Introductory paragraph
Microtubules are polymers of tubulin heterodimers that display dynamic instability: periods of growth followed by periods of shrinkage1. However, the molecular regulation of dynamic instability remains elusive. Here we show that EFA-6, a cortically-localized protein2, limits the growth of microtubules near the cell cortex of early embryonic cells, possibly by inducing microtubule catastrophes. Embryos lacking this protein display abnormally long and dense microtubules at the cell cortex, and growing microtubule plus ends reside at and grow along the cortex for up to five-fold longer times, compared to wild-type embryos. Loss of EFA-6 also caused excess centrosome separation and their displacement towards the cell cortex early in mitosis, and subsequently a loss of anaphase spindle pole oscillations and increased rates of spindle elongation. The centrosome separation phenotype was dependent on the motor protein dynein, suggesting a possible link between the modulation of microtubule dynamics at the cortex and dynein-dependent force production. EFA-6 orthologs activate ARF6-type GTPases to regulate vesicle trafficking3; however, we show that only the C. elegans EFA-6 N-terminus is both necessary and sufficient to limit microtubule growth along the cortex, and that this function is independent of ARF-6.
During mitosis in one-cell stage C. elegans embryos, microtubule (MT) contact with the cell cortex is important for spindle positioning and function4-8. MTs are regulated such that their plus ends grow from centrosomes towards the cell cortex and reside there briefly prior to undergoing catastrophe and shrinkage through plus end depolymerization, as in other systems1, 4, 9. Furthermore, the minus-end directed microtubule motor cytoplasmic dynein (hereafter designated as “dynein”) is thought to capture and pull on MT plus ends at the cortex to influence spindle position and function10, 11. To identify genes that regulate dynein or influence MT dynamics, we previously identified 12 nonessential and conserved C. elegans loci that, when reduced in function, specifically suppress the lethality associated with conditional dynein heavy chain (dhc-1) mutations2.
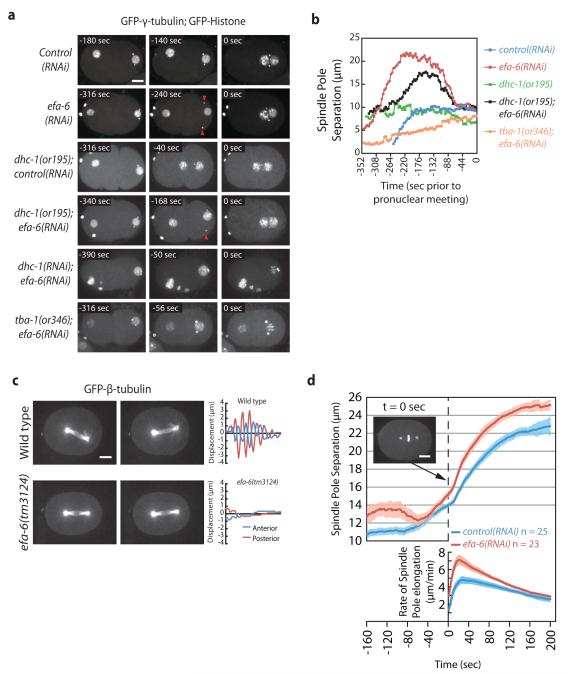
To investigate the requirements for these dynein suppressors, we depleted them and observed the first embryonic mitosis in live embryos. Depletion of one nonessential dynein suppressor, efa-6, resulted in several intriguing anomalies when we imaged the first cell cycle using time-lapse video microscopy (Fig. 1). First, in wild-type embryos, sperm contribute paternal chromosomes and two centrosomes that remain attached to the male pronuclear envelope during pronuclear migration (n = 25). However, in efa-6(RNAi) embryos, one (38%) or both (38%) centrosomes detached from the male pronucleus and moved towards the cell cortex (n = 24; Fig. 1a, Fig. S1, and Movie 1). After the initial detachment, centrosomes always returned to the pronuclei in efa-6(-) embryos with roughly normal separation, mitosis ensued and daughter cells displayed normal size asymmetry. We did notice a weak delay in pronuclear rotation: NEBD occurred at 25 ± 14 ° (n = 16, range: 1-80 °) in wild type and at 40 ± 8 ° (n = 22, range: 5-81 °) in efa-6(tm3124) embryos, however spindles always aligned along the anterior-posterior axis prior to anaphase in both strains. In embryos with reduced dynein function, centrosomes fail to separate or collapse towards each other prior to nuclear envelope breakdown12, 13. After depleting efa-6 in dhc-1(or195ts) embryos, centrosomes again detached from the sperm pronucleus and centrosome separation was rescued at pronuclear meeting (Fig. 1b and Movie 2). However dhc-1(or195ts) is not a null allele and still possesses some activity13, 14. Thus, we used RNAi to more severely deplete dhc-1. as evidenced by the presence of multiple maternal pronuclei, un-separated centrosomes, and no pronuclear migration. In these embryos centrosomes never detached from the male pronucleus, indicating that dynein is required for the detachment observed in efa-6(-) embryos (Fig. 1a and Movie 3). To test if MTs are also required for the forces that pull centrosomes off the male pronucleus, we used a fast-acting temperature-sensitive dominant α-tubulin mutant, tba-1(or346d,ts), to partially disrupt MTs in efa-6-depleted embryos15. This mutant expresses a TBA-1 protein with a single amino acid substitution (S377F) that could alter MT assembly, stability, or interfere with MT-binding proteins; but in contrast to depleting alpha-tubulin with RNAi, robust asters are still present15. We found that the tubulin mutation completely blocked centrosome detachment, even though centrosome separation occurred (Figs. 1a, 1b, and S1). We conclude that both dynein and MTs are required for the centrosome detachment and excess separation that occurs in the absence of efa-6.
Figure 1.
Loss of EFA-6 affects early MT-dependent processes. a, Images from embryos expressing GFP-γ-tubulin and GFP-histone reporter fusions to mark centrosomes and chromosomes. Times indicated are relative to pronuclear meeting, or to paternal pronuclear envelope breakdown in the tba-1(or346d,ts); efa-6(RNAi) and dhc-1(RNAi); efa-6(RNAi) embryos in which pronuclei did not meet. Multiple Z sections were captured every 4-10 sec and merged to give the images. Centrosomes remained attached to the male pronucleus in control(RNAi) one-cell embryos, but detached and moved toward the cortex in efa-6(RNAi) and dhc-1(or195); efa-6(RNAi) embryos (red arrowheads). Centrosome detachment was not observed when efa-6 was depleted in dhc-1(RNAi) embryos with strong dynein depletion, or in the tubulin mutant tba-1(or346d,ts). b, Quantitation of the inter-centrosomal distance over time during pronuclear migration for representative embryos. Spindle poles were tracked in three dimensions over time and the inter-centrosomal distances were plotted. dhc-1(or195ts) and tba-1(or346d,ts) mutant embryos were imaged 30-120 min. after shifting worms from 15 °C to 26 °C. The wild-type and dhc-1(or195ts) strains were grown on control RNAi-expressing bacteria. Time refers to seconds prior to pronuclear meeting. c, Images (left) and plots of spindle pole transverse positions (right) of GFP-β-tubulin-expressing embryos. Three Z sections 1 μm apart were captured every 2 sec and merged. The two images of each embryo represent the extreme transverse distances travelled by the spindle poles during one oscillation. Spindle pole transverse positions were determined every 2 sec for 152 seconds during anaphase and plotted. The x axes represent time. d, Spindle pole to spindle pole distances were plotted over the course of 6 minutes starting prior to anaphase onset (corresponding to nuclear-centrosomal rotation and nuclear envelope breakdown) in a strain expressing GFP-γ-tubulin and GFP-histone. Each of the efa-6(RNAi) embryos displayed normal centrosome attachment to the pronuclei by this time. The time series were aligned at anaphase onset (dashed line). Images were captured every 2 sec. The lower plot displays the rate of spindle elongation with respect to time. The shaded regions represent the S.E.M. with a confidence interval of 0.95. Scale bars, 10 μm.
Loss of efa-6 function also caused two defects during anaphase. First, we observed an absence of anaphase spindle pole oscillations, or spindle rocking, in efa-6 mutant embryos. Whereas wild-type embryos always displayed robust spindle rocking, oscillations were greatly reduced in efa-6(-) embryos, and the spindle pole movements that did occur were aperiodic (Fig. 1c, and Movie 4). Second, we found that the maximum anaphase spindle pole separation rate was 48% faster in efa-6(-) embryos [control(RNAi): 4.80 ± 0.54 μm/min., efa-6(RNAi): 7.10 ± 0.60 μm/min.], and resulted in longer anaphase spindles, compared to wild-type embryos (Fig. 1d).
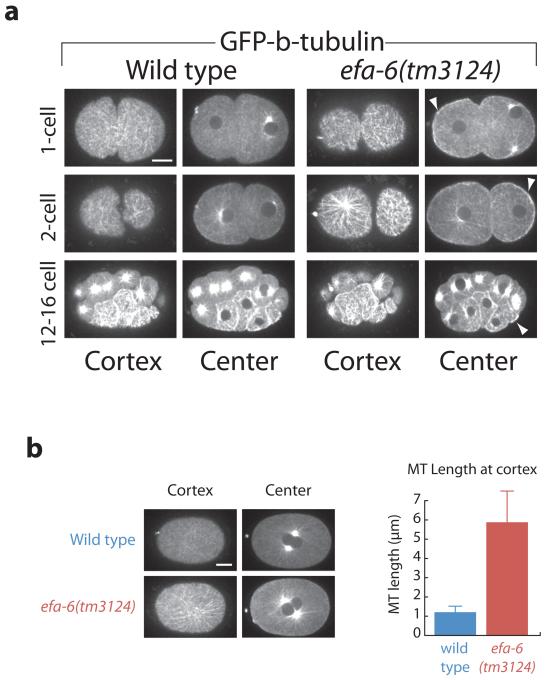
Because centrosome pronuclear detachment in efa-6 mutants required functional MTs, we next used a β-tubulin fusion to Green Fluorescent Protein (GFP) to image MTs in live wild-type and efa-6 mutant embryos. Wild-type one-cell stage embryos displayed very few long MTs at the cell cortex, as reported previously4. However, we observed a dense network of abnormally long MTs at the cell cortex in efa-6 mutants (Fig. 2a and Movie 5). The increase was most pronounced during pronuclear migration (S Phase). Similar to wild type16, efa-6(tm3124) embryos exhibited cell-cycle dependent changes in the density of microtubules, with the number of cortical MTs substantially reduced during nuclear envelope breakdown, metaphase and anaphase. However, at each stage, microtubules were more abundant in efa-6 embryos than in wild type (Supplemental Fig. S2a).
Figure 2.
EFA-6 prevents long cortical MTs in one- and two-cell embryos. Images from wild-type and efa-6(tm3124) embryos expressing GFP-β-tubulin. a, Three Z sections with 0.5 μm spacing were captured and merged to generate the images. Shown for each time point are images captured at the cortical focal plane and the central focal plane. Top row: one-cell stage embryos, middle row: two-cell stage embryos, Bottom row: 12-16 cell stage embryos. Arrowheads indicate examples of enhanced cortical GFP-β-tubulin in central focal planes. b, Cortical MT lengths were determined 100-105 sec after pronuclear meeting (example images at left). 107 MTs were measured from 9 embryos for the wild type, and 118 MTs were measured from 10 embryos for efa-6(tm3124). Error bars represent the S.E.M. with a confidence interval of 0.95. Scale bars, 10 μm.
The efa-6(tm3124) mutation is a deletion allele that removes the C-terminal 456 amino acids in the Y55D9A.1.a isoform and adds a novel 15 amino acid tail. efa-6(tm3124), efa-6(RNAi), and efa-6(RNAi); efa-6(tm3124) embryos each displayed indistinguishable MT phenotypes (not shown). We next examined the length of the cortical MTs in efa-6(-) and wild-type embryos. Because MTs were too dense to easily analyze at earlier stages, we compared their lengths at pronuclear centration and found that MTs were almost 6-fold longer in efa-6(RNAi) embryos compared to wild type (Fig. 2b). A greater abundance of cortical interphase MTs were observed again in two-cell stage embryos, with the mutants displaying substantially longer and denser cortical MTs (Fig. 2a middle row). Interestingly, in four-cell or later stage embryos, dense cortical MTs were present in both mutant and wild-type blastomeres (Fig. 2a, bottom row; Supplemental Fig. S2b). We had previously shown that EFA-6 is enriched at the anterior cortex in one and two-cell embryos but is absent in blastomeres of later stage embryos2. We measured the levels across the embryo cortex and found that GFP-EFA-6 is present throughout the cortex, but at reduced levels in the posterior (Supplemental Fig. S3). We conclude that EFA-6 is present and functions in one- and two-cell stage embryos, but not in later stage embryos, to limit the extent of MT growth throughout the cell cortex.
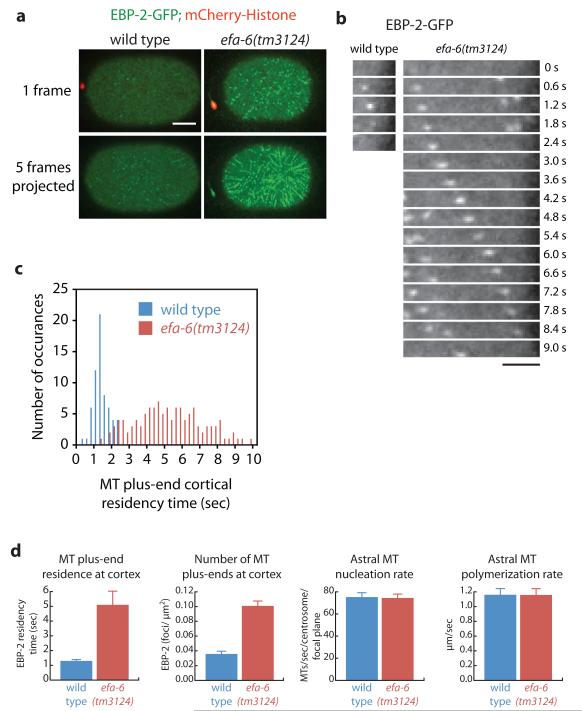
To quantify how EFA-6 influences MT dynamics, we used a GFP fusion to EBP-2, a conserved protein that binds specifically to growing MT plus ends, to measure MT growth and polymerization rates as well as plus end cortical residency times9. In wild-type embryos, EBP-2-GFP foci resided at the cortex for brief periods before the MTs underwent catastrophe or pausing (average: 1.3 ± 0.11 sec, range: 0.25-2.25 sec, n = 63; Fig. 3 and Movie 6; as reported previously4, 9). However, in efa-6 mutant embryos individual EBP-2 foci remained associated with and tracked along the cortex for much longer times (average: 5.1 ± 0.35 sec, range: 1.25-9.75 sec, n = 117; Fig. 3 and Movie 6). We also noted an increased number of plus ends at the cortex in efa-6 mutant embryos [wild type and efa-6(tm3124) = 0.036 ± 0.004 and 0.101 ± 0.009 EBP-2 foci/μm2, respectively], consistent with the increased residency time for plus ends and growth along the cortex. Importantly, EFA-6 activity was limited to the cortex, because astral MT nucleation rates and cytoplasmic MT growth rates were similar in efa-6(-) mutant and wild-type embryos (Fig. 3d). We conclude that EFA-6 limits MT plus end residency time at the cortex, but does not regulate the MT nucleation or polymerization rates of astral MTs. Thus, EFA-6 could either promote MT catastrophe, or pausing of plus end growth17, 18, at the cell cortex.
Figure 3.
EFA-6 limits MT growth at the cortex. a, Cortical views of EBP2-GFP; mCherry-histone-expressing embryos after pronuclear meeting. At the cortical plane, wild-type embryos display EBP-2-GFP puncta which tracked for short times as seen in the merged time points. Embryos lacking EFA-6 show EBP-2-GFP puncta tracking along the cortex for extended periods, which resulted in lines of puncta in the projected image. See Movie 6 for the entire time series. Scale bar, 10 μm. b, Single cortical EBP-2-GFP puncta were followed over time and are presented as a montage. The wild-type EBP-2 spot perdures for less than two seconds at the cortex, but the EBP-2 spot in the efa-6(tm3124) embryo tracked along the cortex for over 8 seconds. Scale bar, 4 μm. c, Histogram showing residency time of EBP-2-GFP puncta at the cortex (number of plus ends = 63 wild-type, n = 117 efa-6(tm3124)). d, Summary of MT dynamics as quantitated by the EBP-2-GFP reporter. Error bars represent the S.E.M. with a confidence interval of 0.95. MT plus end residence at the cortex, astral MT nucleation rate, and the astral MT polymerization rate were determined at pronuclear meeting, the numbers of MT plus ends at the cortex was determined 15 sec after pronuclear meeting.
Cortically localized dynein is thought to pull on MT plus ends to influence mitotic spindle assembly and spindle position in one-cell stage embryos8, 10-12, 19, and embryos with reduced dynein activity lack spindle oscillations10, 13. Moreover, because we identified efa-6 as a dynein suppressor, we tested the possibility that EFA-6 regulates dynein localization at the cell cortex. We found that a GFP-DHC-1 fusion protein20 was localized to puncta at the cell cortex of one-cell stage embryos, similar to EBP-2 puncta (Supplemental Fig. S4a and Movie 7). However, perdurance of DHC-1 foci at the cortex did not depend on EFA-6 (Supplemental Fig. S4b and c). We also assayed cortical MTs in embryos depleted for dhc-1, or both dhc-1 and efa-6 to determine if dynein is required for the dense MT phenotype in efa-6(-) embryos. We found that cortical MTs appeared normal after reducing dhc-1 activity, but they were still long and abundant when dhc-1 and efa-6 were co-depleted (Supplemental Fig. S5). We conclude that EFA-6 does not to regulate MT dynamics by influencing cortical dynein localization, and that dynein is not required for the long cortical MTs in efa-6(-) embryos.
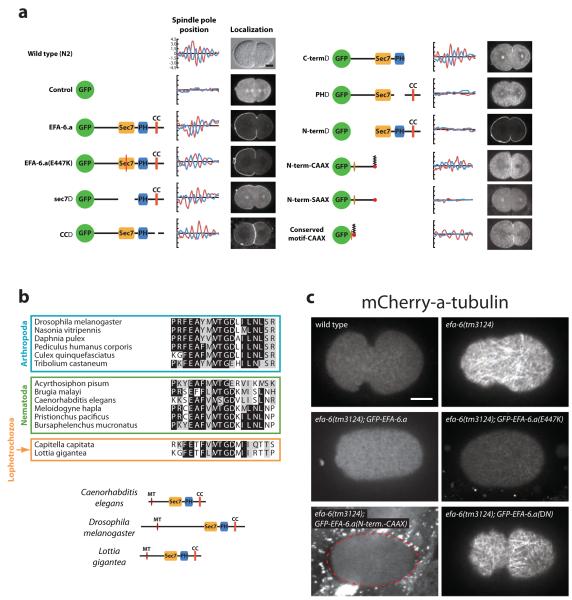
To determine which portions of EFA-6 limit cortical MT growth, we made and expressed a series of EFA-6 mutant constructs. The efa-6 gene encodes a potential guanine nucleotide exchange factor for ARF6-type small G proteins. It contains a sec7 GDP/GTP exchange domain, a pleckstrin homology (PH) domain, and a predicted coiled-coil region. We created a point mutation in a conserved catalytic residue that abolishes exchange factor activity in other systems, and deletion alleles that individually lacked the sec7, PH, and coiled-coil domains, and the N-terminal and C-terminal regions. We expressed GFP alone (as negative control) or these mutant efa-6 constructs in efa-6(tm3124) mutant worms. GFP alone control embryos displayed cytoplasmic fluorescence and spindle oscillations were absent (Fig. 4a). The wild-type EFA-6 GFP fusion protein localized to the cortex and promoted robust spindle rocking, as did the catalytically inactive point mutant, the sec7 deletion mutant, the coiled-coil deletion mutant, and the C-terminal deletion mutant (Fig. 4a). Thus exchange factor activity appears not to be important for MT regulation, and a small G-protein is likely not involved. The PH domain of the mammalian EFA6 mediates attachment to the cortex by interacting with phosphatidylinositol 4,5-bisphosphate lipids21. As expected, the PH domain mutant was exclusively cytoplasmic and spindle oscillations were absent, suggesting that membrane tethering is required for MT regulation. Finally, the N-terminal deletion mutant was localized to the cortex but spindle rocking was absent. We conclude that the PH domain and the N-terminus, but not GTPase activity, are required for spindle oscillations.
Figure 4.
Determination of the EFA-6 MT-regulating residues. a, Domain deletion analysis showing the constructs, graphs of spindle pole movements from one representative embryo (as described for Fig. 1c, except that Nomarski optics were used to track the spindle poles at a rate of one frame/ sec), and localization (1 midembryonic Z section) of the GFP-tagged proteins at telophase/cytokinesis. The fusions are described in the Methods. For the wild-type, a DIC image is presented. b, Alignment of the conserved EFA-6 N-terminal motif from several animal species and phyla, and the position of the domain in the context of the full-length proteins of the nematode C. elegans, the insect D. melanogaster, and the mollusk L. gigantea. c, mCherry-TBA-1-labeled cortical MTs in one-cell embryos were imaged by merging 3 Z sections 0.5 μm apart. Dashed red line indicates the embryo boundary in the bottom left image. Scale bars, 10 μm.
We used chimeric protein fusions to further elucidate requirements for the EFA-6 N-terminus. To test whether the C. elegans EFA-6 N-terminal region can regulate MT dynamics independently of other EFA-6 sequences so long as it is cortically localized, we expressed the EFA-6 N-terminus fused to the C. elegans MIG-2 CAAX box to link it to the plasma membrane independently of its endogenous PH domain22, 23. We found that GFP-EFA-6N-terminus+CAAX was membrane-localized and promoted spindle oscillations in efa-6(-) embryos, but the control GFP-EFA-6N-terminus+SAAX protein was cytoplasmic and did not promote rocking (Fig. 4a). Thus, the EFA-6 N-terminal region regulates spindle function independently of the other domains if it is at the cortex. Additionally, we assayed centrosome detachment from the male pronucleus in the GFP-EFA-6N-terminus+CAAX embryos and found that the fusion protein partially rescued the detachment phenotype (76% of the embryos had two attached centrosomes, 24% of the embryos had one detach, and no embryos had both detach, n = 21). We then identified an 18 amino acid conserved motif present in the N-terminus of EFA-6 proteins in nematodes, arthropods and lophotrochozoa (Fig. 4b). Other than this motif, the N-terminus of EFA-6 shows little sequence conservation among different phyla. We expressed in early embryos a 45 amino acid fragment containing this motif fused to GFP and the CAAX box. This chimeric protein was present at the cortex and promoted periodic spindle oscillations (Fig. 4a). Thus, this limited region is sufficient to regulate spindle function. Consistent with this deletion analysis, we did not find defects in centrosome-pronucleus attachment or spindle rocking in embryos depleted for arf-6 (not shown), which encodes the small GTPase that is regulated by the guanine nucleotide exchange activity of efa-63. We conclude that EFA-6 does not function through ARF-6 to regulate microtubule dynamics.
To determine if the different efa-6 gene constructs rescued spindle rocking by reducing the abundance of MTs at the cortex, we imaged cortical MTs in a number of the strains. We observed short cortical MTs after expression in efa-6(-) embryos of wild-type EFA-6, the catalytic point mutant, and the EFA-6N-terminus+CAAX transgenes, while dense arrays of long cortical MTs persisted after expression of the N-terminal deletion mutant (Fig. 4c). We conclude that the EFA-6 N-terminus, when cortically tethered, is both necessary and sufficient to limit cortical MT abundance. While the EFA-6 N-terminus can inhibit cortical MTs independent of its GTPase activity, it is intriguing that the conserved N-terminal motif we have identified is found only in EFA-6 orthologs. Thus the linkage of the N-terminal and GTPase modules is widely conserved. It will be interesting to learn if either function can, in some circumstances, influence the other.
In summary, we have shown that EFA-6 is a cortical protein in the early C. elegans embryo that limits microtubule length, centrosome separation, and spindle elongation rates by altering MT dynamics at the cell cortex. EFA-6 could regulate the behavior of MTs in a number of ways. First, EFA-6 could promote MT plus end catastrophe or pausing directly. Alternatively, EFA-6 could regulate a distinct factor that acts on the cortical MTs. Finally, EFA-6 could either alter MT tip flexibility or the rigidity of the cell cortex as MTs display enhanced catastrophe events when growing into a surface24.
The long cortical MTs that develop in efa-6(-) embryos appear to generate increased pulling forces on centrosomes, as they are associated with increased centrosome separation both during pronuclear migration and during mitotic anaphase. In contrast, reduced spindle oscillations are usually associated with decreased cortical forces8, 10. Nevertheless, in efa-6(-) embryos either altered MT dynamics or increased MT length also appear to limit oscillations, perhaps due to disruption of feedback regulation that is thought to promote spindle pole rocking4, 8, 10.
Dynein is thought to be a cortical force generator that acts on astral MTs to pull on centrosomes in the early C. elegans embryo8, 10-12, 19, and we have demonstrated that it is in fact present at the cortex in discreet foci that persist for time frames similar to MT plus ends (Supplemental Fig. 4). Moreover, we find that centrosomes do not travel from the male pronucleus toward the cortex in embryos depleted for both dhc-1 and efa-6 (Fig. 1 and Movie 3). Thus, dynein in efa-6(-) embryos appears to be increased in activity, perhaps as a result of either the increased lateral MT-cortex interactions, or the longer residency time of MT plus ends at the cortex.
In a broader context, our findings illustrate the value of studying nonessential conserved genes to better understand essential biological processes. As in other model organisms, most cytoskeletal regulators in C. elegans have been identified in screens for essential genes. However, most eukaryotic genes are not essential. In budding yeast and C. elegans, in which extensive genome-wide screens have been performed, only 7-19% of protein coding genes are required for viability25-27. Nevertheless, nonessential genes like efa-6 can clearly play important roles in modulating essential processes2, 28-30. Identifying functions for nonessential genes is important because many human disease gene orthologs are not essential in model organisms. For example, even in the mouse, 47% of human disease gene orthologs are not essential when assayed with null mutations31.
Methods
C. elegans strains and culture
Strains were maintained according to standard procedures32. Temperature-sensitive strains were maintained at 15 ° C, non-temperature-sensitive strains were maintained at room temperature except that GFP-EFA-6-expressing strains were maintained at 25 ° C. GFP-γ-tubulin; GFP-histone H2b-expressing strains were derived from TH3233, GFP-β-tubulin strains were derived from WH20434, and mCherry-histone H2b strains were derived from OD5735. For transformation of C. elegans, we used the microparticle bombardment method2, 36.
RNAi
E. coli clones expressing dsRNA to deplete dhc-1 and efa-6 were obtained from the MRC Geneservice (Cambridge, UK)26, 37. The oligos used to produce the dhc-1 construct had the sequence AAGGAAGGAGCTCAACGACA and CCTTTCCTTCCTGGGTCTTC, while the efa-6 gene fragment was amplified with CCGTCTTGATGTTGAAGCAA and GGACACTCCGTCGAAACATT; both used C. elegans genomic DNA as template26, 37. We produced the arf-6(RNAi) clone by amplifying N2 genomic DNA with the following two oligonucleotides: AAGATATCGGAGCTCCGGATATTAATGCT and AAGATATCCCCAATTTATCCTGAATTTCGT. After cleavage with EcoRV and ligation to EcoRV cleaved L4440 vector, the plasmid was introduced into the HT115(DE3) RNAi feeding bacterial strain as described37. RNAi to reduce the function of efa-6 was performed by placing L1 larvae synchronized by hypochlorite hatch-off onto 60-mm NGM agar plates with 100 mg/ml ampicillin and 1 mM isopropyl-beta-D-thiogalactopyranoside, and seeded with the double-stranded RNA-expressing E. coli, as described2. For co-depletions, we transferred L4 larvae to plates seeded with an equal mixture of the double-strand RNA-expressing E. coli strains. We examined dhc-1-depleted embryos 27 to 37 hours after the transfer and only analyzed embryos that had multiple maternal pronuclei, un-separated centrosomes, and lacked pronuclear migration, indicating a strong dynein depletion.
Microscopy
Imaging of fluorescent fusion proteins was accomplished by mounting embryos on 3% agar pads on microscope slides covered with a coverslip. Slides were analyzed on a Leica DMI 4000B microscope fitted with a Leica 63X/ 1.40-0.60 HCX Plan Apo oil objective lens in a room maintained at 25 ° C. Time-lapse videos were obtained with a Hammamatsu EM-CCD digital camera using Volocity software (Perkin Elmer Inc.). After recording, the videos were adjusted for position and contrast in ImageJ (http://rsb.info.nih.gov/ij/). For images of GFP-β-tubulin-expressing embryos, we normalized the images such that the nuclear fluorescence in interphase cells was similar.
Analysis of spindle pole positions
We recorded spindle pole positions using the ImageJ package “MTrackJ” (http://www.imagescience.org/meijering/). From these coordinates we calculated the distance between spindle poles. Measurements from multiple embryos were synchronized at anaphase onset to produce the graphs in Fig. 1d. To find the spindle pole separation rates for each time point after anaphase onset, we divided the distance travelled (beginning at anaphase onset) by the time elapsed. The greatest rate for control(RNAi) embryos occurred 30 sec after anaphase onset and the greatest rate for efa-6(RNAi) embryos occurred 22 sec after anaphase onset. The errors given in the text represent the S.E.M. with a confidence interval of 0.95.
Quantitation of MT and DHC-1 kinetics
To calculate MT nucleation rates, we followed a previously published procedure9. Briefly, we captured a single focal plane every 0.5 sec for 1 min while focused on a centrosome beginning at pronuclear meeting. We then used the kymograph function in the ImageJ package “TimeSpacePlot” (http://www.embl.de/eamnet/html/kymograph.html) to draw a semicircle at a distance of 9 μm from the center of the centrosome and produced kymographs corresponding to the 1 min video micrograph. By counting the number of spots, we determined the number of MT plus ends passing the semicircle in one minute. We counted 602 MT plus ends from eight wild-type embryos and 521 MTs from seven efa-6(tm3124) embryos.
We determined the number of EBP-2-GFP and GFP-DHC-1 puncta at the cortex by analyzing an image of the cortex 15 sec after pronuclear meeting in the transgene-expressing embryos. We drew a rectangle that included only cortical regions of the embryo (because the amount of cortical area visible varied in each embryo) and counted the number of puncta present and the area of the rectangle in ImageJ. The rectangle was positioned such that both anterior and posterior regions were equally sampled. We then calculated the number of puncta per μm2 of cortex.
For quantitating MT plus end and GFP-DHC-1 cortical residency times, we captured video micrographs for 30 sec beginning at pronuclear meeting in EBP-2-GFP or GFP-DHC-1-expressing embryos at the cortex at a rate of 4 images/ sec (for EBP-2-GFP) or 3 frames/ sec (for GFP-DHC-1). At least five embryos were analyzed for determining each measurement. The images were then analyzed in ImageJ with the measurement tool. As GFP puncta appeared we counted how many frames they each persisted, giving residency times. For determining the GFP-DHC-1 residency times, we first filtered the images with a Laplacian filter using the default parameters in the ImageJ package “FeatureJ” (http://www.imagescience.org/meijering/).
Determination of astral MT polymerization rates was performed as previously reported9. We tracked over time individual EBP-2-GFP puncta at pronuclear meeting that began at a centrosome and moved towards the cell cortex using the ImageJ package “MTrackJ” (http://www.imagescience.org/meijering/). Dividing the total distance travelled by the number of frames (time) yielded the rate of polymerization. The MT polymerization rate was 1.2 ± 0.09 μm/sec for both wild type and efa-6(tm3124). We tracked 83 MTs from six wild-type embryos and 65 MTs from five efa-6(tm3124) embryos.
We assayed pronuclear-centrosomal complex rotation by measuring the angles of the two centrosomes relative the anterior-posterior axis in wild-type or efa-6(tm3124) embryos expressing GFP-β-tubulin. The angles were determined when cytoplasmic GFP-β-tubulin began to enter the nuclei, indicating nuclear envelope breakdown.
Molecular Biology
For constructing the efa-6 variants, we started with a cDNA clone for Y55D9A.1.a (encoding EFA-6.a) isolated previously and contained in the pGEM-T easy vector (Promega Corp., Madison, WI)2. The glutamic acid codon at amino acid (a.a.) position 447 in EFA-6.a was changed to a lysine codon by using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). After sequencing of the product to detect successful mutagenesis, the cDNA was sub cloned into the pSO26 vector using SgfI (Promega Corp., Madison, WI) and SpeI (New England Biolabs, Ipswich, MA) restriction enzymes. pSO26 contains the maternal pie-1 promoter to express cDNA clones as well as an unc-119(+) gene for selecting C. elegans transformants2. Similarly, we used the QuikChange method to produce the sec7, pleckstrin homology, coiled-coil, C-terminal, and N-terminal deletion mutants in the Y55D9A.1.a cDNA. For expressing the N-terminal fragments with either the CAAX or SAAX box, we amplified the Y55D9A.1.a cDNA with oligonucleotides which included sequence encoding the additional protein tag and cloned them into pSO26, as previously detailed22, 23. The sec7 domain mutant deleted amino acids (a.a.) 354-530, the pleckstrin homology domain mutant deleted a.a. 567-683, the coiled-coil deletion removed a.a. 711-736, the C-terminal deletion mutant lacked a.a. 684-816, and the N-terminal deletion mutant lacked a.a. 2-352. The N-terminus (containing a.a. 1-353) was fused to the MIG-2 CAAX box peptide (KPQKKKKSCNIM) or to a mutant version, “SAAX”, (KPQKKKKSSNIM). Finally, the conserved motif construct present in the N-terminus contained a start codon and a.a. 26-60, followed by the MIG-2 CAAX box. The efa-6 plasmids were then transformed into unc-119(ed3); efa-6(tm3124) mutant worms. At least 10 DIC recordings were made for each construct to monitor spindle oscillations, one representative example is shown for each construct in Fig. 4.
For constructing the mC-TBA-1 and GFP-DHC-1 constructs, we used standard recombineering techniques with SW102 E. coli38 and Moerman lab fosmids containing the entire promoter, all exons and introns, and the 3′ untranslated regions. Fosmids were obtained from the MRC Geneservice (Cambridge, UK). The mCherry and GFP genes containing C. elegans introns35, 39 were inserted at the start codon of the tba-1 and dhc-1 genes. We then gap repaired an amplified C. briggsae unc-119(+)-containing plasmid40 such that we captured the entire tba-1 or dhc-1 genes (promoters, exons, introns, and 3′ untranslated regions) between the flanking open reading frames. These gap-repaired plasmids were used to transform unc-119(-) worms.
Supplementary Material
Acknowledgements
We thank M. Price, G. von Dassow, L. Pelletier, and C. Doe for insightful discussions; M. Goulding and E. Munro for technical advice; S. Schneider for advice on taxonomical and sequence alignments; S. Mitani for isolating and providing efa-6(tm3124); V. Davis Haug for the mC-TBA-1 strain; and C. Doe and C. Cabernard for comments on the manuscript. S.M.O was supported by the Leukemia and Lymphoma Society of America, S.M.O. and B.B. are supported by NIH R01GM058017.
Footnotes
Accession numbers. The following sequences used in Fig. 4b were obtained from the National Center for Biotechnology Information: Drosophila melanogaster: NP_001097874.1, Acyrthosiphon pisum: XP_001943684.1, Nasonia vitripennis: XP_001603899.1, Tribolium castaneum: XP_969387.1, Pediculus humanus corporis:XP_002425811.1, Culex quinquefasciatus: XP_001865870.1, Trichinella spiralis: ABIR01001549, Brugia malayi: XP_001899191.1, Caenorhabditis elegans: NP_001122818.1, and Meloidogyne hapla: ABLG01000531.1_contig530. Sequences for the following proteins were obtained from the Joint Genome Institute:Daphnia pulex: NCBI_GNO_6700040, Capitella capitata: scaffold_117:275944-280349, Lottia gigantea: fgenesh2_pg.C_sca_5000157. We obtained the CJ976727 EST sequence for Bursaphelenchus mucronatus from http://forestgen.ffpri.affrc.go.jp, and the sequence for Pristionchus pacificus PPA21096 was obtained from http://wormbase.org.
Author Contributions S.M.O. and B.B. designed the experiments and wrote the paper. S.M.O. conceived of the project, performed the experiments, and analyzed the data. Microscopy and strain generation were also contributed by S.N.C.
Competing financial interests The authors declare no competing financial interests.
References and Notes
- 1.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 2.O’Rourke SM, Dorfman MD, Carter JC, Bowerman B. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 2007;3:e128. doi: 10.1371/journal.pgen.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco M, et al. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. Embo J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozlowski C, Srayko M, Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129:499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 6.Oegema K, Hyman AA. Cell division. WormBook. 2006:1–40. doi: 10.1895/wormbook.1.72.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr. Biol. 2009;19:R749–761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 9.Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Pecreaux J, et al. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr. Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 11.Severson AF, Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J. Cell Biol. 2003;161:21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gönczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt DJ, Rose DJ, Saxton WM, Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol. Biol. Cell. 2005;16:1200–1212. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 15.Phillips JB, Lyczak R, Ellis GC, Bowerman B. Roles for two partially redundant alpha-tubulins during mitosis in early Caenorhabditis elegans embryos. Cell Motil. Cytoskeleton. 2004;58:112–126. doi: 10.1002/cm.20003. [DOI] [PubMed] [Google Scholar]
- 16.Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J. Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sammak PJ, Borisy GG. Direct observation of microtubule dynamics in living cells. Nature. 1988;332:724–726. doi: 10.1038/332724a0. [DOI] [PubMed] [Google Scholar]
- 18.Schulze E, Kirschner M. New features of microtubule behaviour observed in vivo. Nature. 1988;334:356–359. doi: 10.1038/334356a0. [DOI] [PubMed] [Google Scholar]
- 19.Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gassmann R, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macia E, et al. The pleckstrin homology domain of the Arf6-specific exchange factor EFA6 localizes to the plasma membrane by interacting with phosphatidylinositol 4,5-bisphosphate and F-actin. J. Biol. Chem. 2008;283:19836–19844. doi: 10.1074/jbc.M800781200. [DOI] [PubMed] [Google Scholar]
- 22.Diogon M, et al. The RhoGAP RGA-2 and LET-502/ROCK achieve a balance of actomyosin-dependent forces in C. elegans epidermis to control morphogenesis. Development. 2007;134:2469–2479. doi: 10.1242/dev.005074. [DOI] [PubMed] [Google Scholar]
- 23.Portereiko MF, Mango SE. Early morphogenesis of the Caenorhabditis elegans pharynx. Dev. Biol. 2001;233:482–494. doi: 10.1006/dbio.2001.0235. [DOI] [PubMed] [Google Scholar]
- 24.Janson ME, de Dood ME, Dogterom M. Dynamic instability of microtubules is regulated by force. J. Cell Biol. 2003;161:1029–1034. doi: 10.1083/jcb.200301147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 26.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 27.Sönnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 28.Labbé JC, Pacquelet A, Marty T, Gotta M. A genomewide screen for suppressors of par-2 uncovers potential regulators of PAR protein-dependent cell polarity in Caenorhabditis elegans. Genetics. 2006;174:285–295. doi: 10.1534/genetics.106.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 30.Dorfman M, Gomes JE, O’Rourke S, Bowerman B. Using RNA interference to identify specific modifiers of a temperature-sensitive, embryonic-lethal mutation in the Caenorhabditis elegans ubiquitin-like Nedd8 protein modification pathway E1-activating gene rfl-1. Genetics. 2009;182:1035–1049. doi: 10.1534/genetics.109.104885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park D, Park J, Park SG, Park T, Choi SS. Analysis of human disease genes in the context of gene essentiality. Genomics. 2008;92:414–418. doi: 10.1016/j.ygeno.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strome S, et al. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraser AG, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 38.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheeseman IM, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarov M, et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.