Abstract
Mate selection and maintenance of genetic diversity is crucial to successful reproduction and species survival. Plants utilize self-incompatibility system as a genetic barrier to prevent self pollen from developing on the pistil, leading to hybrid vigor and diversity. In Brassica (canola, kale, and broccoli), an allele-specific interaction between the pollen SCR/SP11 (S-locus cysteine rich protein/S locus protein 11) and the pistil S Receptor Kinase, results in the activation of SRK which recruits the Arm Repeat Containing 1 (ARC1) E3 ligase to the proteasome. The targets of Arm Repeat Containing 1 are proposed to be compatibility factors, which when targeted for degradation by Arm Repeat Containing 1 results in pollen rejection. Despite the fact that protein degradation is predicted to be important for successful self-pollen rejection, the identity of the various proteins whose abundance is altered by the SI pathway has remained unknown. To identify potential candidate proteins regulated by the SI response, we have used the two-dimensional difference gel electrophoresis analysis, coupled with matrix-assisted laser desorption ionization/time of flight/MS. We identified 56 differential protein spots with 19 unique candidate proteins whose abundance is down-regulated following self-incompatible pollinations. The identified differentials are predicted to function in various pathways including biosynthetic pathways, signaling, cytoskeletal organization, and exocytosis. From the 19 unique proteins identified, we investigated the role of tubulin and the microtubule network during both self-incompatible and compatible pollen responses. Moderate changes in the microtubule network were observed with self-incompatible pollinations; however, a more distinct localized break-down of the microtubule network was observed during compatible pollinations, that is likely mediated by EXO70A1, leading to successful pollination.
Recognition of suitable pollen is a key aspect of reproductive success of angiosperms. Despite having little control over the type of pollen that lands on the stigma of the pistil, many plant species have developed elaborate recognition systems to allow the growth of only suitable pollen grains and to reject genetically similar (self-incompatible) and incompatible mates in order to increase the probability of successful reproduction and survival. Adherence of compatible pollen grains to the stigmatic papillae results in early signaling events leading to hydraulic connectivity with the stigma, which facilitates movement of water into the pollen grains. Following hydration, pollen grains germinate and produce tubes that penetrate the stigmatic cell walls and traverse the pistil down to the ovules where fertilization takes place (1, 2). In species of Brassicaceae (canola, broccoli, cabbage, Arabidopsis), which are characterized by dry stigmas, this recognition occurs at the earliest stages of pollen adhesion and hydration. Only compatible pollen is capable of inducing the stigma to release its resources such as water and other factors necessary for pollen growth, whereas incompatible pollen is blocked either prior to hydration or during attempts to penetrate the stigmatic barrier (3). Although multiple genetic screens have identified various compatible pollen factors necessary for the initial recognition mechanisms, the molecules or mechanisms behind delivery of stigma factors necessary to support pollen growth remain largely unknown (3, 4). The lipids of the stigmatic cuticular layer are proposed to interact with the lipids from pollen coat to form a hydraulic conduit for passage of water from the stigma to pollen. For example, Arabidopsis fiddlehead mutants, which are altered in their cuticular properties, support pollen hydration on nonstigmatic tissues (5, 6). Removal of the pellicle, the proteinaceous outermost layer of the papillary cell that functions as the initial site of the pollen-pistil interaction, also results in lack of pollen tube entry into the stigma (7). Two Brassica stigma proteins, S-locus related 1 (SLR1) and S-locus glycoprotein, are implicated in pollen adhesion (8). Both of these bind to specific pollen coat proteins and are proposed to mediate pollen adhesion.
In contrast to the limited knowledge about the signaling mechanism behind compatible interactions, extensive studies have identified the stigmatic factors that regulate the self-incompatibility response (SI)1 in Brassica. Recognition of self-pollen induces a highly ordered signaling mechanism that rejects the self-pollen (i.e. blocks allocation of pistil resources for pollen hydration and germination). The SI response, which is initiated following attachment of the self-pollen to the stigma, involves binding of a secreted pollen ligand SCR/SP11 (9–10) to the S-Receptor Kinase (SRK) localized to the plasma membrane of the stigmatic cells. This binding event activates SRK, which, along with another cytoplasmic receptor kinase, M-locus protein kinase (MLPK) (11), can interact with and phosphorylate the E3 ubiquitin ligase, ARC1 (12), to promote rejection of “self-pollen.” ARC1 acts as a positive regulator of SI as activated SRK can target ARC1 to the proteasome (protein degradation machinery) (13, 14) and suppression of ARC1 in self-incompatible Brassica napus W1 lines leads to breakdown of SI response (13). In addition to the genetic evidence, treatment of W1 stigmas with proteasomal inhibitors resulted in breakdown of SI response leading to pollen germination and growth, indicating the requirement of an intact ubiquitin-mediated, proteasomal degradation pathway in order to confer SI response (14). The present hypothesis is that, following initiation of SI, ARC1 rapidly targets specific substrate proteins for proteasome-mediated degradation. These substrates targeted for destruction are likely compatibility factors, and their degradation results in pollen rejection (3). These compatibility factors potentially play a role in promoting hydration of compatible pollen grains and/or allow loosening of the cell wall for the compatible pollen tubes to penetrate. Recently, EXO70A1, a member of the exocyst complex involved in targeted vesicle trafficking was identified as one of the downstream targets of ARC1 (15). Suppression of EXO70A1 in both compatible Brassica (‘Westar’ canola line) and Arabidopsis resulted in pollen rejection at very early stages of hydration and germination. Consistent with this, overexpression of EXO70A1 in self-incompatible W1 line partially restored compatibility indicating the role of EXO70A1 as a compatibility factor and sufficiency of EXO70A1 to breakdown SI response (15). But the exact mechanism through which EXO70A1 imparts its function is still unknown. Although EXO70A1 functions as one of the compatibility factors, whether there are other early signaling proteins involved in compatibility response remains unanswered. Given the fact that proteasome blockers could restore compatibility in an SI background (14), it may be likely that many compatibility proteins are targeted for degradation following SI response. Identification of these factors could provide us with significant insights into both how compatibility response operates and how SI abrogates this process by targeting these proteins for degradation. Thus, by quantifying changes in protein abundance through analysis of the proteome prior to, and after self-incompatible pollination, we can identify the subset of proteins whose abundance is modified following SI response. These proteins would be predicted to include critical compatibility factors that are targets of SI.
In order to identify changes in stigma protein levels following SI, we utilized a two-dimensional difference gel electrophoresis (2-D DIGE technology). The 2-D DIGE approach has significant advantages over conventional 2-D electrophoresis where the analysis is biased because of gel to gel variations that hinder precise quantification (16). In 2-D DIGE, respective protein samples are labeled with different fluorescent dyes such as Cy2, Cy3, or Cy5 followed by mixing of these samples and electrophoresis through conventional two dimensions. The scans for the various channels will allow precise quantitation of the relative abundance of the perfectly matched spots. Subsequently, these spots can be isolated and protein identified through mass spectrometry. Previously 2D-DIGE technology has been effectively used to identify stress and hormone regulated proteins from various plant species including Brassica and Arabidopsis (16–18). Through using 2D-DIGE and mass spectrometry to identify protein changes following SI response, we have identified 19 unique SI-regulated proteins. Interestingly, all of these proteins exhibited reduced levels following the SI response suggesting that some of these proteins could be rapidly degraded following activation of SI. From the identified list of proteins, one candidate, tubulin, was chosen for further testing, and the role of microtubule (MT) network was explored during SI and compatible pollinations.
EXPERIMENTAL PROCEDURES
Plant Growth Conditions and Treatment
Brassica napus canola cultivars spp., compatible Westar and self-incompatible W1, were germinated on soil and maintained at standard growth conditions (22 °C, 16 h/8 h day/night cycle). For proteomic analysis, W1 flowers were self-pollinated with W1 pollen for different times and pistils were harvested and frozen at −80 °C until protein extraction. Arabidopsis Col-0 and mor1–1 were sterilized with 30% bleach for 10 min and germinated on half strength Murashige and Skoog basal salt mixture pH 5.7, grown for a week, transplanted on to soil and maintained at standard growth conditions. For restrictive temperature treatments (29 °C), Col-0 and mor1–1 flowers were emasculated and moved to chambers preconditioned at 29 °C followed by pollination after 12 h.
Extraction of Total Proteins for 2-D DIGE
Total proteins were extracted using the phenol-methanol method (19). For each biological replicate, ∼100 stigmas were ground in liquid nitrogen with a pestle and mortor, followed by addition of 1 ml of protein extraction buffer (0.5 m Tris (pH7.5), 10 mm EDTA, 2% β-mercaptoethanol and 1 × protease inhibitor) and centrifugation at 15,000 × g for 20 min at 4 °C and the supernatant was collected in 2-ml eppendorf tubes. The supernatant was mixed with equal volume of ice-cold phenol (Tris-buffered, pH 7.5–7.9) and spun down at 15,000 × g for 20min at 4 °C to separate phenol. Following removal of the aqueous phase, the intact interface along with the phenol phase was extracted twice with the extraction buffer. To the interface and phenol phase, 5 volumes of cold 0.1 m ammonium acetate in methanol was added and incubated at −20 °C overnight, followed by centrifugation at 15,000 × g. The supernatant was discarded and the pellet was washed twice with 100 μl of 80% acetone, allowed to dry and resuspended in lysis buffer (7 m urea, 2 m thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate) (CHAPS), and protein concentration determined using the Bio-Rad protein assay mixture using BSA as a standard.
Protein CyDye Labeling
Proteins were labeled with CyDye DIGE Fluor minimal dye (GE Healthcare) according to the manufacturer's instructions with modifications. Briefly, 10 μl (around 3 μg/μl) of proteins were mixed with 0.2 μl 1.5 m Tris-HCl buffer (pH 8.8) to adjust the pH of the protein solution to around 8.8. To the protein solution, 100 pmol CyDye (1 μl of 0.1 mm CyDye stock solution) was added and incubated on ice in the dark for at least 2 h. The reaction was stopped by addition of 0.5 μl of 10 mm lysine and incubated on ice for 10 min.
Two-dimensional Gel Electrophoresis
Pairs of Cy3- and Cy5-labeled protein samples were mixed together. The DIGE sample buffer (7 m urea, 2 m thiourea, 4% CHAPS, 20 mm dithiothreitol, and 1% IPG buffer (GE Healthcare)) was added to bring the volume to 450 μl, and the samples were then applied to 24-cm Immobiline Drystrips (GE Healthcare). IEF was carried out on an Ettan IPGphor II (GE Healthcare) at 20 °C with maximum 50 μA/strip and the following setting: rehydration for 2 h and then 50 V for 10 h, 500 and 1000 V each for 1 h, gradient increase to 8000 V in 3 h, and remaining at 8000 V until reaching the desired total V-h (80,000 for pH 4–7 and 100,000 for pH 3.0–5.6/5.3–6.5/6.3–7.5/6–11 IPG strips). After IEF, IPG strips were equilibrated in equilibration buffer (6 m urea, 30% (w/v) glycerol, 2% SDS, 50 mm Tris-HCl, pH 8.8) first with 0.5% dithiothreitol and then with 2% iodoacetamide each for 15 min. The strips were then transferred to 10% SDS-PAGE gels for the second dimension electrophoresis using the Ettan Dalt Twelve gel system (GE Healthcare). SDS-PAGE was run at 50V for 2 h and then 120V for 12–16 h or until the bromophenol blue dye front reached the gel end.
Image Scanning
The gels were scanned using a Typhoon 8600 scanner (GE Healthcare). Cy3-labeled sample was scanned using a 532 nm laser and a 580/30-nm band pass emission filter, whereas Cy5-labeled sample was scanned with a 633 nm laser and 670/30-nm band pass emission filter. The photomultiplier tube was set to ensure that the maximum pixel intensity was between 60,000 and 90,000 pixels. Post-electrophoretic staining with deep purple (GE Healthcare) fluorescent dyes was performed essentially according to the manufacturer's instructions, and subsequent scanning was performed using the same setting as that used for Cy3-labeled sample.
2-D DIGE Data Analysis Using the DeCyder Software
The DIGE images were analyzed using DeCyder version 6.5 software as described in the user manual (GE Healthcare). Briefly the differential in-gel analysis module was used for spot detection, spot volume quantification, and volume ratio normalization of different samples in the same gel. Then the biological variation analysis module was used to match protein spots among different gels and to identify protein spots that exhibited significant difference. Manual editing was performed in the biological variation analysis module to ensure that spots were correctly matched between different gels and were not contaminated with artifacts, such as streaks or dust. Differentially expressed spots were selected based on the p value of t test (p < 0.05); presence on at least three of the five replicates, including presence on the reverse labeled one; and peak height of over 100 (absolute value prior to subtracting background for normalization) on at least three gels.
Spot Picking
Approximately 30 μg of protein were labeled with 50 pmol of Cy5 dye and mixed with 500 μg of unlabeled protein (250 μg each from pollinated and unpollinated samples) and separated with 2-DE. SDS-PAGE gels were stained with deep purple dye (GE Healthcare) or SYPRO Ruby dye (Bio-Rad). After scanning the gels with Typhoon 8600, spots of interests were selected with DeCyder software and picked by an Ettan spot picker (GE Healthcare) and stored in 1% (v/v) acetic acid until further processing.
In-gel Tryptic Digestion, Mass Spectrometry, and Database Searching and Compilation
Samples were received as excised spots from a two-dimensional polyacrylamide gel and were reduced, alkylated, tryptically digested and extracted using the standard protocol outlined at http://www.biol.yorku.ca/cm/proteomics/home.html. Extracted peptides were desalted and concentrated using Millipore C18 zip-tips®, then spotted in 1.5 μl MALDI matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid (CHCA), in 60% acetonitrile, 0.3% trifluoroacetic acid) onto a 100-well MALDI target plate. The MALDI target plate was then loaded in to a QSTAR XL QqTOF mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA) and MALDI MS spectra were collected for each sample over 20–50 s, with laser energy of 12 uJ and frequency of 20 Hz. Internal trypsin autolysis peptides were used to calibrate the instrument for each sample. MS spectra were then individually examined by hand and up to 17 monoisotopic peaks were selected from each for MS/MS analysis, which was carried out over varied lengths of time using the same laser settings and collision energy settings between 50 and 100 (arbitrary units).
The mascot.dll data processing script from the Analyst QS 1.1 software package (Applied Biosystems/MDS Sciex, Foster City, CA) was used to generate and concatenate peak lists for each sample. Peak lists were searched against the entire NCBI non-redundant database (version NCBInr_20100721 (11480388 sequences; 3917815449 residues)) using Mascot software version 2.3.02 (Matrix Science, Boston, MA). Peptide and MS/MS error tolerances were set to 100 ppm and 0.2 Da respectively. Tryptic peptides having a charge of 1+ and up to 2 missed cleavages were considered. Carbamidomethylation of cysteine and oxidation of methionine were set as fixed and variable modifications respectively. All protein scores exceeding Mascot's 95% significance threshold (score > 56) were accepted (Supplemental data, S1). Identifications based on single peptide identifications therefore required an individual ion score > 56 (Supplemental data, S2).
Because the B. napus proteome is not exhaustively represented in the NCBI database, some tryptic peptides interrogated by MS/MS will yield no protein match regardless of their quality. In other cases, a protein from a source organism other than B. napus is identified because an unannotated protein from B. napus shares some identical amino acid sequence and tryptic peptides with an annotated protein from a close genetic relative (i.e. Arabidopsis thaliana). The assumption here is that the protein being interrogated is similar in sequence, and therefore similar in function to the identified protein. If the top MASCOT hit was not from Arabidopsis, the identified protein was then subjected to a BLAST analysis against the Arabidopsis protein database to find the closest homolog, thereby generating a homogenous list of Arabidopsis proteins. To help organize the list of proteins identified through MS/MS analysis, all proteins were then categorized according to GO ontology; “biological process,” “molecular function,” and “cellular compartment ” (The Gene Ontology Consortium, 2000). To assess whether these classifications are over-represented among Arabidopsis genes as a whole, all genes identified through MS/MS analysis were submitted to enrichment analysis using the DAVID Functional Annotation tool (20).
Treatment of Brassica Stigmas with MT Manipulating Drugs
Taxol and oryzalin were used as MT manipulating drugs. The stigmas of self-incompatible W1, stage 12 flowers from Brassica were used for the study. These flowers actively reject genetically similar W1 pollen (self-pollen) while they allow genetically dissimilar compatible pollen from Westar cultivar to develop on them. Thus the same female can be used to analyze both self-incompatible and compatible traits. The pedicel (flower stalk) of W1 flowers were immersed for 3 h with either the drugs at 1 μm and 10 μm concentration or with DMS0 (1:1000 dilution), followed by pollination for four hours with self-incompatible W1 or compatible Westar pollen. The stigmas were then fixed in 3:1 ethanol and glacial acetic acid, followed by aniline blue assay.
Sample Preparation for Anti-tubulin Staining
Whole mount immunostaining was performed as described earlier (21). In-planta pollination was done on self-incompatible Brassica W1 stigmas with either compatible Westar or incompatible W1 pollen for 30 min (n = 10). The unpollinated and pollinated stigmas were fixed by infiltration with 4% paraformaldehyde for 5 min followed by incubation at RT (room temperature) for 1 h. After fixation, stigmas were washed three times with 1 × MTSB (MT stabilizing buffer; 100 mm PIPES, 2 mm EGTA, 1 mm MgSO4) and subjected to freeze fraction. The stigmas were then permeabilized overnight in 1 × phosphate-buffered saline (PBS) with dithiothreitol and 0.005% Tween 20. The next morning, blocking was performed with 1% bovine serum albumin for 30 min, followed by staining with 1:150 dilution of anti-alpha tubulin antibodies (Sigma). After two hours of incubation with the primary antibodies, the samples were incubated with the secondary antibodies conjugated to Alexa Fluor 594 (Invitrogen) (1:150 in 1 × PBS) for 2 h. The stigmas were then mounted in 50% glycerol and visualized under the Perkin Elmer UltraVIEW VoX spinning disc confocal microscope. The samples were imaged using a 63 × objective lens and Z-stacks collected were subjected to restorative deconvolution to improve signal to noise ratio and visualized following extended focus view of a volume.
Genetic Analysis Using mor1–1 Mutant
A temperature sensitive Arabidopsis mutant of MOR1 (microtubule organization 1) gene was used to study the impact of MT depolymerization on pollen acceptance. At restrictive temperature of 29 °C the microtubule in mor1–1 becomes short, sparse and misaligned (22). Arabidopsis mor1–1 and Col-0 plants were grown under the permissive (22 °C) growth conditions. Stage 11/12 flowers from Col-0 and mor1–1 were emasculated and maintained either at permissive temperature 22 °C or restrictive temperature 29 °C for 12 h. The next morning, these stigmas (permissive and restrictive) were pollinated using wild type Col-0 pollen collected from plants maintained at 22 °C. Pollination was allowed to continue for 15, 30 and 60 min respectively, followed by aniline blue assay to analyze pollen attachment and germination.
Transient Expression of RFP-Exo70A1 in GFP-tubulin BY2 Cells
Biolistic-mediated transformation of log-phase GFP-TUA3 BY2 suspension cultured cells was performed using RFP -EXO70A1 and RFP constructs. Transformations were performed with a total of 10 μg of purified plasmid DNA. Following transformations, BY2 cells were left in the dark and then harvested at 20 h post-bombardment and visualized directly using a Leica DMR epifluorescence microscope.
Aniline Blue Assay
Aniline blue assays were performed as described previously (12). After pollination, Brassica and Arabidopsis pistils were fixed in 3:1 ethanol, glacial acetic acid for 30 min, followed by incubation in 1N NaOH at 60C° for 1 h. After 1 h, the pistils were washed with distilled water and stained with basic aniline blue (0.1 m K3PO4 0.1% aniline blue). The stained samples were mounted in 50% glycerol to visualize pollen germination and pollen tubes under Leica DMR epifluorescence microscope.
RESULTS
Identification of Differentially Expressed Proteins Following Self-Incompatible Pollinations
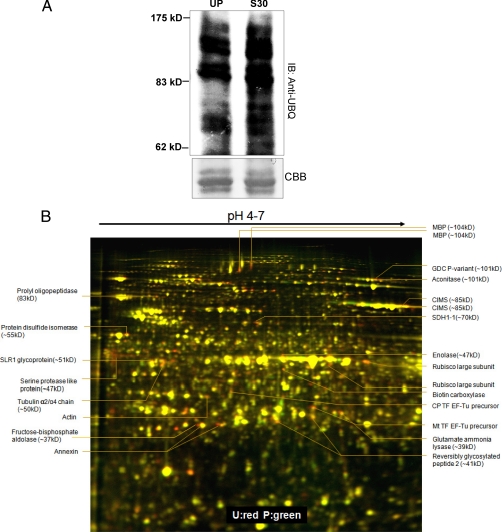
In Brassica, following incompatible pollination, increased ubiquitination has been reported to occur and the proteasome's proteolytic function was shown to be essential for manifestation of successful SI response (14). In order to reconfirm the increased ubiquitination of proteins following the SI response, extracted proteins from unpollinated and SI pollinated stigmas were resolved through either SDS-PAGE or 2D gel electrophoresis followed by Western blotting using anti-UBQ antibodies. The blots revealed basal level of protein ubiquitination in the unpollinated stigma sample and an increased ubiquitination at 30min following SI pollination, consistent with previous observations (Fig. 1A, supplemental Fig. S1A) (14). We hypothesized that the increasing ubiquitinated pools of potential SI targets will likely be degraded at 60 min and thus can be identified as differential spots on a two-dimensional gel. To test this, we adopted the 2-D DIGE technique and resolved differentially expressed proteins from unpollinated stigmas and SI pollinated stigmas, followed by scanning and BVA analysis. Five biological replicates of stigmas harvested from different batches of W1 plants were subjected to DIGE analysis. Initially these samples were processed using various narrow range IPG strips (pH 3.0–5.6/5.3–6.5/6.3–7.5/4–7/6–11) to provide highest resolution. We found that most of the protein spots identified from narrow range 2-DE can also be seen in 2-DE gel using pH 4–7 IPG strips. The five biological replicates were then processed using pH 4–7 strips (Fig. 1B). Interestingly, following DeCyder and BVA analyses (p value of t test (p < 0.05) and presence on at least three of the five replicates, including presence on the reverse labeled gel), we did not observe any proteins that were reproducibly and significantly up-regulated, whereas a total of 56 proteins were consistently and significantly down-regulated displaying 1.24 to 2.13 fold change in protein levels (Fig. 1, Table I).
Fig. 1.
SI response leads to increased ubiquitination and down-regulation of a subset of proteins. A, Increased ubiquitination following SI (30min) was observed through Western blotting using anti-ubiquitin antibodies. B, Two-dimensional DIGE analysis of SI regulated proteins. Proteins from W1 stigmas either unpollinated (U) or pollinated (P) for 60 min with self-incompatible W1 pollen were compared through Two-dimensional DIGE using 24 cm pH 4–7 IPG strips followed by 12.5% SDS-PAGE. Proteins that are down-regulated by SI appear red and that are up-regulated following SI appear green and those that are unaffected are yellow. Interesting spots identified are marked with their respective identifications, the differentials predicted are shown in supplemental Fig. S1B and the fold changes are shown in Table I.
Table I. The pl and molecular mass values shown are theoretical. Score, the Mascot score. Cov., the protein sequence coverage (%). (No. peptide) Number of observable peptides per protein detected through Mass spec.
| Protein ID | Arabidopsis AGI | Gl | pl | Molecular mass (KDa) | Score | Cov. (%) | No. peptide | Fold change | T-test | Function |
|---|---|---|---|---|---|---|---|---|---|---|
| Aconitase | At4g35830 | 599625 | 5.98 | 101.4 | 92 | 6 | 4 | −1.35 | 0.023 | Metabolism |
| Actin | At5g09810 | 45593268 | 5.26 | 19.3 | 129 | 14 | 4 | −1.48 | 0.035 | Development |
| Annexin | At1g35720 | 88659016 | 5.44 | 36.3 | 143 | 19 | 5 | −1.91 | 0.026 | Trafficking |
| Annexin | At1g35720 | 88659016 | 5.44 | 36.3 | 255 | 28 | 7 | −1.32 | 0.02 | Trafficking |
| ATCIMS | At5g17920 | 15238686 | 6.09 | 84.6 | 167 | 8 | 5 | −1.43 | 0.042 | Metabolism |
| ATCIMS | At5g17920 | 15238686 | 6.09 | 84.6 | 70 | 5 | 3 | −1.88 | 0.031 | Metabolism |
| Biotin carboxylase | At5g35360 | 14388188 | 6.52 | 58.9 | 541 | 28 | 12 | −1.46 | 0.033 | Metabolism |
| Enolase | At2g36530 | 34597332 | 5.78 | 47.6 | 85 | 7 | 2 | −1.56 | 0.026 | Metabolism |
| Fructose-bisphosphate aldolase | At4g38970 | 7270880 | 6.78 | 43.1 | 91 | 9 | 3 | −2.13 | 3.40E−06 | Metabolism |
| Glutamate-ammonia ligase | At5g37600 | 599656 | 5.28 | 39.4 | 86 | 3 | 2 | −1.4 | 0.035 | Metabolism |
| Glycine decarboxylase P-protein 1 | At4g33010 | 297798656 | 8.98 | 55 | 83 | 3 | 2 | −1.48 | 0.024 | Metabolism |
| GTP elongation factor Tu family protein | At4g02930 | 297809841 | 5.53 | 51.6 | 374 | 31 | 10 | −1.39 | 0.031 | Translation |
| Myrosinase binding protein | At1g52030 | 1711296 | 5.39 | 104.3 | 153 | 6 | 4 | −1.81 | 0.022 | Metabolism |
| Myrosinase binding protein | At1g52030 | 1711296 | 5.39 | 104.3 | 191 | 8 | 7 | −1.61 | 0.034 | Metabolism |
| Protein disulphide isomerase | At1g77510 | 77999357 | 5 | 55.9 | 117 | 7 | 3 | −1.74 | 0.034 | Chaperone |
| Putative chloroplast translation, EF, EF-TU precursor | At4g20360 | 297804102 | 5.84 | 51.9 | 56 | 3 | 1 | −1.52 | 0.035 | Translation |
| Putative serine protease-like protein | At4g17100 | 5302811 | 5.15 | 47.1 | 58 | 4 | 1 | −1.88 | 0.026 | Metabolism |
| reversibly glycosylated peptide-2 | At5g15650 | 2317731 | 5.93 | 41.9 | 104 | 8 | 3 | −1.27 | 0.031 | Metabolism |
| Reversibly glycosylated peptide−2 | At5g15650 | 2317731 | 5.93 | 41.9 | 272 | 17 | 6 | −1.24 | 0.029 | Metabolism |
| RUBISCO-large subunit | ATCG00490 | 8745521 | 5.88 | 53.4 | 341 | 22 | 10 | −1.93 | 0.022 | Metabolism |
| RUBISCO-large subunit | ATCG00490 | 8745521 | 5.88 | 53.4 | 113 | 11 | 5 | −1.91 | 0.022 | Metabolism |
| SDH1–1 (succinate dehydrogenase subunit) | At5g66760 | 15240075 | 5.86 | 70.2 | 149 | 8 | 4 | −1.67 | 0.031 | Metabolism |
| SLR1 glycoprotein | At3g12000 | 456317 | 6.86 | 51.1 | 55 | 3 | 1 | −1.42 | 1.40E−04 | Metabolism |
| Tubulin alpha-2/alpha-4 chain | At4g14960 | 34733239 | 4.91 | 50 | 57 | 3 | 1 | −1.48 | 0.03 | Structural |
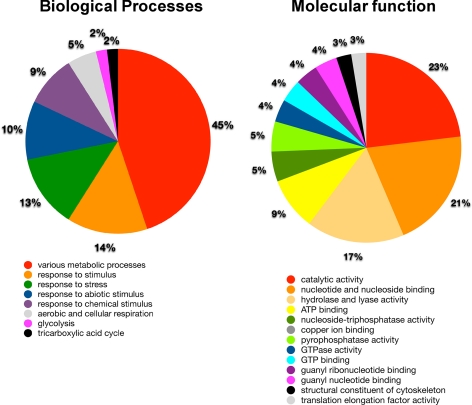
These 56 spots were then isolated with robotic spot picker using preparative gels which were stained by deep purple following electrophoresis. These spots were subjected to MALDI/MS analysis that identified proteins from 28 spots with high confidence, of which19 were unique proteins (Table I, supplements S1 and S2). These 19 proteins can be classified into several functional categories based on GO ontology annotation (Fig. 2). The three most frequently identified biological processes were various metabolic processes, response to stimulus and response to stress at 45%, 14%, and 13% respectively (Fig. 2). The three most frequently identified molecular functions were catalytic activity, nucleotide and nucleoside binding, and hydrolase and lyase activity at 23%, 21%, and 17% respectively (Fig. 2). To assess whether these classifications are over-represented among Arabidopsis genes as a whole, all genes identified through MS/MS analysis were submitted to enrichment analysis using the DAVID Functional Annotation tool (supplemental Table S1) (20). As observed in supplemental Table S1, the identified candidates were strongly enriched relative to the gene content in the Arabidopsis genome.
Fig. 2.
Distribution of SI regulated proteins based on proposed molecular function and biological processes classification. The identified SI regulated proteins were classified according to Gene Ontology (GO) terms relating to biological processes and molecular function, The subcellular localization and biological functions are listed in supplemental Table S1.
Of the 19 unique proteins identified, many of the proteins were predicted to be in metabolic pathways. This may be a reflection of the SI system in the stigma reducing metabolic activities to stop the self-pollen (as the pollen requires resources from the stigma to develop). Another candidate is the S-locus related-1 (SLR1) protein which has previously been found to be involved in the adhesion of compatible pollen grains (23); thus, the down-regulation of SLR1 could be related to an SI response to reduce the adhesion of the SI pollen. Annexin is an interesting candidate as annexins have been implicated in Golgi-mediated secretion (24). Vesicle-mediated secretion is proposed as a mechanism to provide resources to the pollen during compatible pollinations, and we have previously uncovered evidence for this process to be blocked in SI pollinations (15). Both actin filaments and MTs make up the plant cytoskeleton and are required for various developmental processes including, cell division, intracellular trafficking, and controlling cell shape. Previous studies have established a functional role for actin polymerization during both compatible and incompatible interactions in Brassica (21). The reduced abundance of actin in this analysis is consistent with a previous study demonstrating the depolymerization of actin in the stigmatic papilla following SI pollination (21). Actin-based vesicle targeting is a common theme in eukaryotic systems, and it has been previously shown that blocking actin polymerization inside the stigmatic papilla could abrogate compatible pollen acceptance (21). We confirmed that both preventing actin polymerization through cytochalasin D treatment and blocking vesicle trafficking with brefeldin A treatment in the stigma resulted in reduced compatible pollen attachment, mimicking the SI response (supplemental Fig. S2). Thus, the reduced abundance of both annexin and actin would be consistent with the SI response inhibiting vesicle delivery to the pollen attachment site. Finally, alpha 2–4 tubulin, a subunit of the MT network, was another interesting candidate that was identified from this study and was selected for further analyses.
In plants, apart from their function during cell division in the nucleus, there is an extensive network of acentrosomal microtubule arrays in the plant cell cortex that is required for controlling cell expansion, cell morphogenesis, and is proposed to control anisotropic expansion in plant cells. In Arabidopsis genome, alpha tubulins are represented by at least six genes that code for four isoforms with TUA2 and TUA4 coding for the same isoform (alpha 2–4 tubulin) (25, 26). Mutation in TUA4 has been shown to result in left handed helical growth of Arabidopsis hypocotyls (27, 28). Although both actin filaments and MTs make up the cytoskeletal network, information is lacking on the precise role of MT network during pollen responses. Using a multi-faceted approach of pharmacological, genetic and cell biological techniques, we were able to further elucidate the significance of altered alpha 2–4 tubulin levels and the microtubule network following pollination responses.
Localized Destabilization of MT Network Following Compatible Pollinations in Brassica
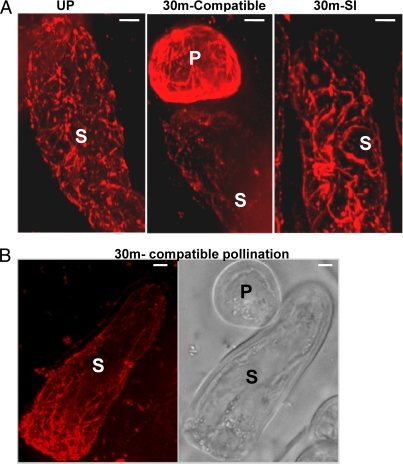
The reduced abundance of α2–4 tubulin in this DIGE analysis indicated that MT dynamics could be altered following the induction of an SI reaction. To visualize in vivo changes in MT dynamics following both SI and compatible pollination, W1 stigmas were pollinated with either W1 pollen (SI) or with Westar pollen (compatible) and subjected to whole mount immuno-staining procedure using anti-tubulin antibodies and observed through a spinning disc confocal microscope. When the extended focus from the Z-stacks obtained from unpollinated W1 stigmatic papilla was examined, an intricate dense cortical MT network could be observed throughout the cell cortex (Fig. 3A, left panel). When samples were examined 30 min following SI pollinations, this intricate cortical network exhibited changes, where the dense network became re-organized into longer bundles, although no MT shortening or depolymerization could be observed (Fig. 3A, right panel). When stigmatic papillae were examined following compatible pollinations, consistently, the dense MT network could not be visualized. In particular, close to the pollen attachment site, the dense cortical network of MTs was largely eliminated or reduced to shorter segments, indicating a depolymerized state of MT (Fig. 3A middle panel, 3B). These observations suggest that while SI pollination resulted in moderate changes in MT organization (resulting in tubulin being identified in the DIGE analysis); compatible pollination appeared to trigger a more dramatic localized depolymerization of MTs.
Fig. 3.
Compatible pollination induces MT break-down. Status of the MT network was observed through whole mounting of unpollinated (W1), 30 min compatible pollinated (Westar pollen) and 30 min SI pollinated (W1 pollen) stigmas, followed by immunostaining using anti-tubulin antibodies (A). Stacked images from a single papillary cell, pollinated for 30 min with compatible pollen showing a general breakdown of MT network close to the site of pollen attachment (B) (n = 10 stigmas/treatment). P, pollen grain; S, Stigmatic papillary cell; Scale: 5 μm.
Pharmacological Modulators of MT Dynamics Reveal a Role for MTs During Compatible Pollinations
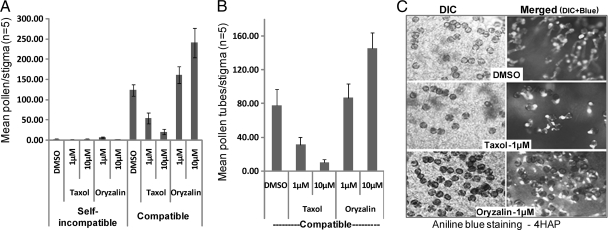
In order to investigate the role of MTs during pollen-pistil interactions, we used pharmacological modifiers of MT stability and tested the ability of these drugs to interfere with both self-incompatible and compatible responses. For both interactions, the W1 pistil was used as the female parent, and following drug treatment, was either pollinated with compatible Westar or self-incompatible W1 pollen. Pistils were treated with either taxol, a microtubule stabilizing drug, or oryzalin, a microtubule depolymerizing agent. Interestingly, neither taxol nor oryzalin affected the self-incompatible phenotype at the level of either pollen attachment or germination (Fig. 4A). In contrast, when the effect of these inhibitors on compatible pollinations was analyzed, treatment with taxol significantly reduced pollen attachment and pollen germination, whereas oryzalin treatment had the opposite effect, promoting pollen attachment and germination (Figs. 4A–4C). Overall, these observations indicated that MT depolymerization in the stigmatic papilla is a requirement for compatible pollen acceptance, which is consistent with the observations in Fig. 3. That is, pretreatment with oryzalin would increase the degree of MT depolymerization leading to increased compatible pollen acceptance; whereas taxol would prevent MT depolymerization by MT stabilization and decrease compatible pollen acceptance. In the case of SI pollinations, if the observations in Fig 3A of longer MT bundles reflect a stabilization of MTs, then taxol's effect would not be detected, as the pollen are already being rejected by the endogenous SI pathway. However, this is unlikely to be a primary response in the rejection of SI pollen as MT depolymerization by oryzalin also had no effect, indicating that the SI outcome is unaffected by alteration in MT dynamics.
Fig. 4.
Pharmacological modulators of MT organization affect compatible pollination. Average number of adhered pollen grains (A) and germinated pollen tubes (B) on W1 stigmas treated with either taxol or oryzalin, followed by compatible or self-incompatible pollinations with Westar and W1 pollen respectively. n = 5; error bars are ± standard error. The representative DIC and aniline blue fluorescence images (to visualize pollen tubes) of stigmas from the compatible treatments are shown in C. HAP, Hours after pollination.
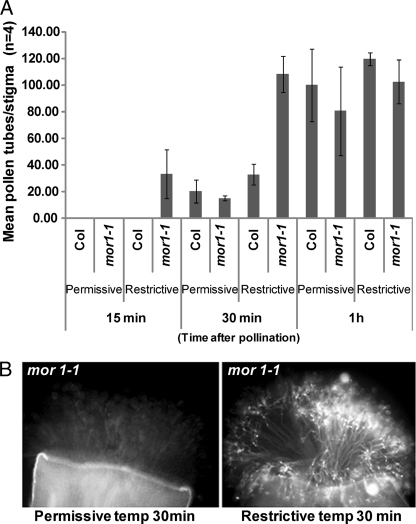
mor1–1 Mutants Maintained at the Restrictive Temperature Exhibit Rapid Pollen Acceptance
To further evaluate the role of MTs during compatible pollinations, we used a genetic approach with the Arabidopsis mor1–1 temperature sensitive mutant. MOR1 (At2g35630) is highly expressed in Arabidopsis stigmas as observed from public microarray databases (29, 30) and thus, mor1–1 mutant plants serve as a useful genetic tool to understand role of MT organization during pollen responses. In mor1–1, the cortical MTs become shortened, mimicking a depolymerized state after exposing the plants to the restrictive temperature (29 °C) for shorter periods (2 h) of time (22). Stage 12 mor1–1 flowers were emasculated in-planta and maintained either at permissive (22 °C) or restrictive (29 °C) temperatures overnight and then were pollinated with pollen from wild-type Col-0 flowers maintained at permissive conditions. When pollinated pistils were left for 24 h at restrictive temperature, no differences could be observed for germination of pollen grains and pollen tube growth, in comparison to wild-type Col-0 pistils (supplemental Fig. S3). Based on our immunostaining and pharmacological evidence, if MT breakdown in the stigma is necessary for compatible pollen interactions, then in mor1–1 mutant (which exhibits constitutive MT depolymerization at restrictive temperature) both pollen germination and growth will be expected to occur more rapidly than the controls maintained at permissive conditions. In order to test this, we performed a shorter temporal pollination profile (15 min–1 h) and examined changes in pollen responses using the aniline blue staining assay. At 15 min after pollination, mor1–1 stigmas at 29 °C allowed pollen germination to occur, whereas very minimal germination could be observed in both wild-type Col-0 (restrictive and permissive) and mor1–1 (permissive) controls (Fig. 5A). By 30 min, the difference was greater with stigmas from mor1–1 maintained at 29 °C allowing much more rapid growth of pollen tubes through them (Fig 5B). By 1 h after pollination, the average number of pollen germinated was insignificant between wild-type Col-0 and mor1–1 stigmas suggesting that within one hour after pollination, the compatible pollen grains on the control stigmas had caught up producing the same number of pollen tubes (Fig 5A). Thus, the results indicate that the switching of MTs from an elaborate network to a shortened depolymerized state following treatment at 29 °C in mor1–1 stigmas, allowed a more rapid compatible pollen response to occur.
Fig. 5.
mor1–1 mutant allows rapid pollination at restrictive temperatures. Average number of pollen tubes on stigmas of Col-0 and mor1–1 plants maintained at either permissive or restrictive temperatures, followed by pollination for various times with Col-0 pollen maintained at permissive temperature (A) n = 4; error bars are ± standard error. The representative aniline blue fluorescence images of pollinated (30 min) stigmas of mor1–1 from the different temperatures are shown in B.
EXO70A1 Expression Leads to Breakdown of the MT Network in Tobacco BY2 Cells
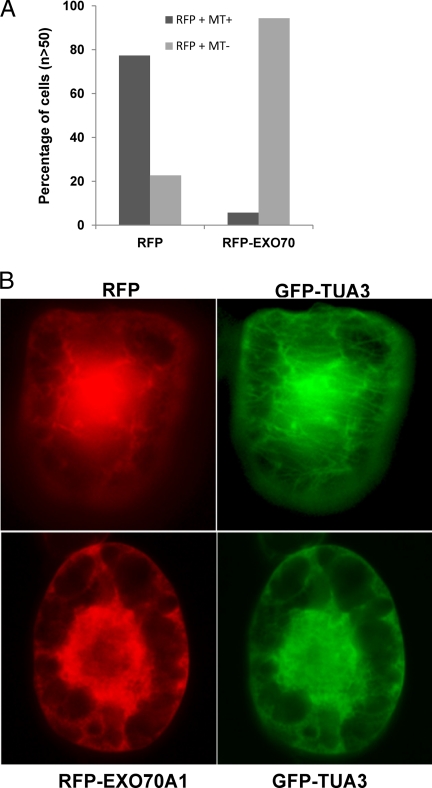
We have recently identified one of the exocyst complex members, EXO 70A1, as a compatibility factor in Brassica and Arabidopsis, but the exact mechanism by which it regulates pollination is still unknown (15). There is precedence for a role of exocyst complex in regulating MT dynamics in mammalian systems. Previous studies have reported that an intact MT network is required for appropriate localization of EXO70 and the exocyst complex, whereas EXO70 and exocyst complex function to cause end instability of MT network (31). Based on these previous findings and our observations that both EXO70 and MT depolymerization are essential for compatible pollinations, we hypothesized Brassica EXO70A1 could cause depolymerization of the MT network. To test this, we used a transgenic tobacco BY2 cell system expressing GFP-TUA3 protein that illuminated the MT network (Fig. 6, supplemental Fig. S4). Through biolistics, we transformed these cells either with an RFP construct or an RFP-EXO70A1 construct and examined the cells for presence of intact MT network. In cells expressing RFP alone, ∼70% of the cells also displayed an intact MT network (Figs. 6A, 6B), whereas in cells that expressed RFP-EXO70A1, the MT network was completely disrupted or absent more than 90% of the time (Figs. 6A, 6B), indicating that EXO70A1 over-expression can destabilize the MT network.
Fig. 6.
EXO70 expression leads to break-down of microtubule network. Transgenic BY2 cells expressing GFP-TUA3 were either bombarded with RFP alone (n > 50) or with RFP-EXO70A1 (n > 50) and observed for intact MT network, 20 h after bombardment. Number of RFP positive cells with MT network (present/absent) was expressed in percentage (A). B, Representative cells expressing either RFP or RFP-EXO70A1 and the status of MT network are shown.
DISCUSSION
Unlike nonspecific, unrelated pollen that are incapable of inducing the compatible response on the dry stigmas of Brassicaceae, SI pollen are fully capable of triggering the compatible pollen response (3, 32, 33). However, the presence of the self-pollen rejection response in the stigmas allows the active inhibition of the compatibility signaling, thus preventing self-pollen germination. During this active rejection process, binding of the pollen ligand SCR/SP11 induces the SRK-mediated SI pathway in the stigmas that stops any compatible signaling events. Thus, instead of being a linear module to self-rejection, the SI system is now proposed to negatively regulate the compatibility responses (3, 33). The identification of ARC1, an E3 ubiquitin ligase that likely targets compatibility factors such as EXO70A1 following activation of SI response, is further evidence for such a theory (14, 15). The fact that blocking ubiquitination-mediated protein turnover in the stigma results in the breakdown of the SI response leading to incompatible pollen acceptance (14) indicates that post-translational targeting through ubiquitin tagging is a predominant mechanism during SI response. This proteasome-mediated regulation could be reflected in changes in protein abundance following SI pollinations. To identify potential targets of this pathway, we analyzed the stigma proteome through 2D-DIGE approach, for differential regulation of protein abundance following activation of the SI response.
From the DIGE analysis, 19 unique proteins were identified with reduced abundance following SI pollinations, and a large component of these proteins was identified as belonging to metabolic pathways. During compatibility responses, there is a requirement of increased energy for supporting the hydration, germination, penetration, and growth of the compatible pollen tube. Thus, to maintain the stigmatic papillary cells in an energy efficient state, the SI system actively works toward stopping any SI pollen from developing on the surface, conserving its resources for the compatible pollen. Because most of this energy is usually derived from increased glycolysis, a block in glycolytic pathway enzymes (enolase, fructose bisphosphate aldolase) could directly result in ATP depletion, blocking various energy-dependent processes. AtCIMS is the cobalamine-independent methionine synthase that is involved in the biosynthesis of methionine, providing methyl groups for various metabolic processes including the production of ethylene and biotin (34, 35). Interestingly, Biotin carboxylase is also down-regulated, which is a subunit of Acetyl-Co A carboxylase involved in production of the precursor malonyl CoA for fatty acid synthesis. Fatty acids are important for successful pollen-pistil interactions as cer mutants defective in long chain fatty acids produce pollen that are unable to interact with the stigmas (1, 36). Arabidopsis fiddlehead mutants possess altered cuticular properties that support pollen hydration on nonstigmatic tissues (6). In Papaver, SI is known to target soluble inorganic pyrophosphatases (sPPases) in pollen, which are required to provide energy for various biochemical pathways (37). Activation of SI in Papaver causes rapid phosphorylation of sPPases resulting in their reduced activity and increased inorganic pyrophosphate levels in the system (37).
Down-regulation of proteins from various compartments such as the cytosol, endoplasmic reticulum, mitochondria, and chloroplast indicates a system under stress, leading to a general shut-down of various processes required for pollen germination on the stigma. If the SI response causes a general shut down of the various energy centers, then how does the stigmatic papillary cell keep itself receptive for the compatible pollen? This is likely reflected in the moderate levels of protein changes observed (1.24-fold to 2.13-fold) following SI pollinations, which suggests that the response is quite localized so that stigmatic papillary sites remaining unoccupied by incompatible pollen can still accommodate compatible pollen. This is quite consistent with the model of polarized secretion and molecular signaling observed exclusively in the foot region of the pollen attachment site (2, 38).
Two other candidate proteins with reduced abundance following SI pollinations are the S-locus related-1 (SLR1) and annexin. SLR1 is a stigma-specific secreted glycoprotein, and its suppression has been shown to result in reduced pollen-stigma adhesion. Treatment of stigmas with anti-SLR1 antibodies resulted in reduced pollen adhesion, confirming the role of SLR1 in mediating compatible responses (23). The observed down-regulation of SLR1 following SI response indicates that suppression of SLR1 protein levels could be one of the modes through which SI operates. Annexins are multifunctional proteins that have been observed to accumulate in growing tips of secretory cells where they have been implicated in Golgi-mediated secretion (24). In Brassicaeae, the stigmatic papillae provide resources to the quiescent pollen to initiate metabolic activity in pollen grains following attachment to compatible stigmas. In this context, a regulatory circuit that controls targeted secretion of cargo necessary for pollen germination, at the site of pollen attachment, should be functioning on the stigmatic papillary side. In mammalian systems, annexins have been found to be associated with actin, exhibiting a calcium-dependent binding to F-actins, and are involved in actin bundling (39). Absence of annexin A2 resulted in the inhibition of actin-dependent apical transport of exocytotic vesicles in polarized epithelial cells (39). This suggests that annexins play an important role in vesicle trafficking along with calcium and actin. A similar role for calcium and actin is also observed in stigmatic papillae following compatible pollinations. A triphasic increase in cytosolic calcium is observed immediately after pollen attachment, and actin polymerization has also been shown to be required for compatible pollen acceptance (40, 21). In contrast, induction of SI response leads to a reduced, single calcium spike and complete de-polymerization of actin at the site of pollen attachment (40, 21).
In Brassica, prior to pollination, a large vacuole that is interconnected and branching into a globular and tubular vacuolar network can be observed at the apical region of stigmatic papillary cells (21). Following compatible pollination, this vacuolar network extended toward the site of pollen attachment, while following SI pollination, the vacuolar network was altered into a more fragmented pattern with few elongated vacuoles near the pollen attachment site (21). Given the fact that the vacuole contains the majority of ions, water and nutrients required for pollen germination, it is likely that a complex involving calcium-induced exocytosis, mediated by both annexin and actin could result in proper delivery of the vacuolar cargo necessary for pollen germination. We recently showed that EXO70A1 functions as a compatibility factor, and at least one mode through which SI response could operate is by targeting EXO70A1 for degradation (15). EXO70A1 is part of the exocyst complex that is involved in polarized secretion and thus, could perform a compatible function along with actin and annexin in delivering vesicles to the site of pollen attachment. Interestingly, during cytokinesis in plants, a tubular-vesicular network is formed at the equatorial plane, and exocyst-like tethering complexes are proposed to initiate the fusion of multiple vesicles to give rise to the tubular-vesicular network (41). The proteomic data presented here, with both annexin and actin showing reduced abundance following SI pollination, suggests that the SI response could potentially target to down-regulate annexin and the actin cytoskeleton, to reject pollen. Our proteomics approach failed to identify EXO70A1 as one of the proteins that is targeted by SI response; this could be because of low abundance of EXO70A1 in the system or because of the basic nature of EXO70A1 protein, which would prevent it from resolving properly on a two-dimensional gel.
Our pharmacological, cell biological and genetic approaches provide evidence that MT depolymerization is a prerequisite for a successful compatible pollen acceptance to occur. The Arabidopsis mor1–1 mutant was used to examine the effects of MT depolymerization on the acceptance of wild-type compatible pollen. MOR1 (microtubule organization 1) belongs to the MAP215 family of microtubule-associated proteins (42). Point mutations that result in single amino acid substitutions in the N-terminal HEAT repeat of MOR1, result in, sparse, misaligned, and shortened microtubules following exposure of these mor1–1 mutants to restrictive temperature of 29 °C (42). When maintained at 22 °C, the MT organization in these mutants is similar to wild-type. The MT shortening induced by the 29 °C treatment of mor1–1 plants allowed for a much more rapid pollination to occur compared with the respective controls (Fig. 5). This indicates that priming of the stigmatic tissue by prior MT depolymerization results in rapid pollen germination, likely because of a much faster rate of delivery of resources from stigmatic papillary cells to the site of pollen attachment. This is also consistent with the pollination data from the Brassica W1 pistils pre-treated with the microtubule depolymerizing drug, oryzalin (Fig. 4).
We also observed that the overexpression of EXO70A1 in tobacco BY-2 cells resulted in the break-down of the MT network (Fig. 6) and this corroborates well with previously published work in mammalian systems. In rat kidney cells, overexpression of EXO70 led to a breakdown of MT network and resulted in increased membrane additions (43). In addition, localized MT depolymerization through MT disrupting drugs was also shown to induce plasma membrane additions (44). This study proposed that EXO70 and the exocyst complex could mediate transfer of Golgi-derived vesicles from MT ends to actin and to the plasma membrane through regulating MT dynamics (43). In Arabidopsis, delivery of cellulose synthase (CESA) complexes were unaffected by depolymerization of microtubules, but MTs were required for guiding these complexes to particular sites on the plasma membrane (45). It was also shown that the motility of the small CESA compartment was mediated by MT depolymerization (45). Thus, a localized depolymerization of MTs following compatible pollination could be an essential event for targeted secretions, which are required for stimulating the metabolic activity of the pollen.
Although a reduced abundance of tubulin (alpha 2–4) was observed following SI pollinations, this apparently did not result in any drastic change in MT dynamics. The dense network of MTs was altered into a more helical network, suggesting that the reduction in the relative abundance of tubulin could be related to altered MT morphology (Fig 3A). Consistent with this, neither taxol nor oryzalin was able to alter the Brassica SI response indicating that the regulation of SI is independent of the MT organization status. Nevertheless, the down-regulation of actin along with a previous study (21) documenting the depolymerization of actin at the site of SI pollen attachment, indicate that, a general alteration of the cytoskeleton could be partly responsible for manifestation of the SI response. In conclusion, through DIGE analysis of the stigma proteome for SI regulated proteins, we identified a change in the levels of a tubulin isoform which allowed us to uncover a novel and essential role for MT depolymerization in the stigma during compatible pollen acceptance response. This analysis has also revealed a number of previously unknown candidate proteins which are being further investigated for their respective role during pollen-pistil interactions.
Acknowledgments
We thank Dr. Edward Yeung for his valuable input in sectioning and observation of MT dynamics. We thank Dr. Kevin Quick (Perkin Elmer) for his technical assistance with the spinning disk confocal microscope and Volocity software.
Footnotes
* This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to M.A.S. and D.R.G., University start-up grants to M.A.S., and a Canada Research Chair to D.R.G.
 This article contains supplemental Figs. S1 to S4, Table S1, and Supplements S1 and S2.
This article contains supplemental Figs. S1 to S4, Table S1, and Supplements S1 and S2.
1 The abbreviations used are:
- SI
- Self-incompatibility
- MT
- Microtubules
- SRK
- S-locus receptor kinase
- SCR
- S-locus cysteine rich protein
- ARC1
- ARM-repeat containing protein.
REFERENCES
- 1. Swanson R., Edlund A. F., Preuss D. (2004) Species specificity in pollen-pistil interactions. Annu. Rev. Genet. 38, 793–818 [DOI] [PubMed] [Google Scholar]
- 2. Dickinson H. (1995) Dry stigmas, water and self-incompatibility in Brassica. Sexual Plant Reproduction 8, 1–10 [Google Scholar]
- 3. Samuel M. A., Yee D., Haasen K., Goring D. R. (2008) ‘Self’ Pollen Rejection Through the Intersection of Two Cellular Pathways in the Brassicaceae: Self-Incompatibility and the Compatible Pollen Response, Self-Incompatibility in flowering plants, Berlin, Germany:Springer-Verlag, 173–191 [Google Scholar]
- 4. Chapman L. A., Goring D. R. (2010) Pollen-pistil interactions regulating successful fertilization in the Brassicaceae. J. Exp. Bot. 61, 1987–1999 [DOI] [PubMed] [Google Scholar]
- 5. Zinkl G. M., Zwiebel B. I., Grier D. G., Preuss D. (1999) Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126, 5431–5440 [DOI] [PubMed] [Google Scholar]
- 6. Lolle S. J., Cheung A. Y. (1993) Promiscuous germination and growth of wildtype pollen from Arabidopsis and related species on the shoot of the Arabidopsis mutant, fiddlehead. Dev. Biol. 155, 250–258 [DOI] [PubMed] [Google Scholar]
- 7. Roberts I. N., Harrod G., Dickinson H. G. (1984) Pollen-stigma interactions in Brassica oleracea, II. The fate of stigma surface proteins following pollination and their role in the self-incompatibility response. J. Cell Sci. 66, 255–264 [DOI] [PubMed] [Google Scholar]
- 8. Doughty J., Dixon S., Hiscock S. J., Willis A. C., Parkin I. A., Dickinson H. G. (1998) PCP-A1, a defensin-like Brassica pollen coat protein that binds the S locus glycoprotein, is the product of gametophytic gene expression. Plant Cell 10, 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kachroo A., Schopfer C. R., Nasrallah M. E., Nasrallah J. B. (2001) Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293, 1824–1826 [DOI] [PubMed] [Google Scholar]
- 10. Takayama S., Shimosato H., Shiba H., Funato M., Che F. S., Watanabe M., Iwano M., Isogai A. (2001) Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413, 534–538 [DOI] [PubMed] [Google Scholar]
- 11. Kakita M., Murase K., Iwano M., Matsumoto T., Watanabe M., Shiba H., Isogai A., Takayama S. (2007) Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in. Brassica rapa. Plant Cell 19, 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gu T., Mazzurco M., Sulaman W., Matias D. D., Goring D. R. (1998) Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. U.S.A. 95, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stone S. L., Arnoldo M., Goring D. R. (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286, 1729–1731 [DOI] [PubMed] [Google Scholar]
- 14. Stone S. L., Anderson E. M., Mullen R. T., Goring D. R. (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Samuel M. A., Chong Y. T., Haasen K. E., Aldea-Brydges M. G., Stone S. L., Goring D. R. (2009) Cellular Pathways Regulating Responses to Compatible and Self-Incompatible Pollen in Brassica and Arabidopsis Stigmas Intersect at Exo70A1, a Putative Component of the Exocyst Complex. Plant Cell 21, 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng Z., Zhang X., Tang W., Oses-Prieto J. A., Suzuki N., Gendron J. M., Chen H., Guan S., Chalkley R. J., Peterman T. K., Burlingame A. L., Wang Z. Y. (2007) A proteomics study of brassinosteroid response in Arabidopsis. Mol. Cell. Proteomics 6, 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang W., Deng Z., Oses-Prieto J. A., Suzuki N., Zhu S., Zhang X., Burlingame A. L., Wang Z. Y. (2008) Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics 7, 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alvarez S., Berla B. M., Sheffield J., Cahoon R. E., Jez J. M., Hicks L. M. (2009) Comprehensive analysis of the Brassica juncea root proteome in response to cadmium exposure by complementary proteomic approaches. Proteomics 9, 2419–2431 [DOI] [PubMed] [Google Scholar]
- 19. Hurkman W. J., Tanaka C. K. (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81, 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 21. Iwano M., Shiba H., Matoba K., Miwa T., Funato M., Entani T., Nakayama P., Shimosato H., Takaoka A., Isogai A., Takayama S. (2007) Actin dynamics in papilla cells of Brassica rapa during self- and cross-pollination. Plant Physiol. 144, 72–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whittington A. T., Vugrek O., Wei K. J., Hasenbein N. G., Sugimoto K., Rashbrooke M. C., Wasteneys G. O. (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613 [DOI] [PubMed] [Google Scholar]
- 23. Luu D. T., Marty-Mazars D., Trick M., Dumas C., Heizmann P. (1999) Pollen-stigma adhesion in Brassica spp involves SLG and SLR1 glycoproteins. Plant Cell 11, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clark G. B., Lee D., Dauwalder M., Roux S. J. (2005) Immunolocalization and histochemical evidence for the association of two different Arabidopsis annexins with secretion during early seedling growth and development. Planta 220, 621–631 [DOI] [PubMed] [Google Scholar]
- 25. Meagher R. B., Fechheimer M. (2003) In: The Cytoskeletal Proteome of Arabidopsis, Arabidopsis. Meyerowitz E, Somerville C. editors. Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- 26. Kopczak S. D., Haas N. A., Hussey P. J., Silflow C. D., Snustad D. P. (1992) The Small Genome of Arabidopsis Contains at Least 6 Expressed Alpha-Tubulin Genes. Plant Cell 4, 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsumoto S., Kumasaki S., Soga K., Wakabayashi K., Hashimoto T., Hoson T. (2010) Gravity-induced modifications to development in hypocotyls of Arabidopsis tubulin mutants. Plant Physiol 152, 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abe T., Thitamadee S., Hashimoto T. (2004) Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol. 45, 211–220 [DOI] [PubMed] [Google Scholar]
- 29. Swanson R., Clark T., Preuss D. (2005) Expression Profiling of Arabidopsis Stigma Tissue Identifies Stigma-Specific Genes. Sexual Plant Reproduction 18, 163–171 [Google Scholar]
- 30. Toufighi K., Brady S. M., Austin R., Ly E., Provart N. J. (2005) The Botany Arraye-Northerns, Expression Angling, and promoter analyses. Plant J. 43, 153–163 [DOI] [PubMed] [Google Scholar]
- 31. Vega I. E., Hsu S. C. (2001) The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J. Neurosci. 21, 3839–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ivanov R., Fobis-Loisy I., Gaude T. (2010) When no means no: guide to Brassicaceae self-incompatibility. Trends Plant Sci. 15, 387–394 [DOI] [PubMed] [Google Scholar]
- 33. Tantikanjana T., Nasrallah M. E., Nasrallah J. B. (2010) Complex networks of self-incompatibility signaling in the Brassicaceae. Curr. Opinion Plant Biol. 13, 520–526 [DOI] [PubMed] [Google Scholar]
- 34. Kende H. (1993) Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 283–307 [Google Scholar]
- 35. Ravanel S., Gakière B., Job D., Douce R. (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 95, 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fiebig A., Mayfield J. A., Miley N. L., Chau S., Fischer R. L., Preuss D. (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12, 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Graaf B. H., Rudd J. J., Wheeler M. J., Perry R. M., Bell E. M., Osman K., Franklin F. C., Franklin-Tong V. E. (2006) Self-incompatibility in Papaver targets soluble inorganic pyrophosphatases in pollen. Nature 444, 490–493 [DOI] [PubMed] [Google Scholar]
- 38. Elleman C. J., Franklin-Tong V., Dickinson H. G. (1992) Pollination in Species with Dry Stigmas - the Nature of the Early Stigmatic Response and the Pathway Taken by Pollen Tubes. New Phytol. 121, 413–424 [DOI] [PubMed] [Google Scholar]
- 39. Hayes M. J., Shao D., Bailly M., Moss S. E. (2006) Regulation of actin dynamics by annexin 2. EMBO J. 25, 1816–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwano M., Shiba H., Miwa T., Che F. S., Takayama S., Nagai T., Miyawaki A., Isogai A. (2004) Ca2+ dynamics in a pollen grain and papilla cell during pollination of Arabidopsis. Plant Physiol. 136, 3562–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fendrych M., Synek L., Pecenková T., Toupalová H., Cole R., Drdová E., Nebesárova J., Sedinová M., Hála M., Fowler J. E., Zarsky V. (2010) The Arabidopsis Exocyst Complex Is Involved in Cytokinesis and Cell Plate Maturation. Plant Cell 22, 3053–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kawamura E., Himmelspach R., Rashbrooke M. C., Whittington A. T., Gale K. R., Collings D. A., Wasteneys G. O. (2006) MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140, 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang S., Liu Y., Adamson C. L., Valdez G., Guo W., Hsu S. C. (2004) The mammalian exocyst, a complex required for exocytosis, inhibits tubulin polymerization. J. Biol. Chem. 279, 35958–35966 [DOI] [PubMed] [Google Scholar]
- 44. Zakharenko S., Popov S. (1998) Dynamics of axonal microtubules regulate the topology of new membrane insertion into the growing neurites. J. Cell Biol. 143, 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gutierrez R., Lindeboom J. J., Paredez A. R., Emons A. M., Ehrhardt D. W. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806 [DOI] [PubMed] [Google Scholar]