisks designate conserved nucleotide sequences, and the Fur binding site and the IrgB binding site previously identified in V. cholerae (Watnick et al. 1998) are shown in the box
Fig. 3.
Iron and Fur regulation of fetA and fetR. The expression of fetA (a) and fetR (b) was evaluated by qRT-PCR. 775, 775 (pJM1) grown in CM9; 775Fe, 775 (pJM1) grown in CM9 with 10 µg/ml ferric ammonium citrate (FAC); Met11, 775MET11 (a fur mutant of 775) grown in CM9; Met11Fe, 775Met11 grown in CM9 with 10 µg/ml FAC. aroC was used as an internal control, and expression levels are expressed relative to 775 samples. The data represents mean value of at least three independent experiments (error bars indicate the standard error of the mean)
Fig. 4.
FetR positively regulates the expression of fetA. 775, 775 (pJM1) carrying pMMB208; 775ΔfetR, HNVA-10 carrying pMMB208; 775ΔfetR(fetR), HNVA-10 carrying pHN10. RNAs were extracted from exponential phase cultures of V. anguillarum strains grown in CM9. RT reactions were performed using random primers. aroC was used as an internal control. Data were normalized to aroC and compared with 775 data. The data represents mean value of at least three independent experiments (error bars indicate the standard error of the mean)
Discussion
Little is known about the mechanisms by which V. anguillarum cells recognize the ferric enterobactin complexes and subsequently internalize the essential iron into the cell cytosol. In this work, we initiated the identification and characterization of the components of the ferric enterobactin transport system. The experimental evidence indicates that FetA must be a receptor for ferric enterobactin transport. However it was also clear that the inability to grow in the presence of enterobactin under iron limitation conditions only occurred if the V. anguillarum cells were impaired in the expression of both fetA and fvtA. When these double mutants were complemented with either fetA or fvtA, ferric enterobactin transport proficiency was resumed indicating that FetA and FvtA can both operate as ferric enterobactin outer membrane receptors. Furthermore, our bioassay experiments demonstrated that FetA does not act as a vanchrobactin receptor at the chromosomal expression level. Our recent whole genome sequencing study revealed that both FvtA and FetA exist in all of sequenced V. anguillarum strains such as O1 serotype pJM1-carrying strain 775 (pJM1), O1 serotype pJM1-less strain 96F and O2 serotype pJM1-less strain RV22 while V. ordalii, that is a very closely related species to V. anguillarum only harbors fvtA but not fetA (Naka et al. 2011).
The comparative analysis of the intergenic sequences between V. anguillarum fetA and fetR and V. cholerae irgA and irgB revealed that the IrgB binding site as well as Fur binding site identified in V. cholerae (Watnick et al. 1998) are highly conserved in V. anguillarum suggesting similar expression mechanisms in those two systems. qRT-PCR assays using V. anguillarum 775 (pJM1) confirmed that transcription of the fetA gene is regulated by FetR, which is encoded upstream and in reverse orientation from fetA and that fetA as well as fetR are iron regulated in a Fur dependent manner. Based on the similarity of genes as well as possession of similar regulation mechanisms, we can speculate on the vertical transmission of the fetA–fetR cluster from V. cholerae to V. anguillarum.
It is intriguing that strain 775 (pJM1) is able to transport ferric vanchrobactin although this strain lost the ability to biosynthesize this ancestral siderophore due to interruption of the vanF gene (Naka et al. 2008). The FvtA receptor also can be used as a ferric enterobactin receptor, and V. anguillarum carries the additional enterobactin receptor FetA. We speculate that V. anguillarum 775 (pJM1) can utilize ferric vanchrobactin or ferric enterobactin in environmental conditions where those exogenous ferric siderophores are available. It has been demonstrated that some Aeromonas strains produce enterobactin, and V. anguillarum pJM1-less O1 and O2 serotype strains and Vibrio sp. DS40M4 produce vanchrobactin (Andrus and Payne 1983; Lemos et al. 1988; Massad et al. 1991; Sandy et al. 2010). These bacteria could be candidates for the source of the two exogenous siderophores since they are inhabitants of the aquatic environments where V. anguillarum pJM1-carrying strains can be found. We are currently performing experiments to assess the particular environmental conditions in which FvtA and/or FetA are necessary to enhance the growth of V. anguillarum pJM1-carrying strains and how this organism can interact with the enterobactin and/or vanchrobactin-producing bacteria.
Acknowledgment
This work was supported by a Public Health Service grant (AI19018) from the National Institute of Allergy and Infectious Diseases to J.H.C.
Contributor Information
Hiroaki Naka, Department of Molecular Microbiology and Immunology, Oregon Health and Science University, 3181 SW Sam Jackson Park Road, Portland, OR 97239, USA.
Jorge H. Crosa, Email: crosajor@ohsu.edu, Department of Molecular Microbiology and Immunology, Oregon Health and Science University, 3181 SW Sam Jackson Park Road, Portland, OR 97239, USA.
References
- Actis LA, Tolmasky ME, Crosa JH. Vibriosis. In: Woo PTK, Bruno DW, editors. Fish diseases and disorders, vol 3: viral, bacterial, and fungal infections. 2nd edn. Oxfordshire: CABI International; 2011. pp. 570–605. [Google Scholar]
- Alice AF, Lćpez CS, Crosa JH. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J Bacteriol. 2005;187:2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrus C, Payne SM. Siderophores and iron regulated proteins in Vibrio and Aeromonas species. Abstr Annu Meet Am Soc Microbiol D. 1983;13:61. [Google Scholar]
- Balado M, Osorio CR, Lemos ML. FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: utility as a route of entry for vanchrobactin analogues. Appl Environ Microbiol. 2009;75:2775–2783. doi: 10.1128/AEM.02897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature. 1980;284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- Crosa JH, Hodges LL. Outer membrane proteins induced under conditions of iron limitation in the marine fish pathogen Vibrio anguillarum 775. Infect Immun. 1981;31:223–227. doi: 10.1128/iai.31.1.223-227.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo M, Stork M, Tolmasky ME, Actis LA, Farrell D, Welch TJ, Crosa LM, Wertheimer AM, Chen Q, Salinas P, Waldbeser L, Crosa JH. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J Bacteriol. 2003;185:5822–5830. doi: 10.1128/JB.185.19.5822-5830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Boyko SA, Calderwood SB. Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli fur system. J Bacteriol. 1990a;172:6863–6870. doi: 10.1128/jb.172.12.6863-6870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, DiRita VJ, Calderwood SB. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect Immun. 1990b;58:55–60. doi: 10.1128/iai.58.1.55-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Boyko SA, Calderwood SB. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos ML, Salinas P, Toranzo AE, Barja JL, Crosa JH. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J Bacteriol. 1988;170:1920–1925. doi: 10.1128/jb.170.4.1920-1925.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad G, Arceneaux JE, Byers BR. Acquisition of iron from host sources by mesophilic Aeromonas species. J Gen Microbiol. 1991;137:237–241. doi: 10.1099/00221287-137-2-237. [DOI] [PubMed] [Google Scholar]
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Naka H, Crosa JH. Genetic determinants of virulence in the marine fish pathogen Vibrio anguillarum. Fish Pathol. 2011;46:1–10. doi: 10.3147/jsfp.46.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, López CS, Crosa JH. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ Microbiol. 2008;10:265–277. doi: 10.1111/j.1462-2920.2007.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, López CS, Crosa JH. Role of the pJM1 plasmid-encoded transport proteins FatB, C and D in ferric anguibactin uptake in the fish pathogen Vibrio anguillarum. Environ Microbiol Rep. 2010;2:104–111. doi: 10.1111/j.1758-2229.2009.00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, Dias GM, Thompson CC, Dubay C, Thompson FL, Crosa JH. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect Immun. 2011;79:2889–2900. doi: 10.1128/IAI.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sandy M, Han A, Blunt J, Munro M, Haygood M, Butler A. Vanchrobactin and anguibactin siderophores produced by Vibrio sp. DS40M4. J Nat Prod. 2010;73:1038–1043. doi: 10.1021/np900750g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake SD, Brian DA. Precise large deletions by the PCR-based overlap extension method. Mol Biotechnol. 1995;4:13–15. doi: 10.1007/BF02907467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system in vivo genetic engineering transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:787–796. [Google Scholar]
- Tolmasky ME, Actis LA, Crosa JH. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J Bacteriol. 1988;170:1913–1919. doi: 10.1128/jb.170.4.1913-1919.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky ME, Wertheimer AM, Actis LA, Crosa JH. Characterization of the Vibrio anguillarum fur gene: role in regulation of expression of the FatA outer membrane protein and catechols. J Bacteriol. 1994;176:213–220. doi: 10.1128/jb.176.1.213-220.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MA, Potter SA, Crosa JH. Iron uptake system medicated by Vibrio anguillarum plasmid pJM1. J Bacteriol. 1983;156:880–887. doi: 10.1128/jb.156.2.880-887.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Butterton JR, Calderwood SB. The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes, irgA and irgB. Gene. 1998;209:65–70. doi: 10.1016/s0378-1119(98)00018-3. [DOI] [PubMed] [Google Scholar]