Abstract
Preeclampsia (PE) adversely impacts ∼5% of pregnancies. Despite extensive research, no consistent biomarkers or cures have emerged, suggesting that different molecular mechanisms may cause clinically similar disease. To address this, we undertook a proteomics study with three main goals: (1) to identify a panel of cell surface markers that distinguish the trophoblast and endothelial cells of the placenta in the mouse; (2) to translate this marker set to human via the Human Protein Atlas database; and (3) to utilize the validated human trophoblast markers to identify subgroups of human preeclampsia. To achieve these goals, plasma membrane proteins at the blood tissue interfaces were extracted from placentas using intravascular silica-bead perfusion, and then identified using shotgun proteomics. We identified 1181 plasma membrane proteins, of which 171 were enriched at the maternal blood-trophoblast interface and 192 at the fetal endothelial interface with a 70% conservation of expression in humans. Three distinct molecular subgroups of human preeclampsia were identified in existing human microarray data by using expression patterns of trophoblast-enriched proteins. Analysis of all misexpressed genes revealed divergent dysfunctions including angiogenesis (subgroup 1), MAPK signaling (subgroup 2), and hormone biosynthesis and metabolism (subgroup 3). Subgroup 2 lacked expected changes in known preeclampsia markers (sFLT1, sENG) and uniquely overexpressed GNA12. In an independent set of 40 banked placental specimens, GNA12 was overexpressed during preeclampsia when co-incident with chronic hypertension. In the current study we used a novel translational analysis to integrate mouse and human trophoblast protein expression with human microarray data. This strategy identified distinct molecular pathologies in human preeclampsia. We conclude that clinically similar preeclampsia patients exhibit divergent placental gene expression profiles thus implicating divergent molecular mechanisms in the origins of this disease.
Preeclampsia (PE)1 is a serious disorder of human pregnancy that adversely impacts ∼5% of pregnancies. Despite extensive research, no consistent biomarkers of disease, no well-defined causes, and no cures have emerged. This failure may indicate that different genetic and environmental insults can lead to similar clinical outcomes (1). Establishing whether different molecular mechanisms lead to PE in specific patient subgroups is therefore an important objective. Subgroup classification is likely to improve the accuracy of predictions for individual pregnancy outcomes, and to permit the development of targeted treatments. This approach, for example, has proven effective in individualizing breast cancer prognosis and care (2).
PE is defined by gestational hypertension and proteinuria, but can also be found with other clinical features including fetal intrauterine growth restriction (IUGR), chronic prepregnancy hypertension, abnormal uterine and/or umbilical arterial Doppler blood velocity waveforms, and a variety of characteristic placental pathologies (3). The large variation in clinical signs and associated pathologies is consistent with PE being a family of related disorders rather than a single disease (4). However even among clinically similar groups of PE patients there is wide variation, leading to the hypothesis that different genetic, environmental or allelic variations can all lead to similar clinical outcomes. Further support for this hypothesis comes from the variety of mechanisms resulting in PE-like symptoms in mouse models. PE mouse models have been generated genetically from spontaneous hypertensive mice (5), by excessive angiotensin pathway activation (6), and by mutations affecting trophoblast cell cycle regulation (7). Interestingly, the anti-angiogenic factor, sFLT1, is present in elevated levels in the maternal circulation in most human PE pregnancies, but not all (8), and in some mouse models of PE (e.g. PAH mouse (6)), but not all (e.g. BPH/5 mouse (9)). Thus variations in molecular mechanisms of PE in mouse models, and in the clinical signs in human PE all point to multiple underlying molecular pathologies.
One common feature is that abnormal placental function is believed to underlie the majority, if not all, cases of PE (1). The placenta is a critically important organ essential for a successful pregnancy. It is responsible for maternal-fetal exchange, it protects the fetus from maternal immune rejection, and it secretes factors required for maternal physiological adaptations to pregnancy and lactation. It is likely that dysfunction in one or more of these roles result in PE and its common and variable clinical phenotypes. During a PE pregnancy the trophoblast is believed to be the source of factors released into the maternal circulation that damage the endothelium thereby causing maternal clinical signs of disease (10, 11). Abnormal trophoblast invasion into the maternal decidua has been intensely studied in PE (11, 12), and there is growing evidence that trophoblast in the extensive layer in direct contact with maternal blood in the exchange region of the placenta is also abnormal (11, 12). In the current study, we focus on trophoblast in the exchange region of the placenta. The trophoblast has an extremely large surface area in contact with maternal blood and it exhibits abnormalities in PE, but as it has not been extensively studied it is poorly characterized on the molecular level.
To understand the molecular mechanisms of PE, two steps are required. First, the separation of the different cell types of the placenta or a definition of the molecular signature of the trophoblast cell type and, second, the application of a model based approach to identify a hidden (or unknown) number of patient subgroups using these cell specific gene signatures.
To achieve the first step we utilized the mouse placenta to label the surface proteomes of the syncytial trophoblast and the fetal endothelial cells. Although the analysis of maternal endothelial cells would be of interest, the goal of identifying a syncytial enriched gene expression profile required comparison against the endothelial cells of the placenta. Analysis of the mouse enabled access to the placenta prior to birth, and established a launching point for the development of new mouse models of PE. Investigation of the mouse surface proteomes identified a large set of genes enriched in expression to the mouse trophoblast lineage. This was validated in human by a comparison to the Human Protein Atlas (13) where a 70% conservation of expression of one-to-one orthologs was observed in keeping with previous analyses (14). In addition, a detailed analysis of the endothelial and trophoblast proteomes highlighted the high degree of similarity of gene expression likely driven by their functional similarities.
To address our second goal, that clinically similar PE may have diverse molecular causes leading to different, hidden patient subgroups, we used the syncytial trophoblast signature identified in mouse and selected these genes from a human PE patient microarray. We then applied expectation maximization in combination with the Baysian information criterion to statistically determine the optimum number of human patient subgroups. Application of the patient models to the entire microarray data was used to identify three distinct subgroups of PE patients, each with different Gene Ontology and KEGG pathway abnormalities. To validate the existence of subgroups we analyzed an independent PE patient cohort. We focused in particular on a subgroup that showed changes in genes involved in MAPK signaling and was the only subgroup that did not overexpress FLT1 and ENG. We confirmed the existence of this subgroup via Western blotting and found high correlation with pre-existing chronic hypertension in the mother.
EXPERIMENTAL PROCEDURES
Detailed Experimental Procedures are provided in the supplementary Experimental Procedures.
Vascular Perfusion with Colloidal Silica Beads
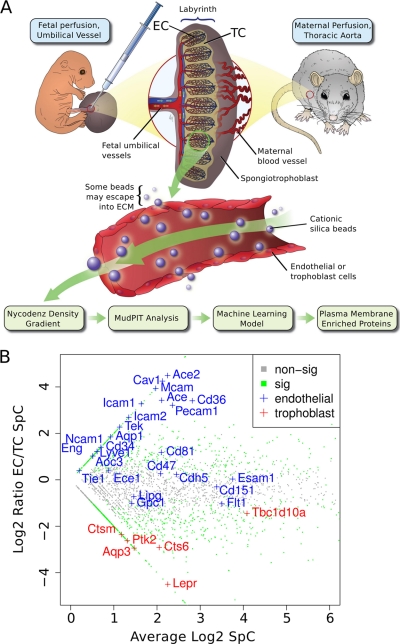
Plasma membrane proteins were isolated using the silica-bead coating strategy as previously described (15). In brief, we perfused the placentas of 10 pregnant C57BL/6J mice at 17.5 days of gestation with cationic colloidal silica beads via the maternal aorta or fetal umbilical cord using published methods (16). Silica beads in the trophoblast lined maternal blood spaces or the endothelial lined fetal capillaries of the labyrinth (Fig. 1A) were isolated from each litter's pooled labyrinthine tissue (∼eight placentas per litter were pooled).
Fig. 1.
Proteomics strategy to identify novel proteins at the fetal and maternal blood-tissue interfaces in the mouse placenta. A, Scheme of our applied workflow. Cationic silica-beads were perfused via the maternal aorta to reach trophoblast lined maternal blood spaces or via the umbilical cord to reach fetal endothelial-lined vessels in the placental labyrinth. Silica-beads were isolated from labyrinth tissue to obtain in vivo surface-associated proteins. Proteins isolated from beads were analyzed by MudPIT-based proteomics. B, A zoomed-in scatter plot representation of the entire dataset highlighting known markers of endothelial cells (EC; blue text) and trophoblast cells (TC; red text). Green data points were significantly associated with either the EC or TC surfaces (FDR p value ≤0.1), whereas gray data points did not significantly differ (FDR p value >0.1).
Protein Extraction, In-Solution Digestion, MudPIT Analyses and Protein Identification
For protein extraction isolated silica-bead pellicles from each litter pool (n = 5 pools of ∼8 placentas/pool) were resuspended in ∼300 μl of ice-cold NE buffer (400 mm NaCl, 25 mm HEPES, pH 7.4, 1% Triton X-100), and incubated at 4 °C with shaking for 30 min followed by centrifugation at 13,000 rpm. The supernatant containing the extracted proteins was collected and precipitated overnight with five volumes of ice-cold acetone with 10% trichloroacetic acid at −20 °C. For an independent litter pool (n = 5 pools of ∼8 placentas/pool), silica-bead pellicles were resuspended in 60 μl of 8 m urea to extract bound proteins, these were then reduced and alkylated with 2 mm dithiothreitol and 8 mm iodoacetamide, digested and purified. A fully automated 9-cycle, 18-h MudPIT procedure was set up similar as previously described (14, 17, 18). Raw files were converted to m/zXML using ReAdW (version 1.1) and searched by X!Tandem (version 06.06.01) against a mouse (version 3.28) IPI (International Protein Index) protein sequence database (51,479 protein sequences). The search was performed with a fragment ion mass tolerance of 0.4 Da and a parent ion mass tolerance of 4 Da. Complete tryptic digest was assumed and one missed cleavage site was accepted. Carbamidomethylation of cysteine was specified as fixed and oxidation of methionine as variable modification. To estimate and minimize our false positive rate the protein sequence database also contained every IPI protein sequence in its reversed amino acid orientation (target-decoy strategy; total sequences in the database, 102,958 IPI sequences plus bovine trypsin). A conservative false discovery rate was set to 0.5% on the peptide level, as recently described (14, 19, 20). Only fully tryptic peptides ≥7 amino acids matching these criteria were accepted to generate the final list of identified proteins. We only accepted proteins identified with two unique peptides per analyzed sample type. To minimize protein inference, we developed a database-grouping scheme and only report proteins with substantial peptide information, as recently reported (14, 19, 20). This resulted in an estimated false discovery rate (FDR) of ∼1.6% on the protein level (63 decoy proteins in the final data set).
Machine Learning Algorithm
Machine learning was performed using the WEKA software package (21). The performance of the training set was assessed using 10-fold cross validation using, Bayes Network, Naïve Bayes, Random Forest, and J48 machine learning algorithms. Algorithm performance was compared using F score, kappa statistic, True and False positive rates, as well as the ROC area. The training set was a merge of the current data with our previously published data set of subcellular factions of the mouse labyrinth (14). The spectral counts were normalized and scaled as a ratio of the maximum spectral counts. Next this list was annotated with the GO cellular component terms, plasma membrane, nuclear, cytosol, mitochondrion, endoplasmic reticulum, and Golgi bodies. Only annotations that had evidence codes for inferred from direct assay and where the cellular component term was unique to a single component were used as training examples. For the determination of enrichment to either the trophoblast or endothelial proteome a table containing the normalized, replicate MudPIT spectral counts was generated. Spectral counts for the five TX-100 and the five urea extractions replicates were summed. A p value was calculated and a correction for false discovery rate (Benjami-Hochberg (22)) was applied.
Statistical Analysis
Data was processed in R (23). MvA scatter plot and FDR for fold change between the trophoblast and endothelial compartments were generated using the samr library (24). Expectation Maximization clustering was performed with the mclust library (25). Principal component analysis was done using the made4 library (26). Heat maps were generated using the gplots library. The GNA12, GPR50 and clinical patho-physiology data correlation plot was made using the arm library. Gene expression difference between subgroups and controls were calculated using the limma library (27). Venn diagrams were generated using the limma library (27).
Calculation of Functional Enrichment
Tables of KEGG (http://www.genome.jp/kegg/) (28), Mouse Mutant Phenotype (http://www.informatics.jax.org/) and Gene Ontology (http://geneontology.org/) data were downloaded from online sources. OBO style files were re-formatted into flat files using a Java program (available upon request). The hypergeometric distribution and corrected p values were calculated using the Bingo plug-in (29) for Cytoscape (30).
Human Patient Sample Collection
Human specimens and their associated clinical data were obtained from the archive of the Research Centre for Women's and Infants' Health BioBank program of Mount Sinai Hospital (http://biobank.lunenfeld.ca) in accordance with the policies of the Mount Sinai Hospital Research Ethics Board. Specimens contained 4 villous cores (one from each patient quadrant) that were flash-frozen and stored in liquid nitrogen.
RESULTS
Selective Enrichment of Surface-associated Proteins
Toward our goal of developing better mouse models of PE, we built on our comparison of human and mouse molecular expression in the placenta (14) by studying the membrane specific expression of TC (trophoblast cells exposed to maternal blood) versus EC (endothelial cells exposed to fetal blood) in the labyrinth, the placental exchange region in mice (31). We used a silica-bead coating strategy as previously described (15) followed by proteomic analysis. Briefly, colloidal silica-beads were perfused into the placenta through the maternal or fetal vascular systems to reach the blood-tissue membrane interface of fetal TC or EC, respectively (Fig. 1A). The surface-associated silica-bead pellicles were then isolated using ultracentrifugation in Nycodenz density gradients, resulting in enrichment for plasma membrane proteins (15). Western blots against the plasma membrane proteins Nos3 (eNOS), Pecam-1, and caveolin-1 all showed enrichment to the membrane fraction. Conversely, Western blots of proteins localizing to other intracellular organelles (endoplasmic reticulum, calreticulin; cytosol, α-transketolase; mitochondria, cytochrome-c; nucleus, nuclear transportin-1) showed a marked depletion of the membrane fraction (supplemental Fig. S1). In summary, this suggests both a successful enrichment of membrane proteins and a depletion of non-membrane proteins. We then employed shotgun proteomics (17, 18) to systematically analyze, for the first time, the similarities and differences in the surface proteome of cells at the maternal blood-TC and the fetal blood-EC interfaces (Fig. 1A).
MudPIT-based analysis of five individual isolates (biological replicates), extracted with either nonionic detergent (Triton-X-100; TX-100) or urea resulted in the confident identification of 3917 proteins (see Experimental Procedures; supplemental Tables S1–S5; for protein and peptide-centric information). To further increase our stringency, we only accepted proteins also detected by our recent mRNA microarray analysis of the same tissues (14), resulting in a panel of 3444 (supplemental Table S6) surface-enriched proteins of the mouse labyrinth. Interestingly, direct comparison of the surface proteomes identified 3087 proteins at the blood-TC tissue interface and 2497 proteins at the blood-EC interface with the majority of proteins (∼62%) in common (supplemental Table S6). Next, we compared the abundance of measured proteins between both tissue interface proteomes using a table of normalized spectral counts (SpC) for all maternal side and fetal side replicates. A Benjamini-Hochberg corrected p value (22) was used to determine if there was a significant difference in protein expression level between the two compartments. The log2-transformed ratio of the mean EC over mean TC SpCs for these proteins were plotted versus the average SpC to visually inspect the separation of known EC and TC markers (Fig. 1B). We found that marker proteins with known cell-type expression were appropriately localized (Fig. 1B). However there were numerous EC markers that did not have a significant difference in expression between the two membranes. This result suggested that selective enrichment of surface-associated proteins at the maternal versus fetal blood-tissue interface was successful and reveals an overlap of the biology of the EC and TC.
Prediction of Plasma Membrane Proteins Using Machine-learning Algorithms
The silica-bead coating technology enables the isolation of enriched membrane-associated proteins (supplemental Fig. S1) although some contamination from other subcellular compartments was noted in the current work and previous publications (15). To compensate for this, we utilized a machine-learning approach as in our previous report (17). First, a smaller subset of our data (472 proteins) containing only highly annotated proteins with known subcellular locations were used as a training set to build a model of subcellular localization (see Experimental Procedures) and the remaining proteins with poor or mixed annotation were used for prediction. Multiple machine learning algorithms were tested and BayesNet gave the best overall performance as measured by precision and recall. BayesNet assigns a probability that a protein belongs to one of the classes represented in the training data. We set a false positive rate cutoff of ≤10%. At this level only those proteins with a ≥75% probability of being plasma membrane were included. This generated a set of 1181 known and predicted plasma membrane-associated proteins.
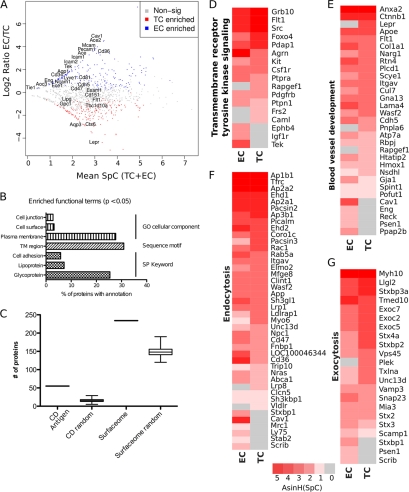
Of the 1181 predicted and known plasma membrane proteins, we identified 171 proteins enriched in the TC and 192 enriched at the EC surface (p ≤ 0.1; supplemental Table S7; Fig. 2A). EC-enriched proteins included numerous markers for EC cells such as Ace, Ace2, Aqp1, Cav1, Cd34, Cd36, Eng, Icam1, Icam2, Lyve1, Mcam, Pecam1, Tek, and Tie1. Many other well-known EC surface markers were expressed at both surfaces such as Flt1, Cdh5, Ece1, Esam, Lipg, Cd151, Cd47, Cd81, Gpc1, Ncam1, Aoc3, as well as many other known plasma membrane markers, annexins, G-proteins, integrins, and solute carriers (supplemental Table S7). The maternal-exposed TC surface is relatively poorly annotated, and only a few markers are currently known. These markers include Aqp3 (32), Cts6 (33), Lepr (34), and Tbc1d10a (35), all of which were preferentially detected in the TC surface proteome (Fig. 2A and supplemental Table S7). Systematic analysis of the 1,181 plasma membrane-enriched proteome revealed significant enrichment for a variety of functional annotation terms with expected plasma membrane expression (Figs. 2B, 2C). Given the common and unique expression of the known markers we next determined if these represented common and distinct biological roles of the two cell types.
Fig. 2.
Plasma membrane markers of the blood-tissue interface in the mouse placenta. A, A plot of the log2 transformed ratio of SpC in the EC over the TC versus the mean SpC of the EC and TC of proteins predicted to be plasma membrane associated by BasesNet machine learning. Proteins are color coded to represent those that were significantly enriched to the TC fraction (n = 171; red), EC fraction (n = 191; blue), or not statistically enriched (n = 891; gray). The plot is annotated with the same known markers presented in Fig. 1B. B, A chart showing selected significantly enriched annotation terms in the membrane predicted data set. Represented are the percentages of genes linked to an enriched term. C, Box plot showing that the predictive membrane data set was enriched for annotation terms CD antigen and surfaceome (36) significantly better than random sampling of the entire dataset. D–G, Heat map displays of selected, significantly over-represented functional classes in the plasma membrane enriched proteins showing differences in expression between TC and EC fractions. Heat maps are sorted in order of highest to lowest expression in TC. Although there is a high degree of similarity between the trophoblast (TC) and endothelial (EC) compartments there are some key differences in protein expression.
Functional Enrichment of Endothelial and Trophoblast Surfaces
To assess the functional similarities and differences between the TC and EC we first tested the entire data set for enriched protein functional classes (see Experimental Procedures). First, we assessed annotation terms associated with plasma membrane localization and found that they were highly enriched as predicted (p < 0.05; Fig. 2B). However these terms were used in model building so we made a more independent assessment by testing for enrichment of the CD (cluster of differentiation) antigens and members of the surfaceome (36). We tested for enrichment against a random draw of the same size as the predicted set of membrane proteins. In both cases our final filtered data set was enriched in CD antigens and known surfaceome proteins (Fig. 2C). We next tested for enrichment of Gene Ontology Biological process terms. Among the significantly enriched terms we identified Receptor tyrosine kinase signaling (Fig. 2D), Blood vessel development (Fig. 2E), Endocytosis (Fig. 2F), and Exocytosis (Fig. 2G) as being of immediate interest as they involve the primary function of these tissues in the transport of biomolecules between the maternal and fetal blood systems and the signaling mechanism involved. We generated heat maps of the SpC information for each of the proteins of these enriched functional groups to visually identify differences in their expression between TC and EC (Figs. 2D–2G). Although many proteins were expressed in common, there were some key differences in expression in each group.
Proteins of interest were Pnpla6, Rapgef1 and Lepr, which are all detected uniquely in TC (Fig. 2E). Rapgef1 is involved in regulation of Fgf signaling (37) and its expression implies a unique role in the trophoblast, although Fgf signaling is also involved in some aspects of endothelial development (38). In contrast, Eng, Psen1, Ppap2b, Reck, and Cav1 were all detected uniquely in EC (Fig. 2E). Eng is involved in artery versus vein specification of endothelial cells (39). However, human data indicates ENG protein is expressed in the TC (www.proteinatlas.org/ENSG00000106991/normal/placenta), but published mouse data supports EC only expression (40) and this is in accord with our current proteomics results. The unique expression of Lepr (34) in TC and Cav1 (41) in EC, has been previously observed in the human placenta.
In the subset of receptor tyrosine kinase signaling pathway-enriched proteins, we observed Cam1, Frs2 and Rapgef1 in TC and Igfr1, EphB4 and Tek uniquely in the EC (Fig. 2D). As noted above, Rapgef1 (37) is involved in Fgf signaling, as is Frs2 (42). The presence of both proteins uniquely at the TC surface highlights a potentially stronger role of Fgf signaling in trophoblast development at this developmental stage. Cam1 is involved in Egfr recycling and Egfr has previously been noted at the trophoblast surface (14). EphB4 is involved in artery versus vein differentiation (39, 43) and similar to Eng was found only in the EC.
We noted enrichment for the functional terms endocytosis and exocytosis (Figs. 2F, 2G). These processes play important roles in transport of products from blood plasma into cells and the secretion of factors from cells into blood plasma. Of interest is the unique detection of Lrp8 (Apoer2) and Vldlr in the TC (Fig. 2F). These two proteins are independent receptors for the extracellular protein Reelin (44). This suggests a role for the Reelin signaling pathway in TC although no placental phenotype has been described in either the Reelin, Lrp8 or Vldlr mouse mutants (45, 46). Lrp8 (Apoer2) and Vldlr are also involved in very low-density lipoprotein endocytosis (47) and triglyceride homeostasis (46). Scrib (Scribbled) was uniquely detected on EC cells (Fig. 2F) where it is important for maintenance of planar cell polarity (48).
In summary, shotgun proteomics in combination with computational machine-learning algorithms resulted in the assignment of 1181 proteins to the plasma membrane compartment of the mouse placental blood-tissue interfaces. Comprehensive analysis of both blood-tissue surfaces enabled us to distinguish the TC (i.e. maternal) and the EC (i.e. fetal) surfaces, based on the expression of identified plasma membrane proteins. This has revealed differences in the cell polarity organization, signaling pathways (Egfr, Fgf) as well as functional differences in the transport of nutrients (Vldlr and Lrp8). This resource will be of future use in the delineation of the functional similarities and differences between these two important cell types involved in blood transport and nutrient exchange.
Validation of Markers of the Maternal Blood-Trophoblast Tissue Interface
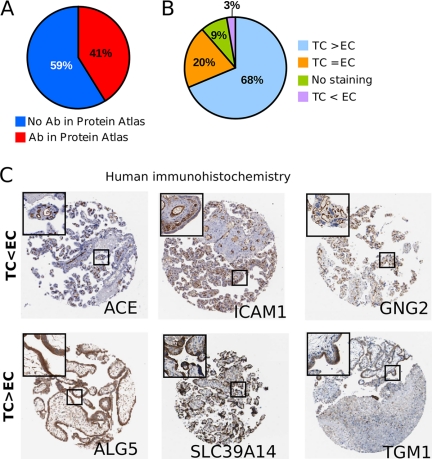
There are currently few published markers of the placental trophoblast. Using our extensive mouse proteomics data we directly addressed this shortcoming and identified a panel of 171 proteins enriched at the TC interface (Fig. 2A). We translated our mouse model findings to human biology using immunohistochemistry (IHC) available the Human Protein Atlas (13). As shown in Fig. 3A, 41% (70/171 proteins) had an antibody available at Human Protein Atlas (version: June 2010). Next, we downloaded all placental IHC images associated with these proteins and evaluated their histological expression. Antibodies were scored according to four categories: “strongest staining in the fetal trophoblast cells ” (TC > EC), “staining equally strong in endothelium and trophoblast cells ” (TC = EC), “staining stronger in the fetal endothelium ” (TC < EC), or “no staining observed ” (Fig. 3B). For proteins with available IHC placental images, ∼70% could be validated in humans with a similar differential expression pattern between the EC and trophoblast compartments. This was considered an excellent correspondence given differences in methods and species. There are notable caveats to this approach because antibody staining can be nonspecific, possibly because of high expression of Fc receptor in the syncytial trophoblast. However, a similar correspondence (70%) was obtained in our recent direct organellar proteomics comparison of human villus and mouse labyrinth tissues (14), which suggested that the 70% concordance observed between mass spectrometry and IHC images is highly significant. As examples we present the IHC results from Human Protein Atlas for the trophoblast enriched genes ALG5, SLC39A14, and TGM1 and EC enriched genes ACE, ICAM1, and GNG2 (Fig. 3C). In our study we also identified GPR50 as a novel TC marker and show specific expression in the TC using IHC staining (supplemental Fig. S2). Thus the mouse and human show a high concordance in expression between our mouse proteomic data and the human IHC images form the Human Protein Atlas database.
Fig. 3.
Immunohistochemistry validation of mass spectrometry-based proteomics data using the Human Protein Atlas database. A, Availability of antibodies in the Human Protein Atlas for 171 trophoblast enriched plasma membrane proteins identified in the mouse placenta. B, Systematic scoring results from all IHC images available at the Human Protein Atlas. Images were scored as “strongest staining in the fetal trophoblast” (TC>EC), “staining equally in the endothelium and trophoblast cells” (EC = TC), “no staining observed” or “staining stronger in the fetal endothelium” (TC<EC). C) Representative images from Human Protein Atlas (ProteinAtlas.org) for proteins enriched to either the trophoblast (ALG5, SLC39A14, TGM1) or endothelial (ACE, ICAM1, GNG2) cells.
Molecular Models of Preeclampsia
Toward our second goal of identifying patient subgroups we use this dataset of 171 TC enriched genes as our starting point. The TC is believed to be the source of factors that impair maternal EC function in PE (10). We therefore hypothesized that molecular heterogeneity in TC may directly correlate with different etiologies or subgroups of PE. To examine this, we selected a microarray cohort study of PE (n = 16) and normal placentas (n = 22) (49). This provided a relatively large number of patients with severe PE pathologies, all analyzed on a single array platform and performed by the same group. As mentioned in the introduction, mRNA may not predict protein expression therefore we linked our mouse protein data to the human microarray via a table of orthologs (www.ensembl.org; data not shown) to establish a set of microarray probes that had supporting evidence for protein expression.
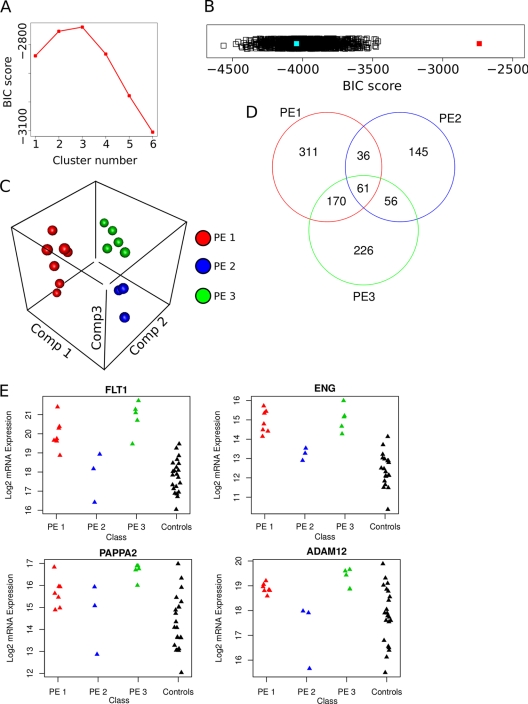
With this translated and integrated data in hand we set out to assess if there are different molecular groups in this phenotypically similar set of human PE patients. We could link the 171 TC enriched mouse plasma membrane proteins to 143 probes on the human array. We applied an expectation maximization (EM) clustering method to build data models ranging from 1 to 9 patient clusters using 143 high confidence TC enriched probes (see Experimental Procedures). We calculated the Bayesian Information Criterion (BIC) to estimate the fit of each of the nine models and found that a model with 3 clusters was optimal (Fig. 4A). This three-cluster model had a significantly better (p < 0.001) fit as determined by BIC score (Fig. 4B, red square) than models generated from a random draw of 143 probes (n = 1100; Fig. 4B, black squares) or the selection of the top 143 differentially regulated probes between all samples (Fig. 4B, light blue square). The latter comparison is critical as it validates our assumption that focusing on the TC cells lead to robust modeling of PE subgroups. Similar data could not be obtained when using data from multiple cell types of the placenta, such as in the published microarray study (49). A three-dimensional plot of a principal component analysis of the top model for the patient samples shows good separation of the three identified subgroups (Fig. 4C).
Fig. 4.
Classification of human PE patient mRNA expression data into molecular-based subgroups. A, Plot of Bayesian Information Criterion (BIC) scores versus models with increasing numbers of subgroups using gene expression data of human preeclamptic placentas (49) based on 143 trophoblast enriched proteins. B, Strip chart of BIC scores for the optimal model solution for the 143 trophoblast enriched proteins (red filled square) and 1150 random draws of 143 probes (black open squares) and the top 143 differentially regulated probes on the arrays (blue filled square). C, 3D plot of first three principal components of patient samples colored based on their subgroup membership (1 is red, 2 is blue and 3 is green). Plot shows strong segregation between subgroups and clustering within subgroups. D, Venn diagram of the overlap of differentially regulated genes in each group. Of note is the higher degree of overlap between groups 1 and 3. E, Strip charts of the microarray probe signal for individual patient samples showing four known markers of PE (FLT1, ENG, PAPPA2, ADAM12). Note that in each case the samples in subgroup 2 have a signal that is in the range of the controls. Subgroups 1 and 3 have a mean expression that is significantly higher (p < 0.01) than controls for all PE markers, whereas for subgroup 2 these markers are not significantly different than controls.
We next applied the patient model classification to the entire microarray data set (∼18,000 probes) and used linear mixed models (27) to assess the differences in gene expression between the patient subgroups and the controls (see Experimental Procedures). Greater than 50% of genes with significantly different expression in a subgroup compared with controls were also uniquely enriched to a single subgroup (Fig. 4D).
Finally, we assessed the genes that were unique and in common to the three subgroups. First we compared expression levels of the known PE markers (50). Of those on the array FLT1, ENG, APPAP2, and ADAM12 all showed significant change (p < 0.01) in expression in subgroups 1 and 3. Subgroup 2 did not have significantly different expression in these known PE markers compared with the controls (Fig. 4E). Clearly subgroup 2 represents a unique patho-physiology of PE. To assess these differences, we calculated enrichment of Gene Ontology terms, KEGG pathways and mouse mutant phenotypes in each subgroup using the Bingo plug-in for Cytoscape (see Experimental Procedures) (29, 30).
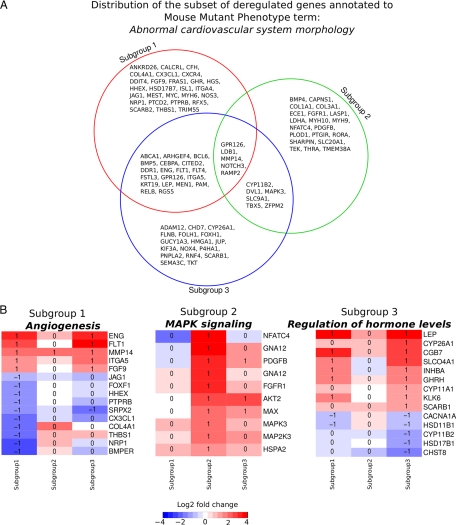
Results showed that each subgroup had many unique functional enrichment categories, and unique associations with mouse mutant phenotypes (Table I). Subgroup 1 has dis-regulation of genes involved in angiogenesis (Fig. 5B), vascular morphology, and development, VEGFR signaling, Jak-STAT signaling and regulation of the circulating levels of estrogen and progesterone. Subgroup 2 is enriched in genes with roles in actin binding, actin regulation, and signaling pathways of PDGF, MAPK (Fig. 5B), and TGF-beta signaling. Subgroup 3 is enriched for genes involved in both peptide and steroid hormone biosynthesis and metabolism (Fig. 5B). Of additional interest was the enrichment of phenotype terms involving abnormal placenta vasculature in subgroup 2. This may indicate that reassessment of these mouse mutant models may be warranted to identify PE like phenotypes.
Table I. Selected functionally related gene sets with enrichment (p < 0.05, FDR corrected) to one or more PE patient subgroups. Blank cell is a p value of >0.05.
| Term | Subgroup 1 | Subgroup 2 | Subgroup 3 |
|---|---|---|---|
| Gene Ontology | |||
| Angiogenesis | 0.0002 | ||
| Blood vessel morphogenesis | 0.0002 | ||
| Regulation of vascular endothelial growth factor receptor signaling pathway | 0.0000 | ||
| Morphogenesis of a branching structure | 0.0003 | ||
| Circulatory system development | 0.0003 | 0.0003 | |
| Platelet-derived growth factor binding | 0.0003 | ||
| Actin binding | 0.0004 | ||
| Hormone biosynthetic process | 0.0000 | ||
| Hormone metabolic process | 0.0000 | ||
| Regulation of hormone levels | 0.0000 | ||
| KEGG | |||
| Cytokine-cytokine receptor interaction | 0.0032 | ||
| Hedgehog signaling pathway | 0.0140 | ||
| Jak-STAT signaling pathway | 0.0456 | ||
| Regulation of actin cytoskeleton | 0.0004 | ||
| MAPK signaling pathway | 0.0080 | ||
| TGF-beta signaling pathway | 0.0239 | 0.0354 | |
| Steroid hormone biosynthesis | 0.0071 | ||
| Galactose metabolism | 0.0201 | ||
| Glutathione metabolism | 0.0227 | ||
| Arachidonic acid metabolism | 0.0367 | ||
| Mouse Mutant Phenotypes (MGI) | |||
| Abnormal placenta morphology | 0.0006 | ||
| Abnormal placenta vasculature | 0.0020 | ||
| Abnormal placenta labyrinth vasculature morphology | 0.0036 | ||
| Increased trophoblast giant cell number | 0.0006 | ||
| Abnormal cardiovascular system morphology | 0.0000 | 0.0000 | 0.0000 |
Fig. 5.
Differential enrichment of gene functional categories among human PE subgroups. A, Distribution of the 94 genes deregulated in the PE patient mRNA expression dataset annotated with the mouse mutant phenotype term abnormal cardiovascular systems morphology. Fewer than 30% show overlap with at least one other patient group although each group was independently assessed as enriched in this functional category. This suggests different molecular pathways are affected in each subgroup leading to a similar phenotypic outcome. B, Specific functional categories are enriched in each subgroup. Heat maps show similar expression for some members of each category across PE subgroups, but the majorities are uniquely deregulated in a single subgroup. Red intensity is increasing and blue is decreasing fold change of a subgroup versus controls. Numbers in the individual cells of the heat map indicate if the observed change is significant at a FDR corrected p value ≤ 0.05: −1 significantly down-regulated, 0 no significance and 1 significantly up-regulated.
Interestingly even when functional terms were in common, they had low levels of overlap in the specific genes affected within these terms. For example, of the 94 genes annotated to the mutant phenotype term Abnormal cardiovascular system morphology (Table I; Fig. 5A) 69% were misexpressed uniquely by one subgroup, 20% by subgroups 1 and 3, 6% by subgroups 2 and 3 and 5% by all three subgroups. This indicates that each group has alterations in a largely unique underlying set of genes anticipated to lead to Abnormal cardiovascular system morphology. A similar situation was observed for other functional enrichment categories (Table I). Thus even when a functional term was found to be enriched to more than one subgroup the associated deregulated genes were largely unique to each subgroup.
Protein extracts were prepared for Western blotting from human placental samples obtained at similar gestational ages; 10 PE, 21 with PE and IUGR, and 10 controls (supplemental Table S8). For our initial validation, we chose GNA12 because this protein had higher expression in subgroup 2 versus controls, whereas subgroups 1 and 3 showed no significant difference compared with controls. GNA12 and its mouse homologue Gna12 are expressed at the maternal blood-trophoblast tissue interface in both species (Human Protein Atlas and current study) and it has an available antibody that was highly rated for Western blotting (Human Protein Atlas). As a control we also tested the expression of the G-coupled protein receptor GPR50, which was identified in our study as a novel trophoblast membrane protein but did not show significant change in expression in any PE subgroup.
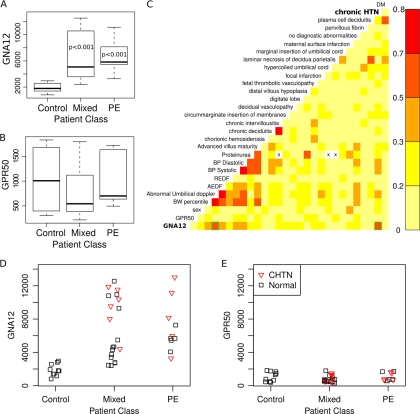
Boxplots of the signal intensity of protein expression for PE, mixed PE pathology and control placentas showed that the abnormal samples had significantly higher levels of GNA12 expression compared with controls (Figs. 6A) (p < 0.05; Wilcoxson test). The control protein GPR50 did not show significant differences in expression (Fig. 6B and supplemental Fig. S3).
Fig. 6.
Correlation of GNA12 expression with human PE and chronic hypertension. A–B, Box plot representation of densitometry readings from Western blot results for two selected trophoblast surface markers identified in the current study versus the patient groups: Control, IUGR-preeclampsia (Mixed) and preeclampsia without IUGR (PE). Statistical differences between groups were calculated using the Wilcoxson test for nonparametric data. Significant differences between controls and either Mixed or PE patients were observed for GNA12. A control protein GPR50, for which we did not observe any significant difference by microarray, had no significant difference by proteins. p values are reported at the 95% confidence level. C, Pair wise correlation plot of GNA12 and GPR50 protein expression and mother/fetus/child clinical data showing significant correlation of GNA12 with chronic hypertension (CHTN; green box bottom row on right). Also of note is the correlation with other signs of PE such as abnormal umbilical Doppler (green box bottom row on left). D–E, strip chart of GNA12 and GPR50 protein expression of individual patients colored by presence of (red triangles) or absence (black boxes) of chronic hypertension. GNA12 expression was statistically higher in the chronic hypertension group (p < 0.05) versus the non-chronic hypertension group in either the mixed or PE groups. None of the controls had chronic hypertension.
We generated a correlation matrix of the expression of GNA12 and GPR50 by Western blot versus clinical measurements of the mother and fetus, as well as detailed placental pathology and co-morbidity data for the mothers (Fig. 6C). We noted that there was a correlation between the level of GNA12 expression and the presence of chronic hypertension (CHTN) in the mothers (Figs. 6C; Pearson = 0.55, p < 0.001). There was also a correlation between GNA12 and abnormal umbilical artery Doppler wave-forms in the fetus, however this correlation was because of the much stronger association of abnormal umbilical Doppler with PE as only 4/24 PE cases had normal umbilical Dopplers reported. A strip chart of individual sample values for GNA12 (Fig. 6D) shows that PE samples with CHTN (Fig. 6D, red triangles) have higher levels of GNA12 expression (p < 0.01) versus PE samples without CHTN (Fig. 6D, black squares). GPR50, which was used as a control for these experiments, had no significant associations (Fig. 6E). We observed that there are several samples without CHTN that also show a high level of GNA12 expression similar to those with CHTN. It is important to note that the data of Sitras et al. (49), from which we built our model, specifically excluded patients with CHTN from their study. Our data would suggest that it may be difficult to accurately assess CHTN or that many women with CHTN are undiagnosed prior to entering into prenatal care of an OBGYN or midwife.
DISCUSSION
Patient classification is critical for prognosis and for designing diagnostic and treatment regimes. However, to date preeclampsia studies have sought sensitive and specific molecular markers to predict later onset of disease rather than focusing on markers for patient classification. In the current study, we successfully subclassify PE patients into three distinct patient subgroups, each with a strongly unique molecular difference. We did this using a translational analysis integrating TC enriched proteins identified in mice with microarray data from human PE placentas. Selecting microarray probes based on protein enrichment in TC was critical for successful classification and it out-performed selecting probes based on high differential mRNA expression in PE samples. Our translational analysis successfully identified distinct molecular subgroups of PE patients in a published microarray data set. By focusing our microarray data analysis on proteins significantly enriched at the TC surface, we focused our analysis on the putative primary source of disease in PE, the trophoblast and effectively only included microarray probes validated at the level of protein expression. Numerous genes are regulated by post-transcriptional and post-translational mechanisms and as such the array signal for the mRNA may not directly correlate with protein expression levels (17, 51, 52).
Using this novel analysis strategy, we identified three PE subgroups that contained numerous genes not previously linked to this disease. Previous studies may have missed these genes because prior microarray analysis of gene expression in PE has assumed that all patients with similar clinical signs have similar molecular pathologies. We show for the first time that this assumption is incorrect. This was even true for PE patients that had similar deregulated expression of know PE markers. Interestingly, even for functional terms enriched to two or all three subgroups, there was low overlap of the associated deregulated genes, indicating that different molecular pathologies lead to similar systemic phenotypes (i.e. abnormal placenta or cardiovascular system morphology).
An important aspect of the current work was the creation of a large resource of proteins expressed in common between mouse and human at the trophoblast maternal blood interface. Using Human Protein Atlas, we found that ∼70% of proteins identified as preferentially expressed at the maternal blood-trophoblast interface in mouse also showed preferential expression at this interface in humans. This high correlation was observed despite differences in methods, species, and anatomy of the trophoblast interface (31, 53). Knowledge of mouse-human similarities at this interface is important for creation of clinically-relevant mouse models of PE. There are over 200 genes with a placental phenotype when deleted in mice (MGI). These include seven genes involved in human placental pathologies (OMIM), which we have observed with conserved placental expression between humans and mice in a previous study (14). We note that we observed many human homologues of mouse genes associated with mouse placental phenotypes enriched in specific patient subgroups (Table I). These mouse models are good candidates for re-evaluation, to seek PE-like phenotypes. As well, generation of novel gain of function mouse mutants targeting the many genes identified here may reveal PE-like phenotypes and novel disease models.
Analysis of these three subgroups differed in genes misexpressed within specific GO, KEGG and Mouse Mutant Phenotype categories. For example, enrichment of altered angiogenesis genes in subgroup 1, MAPK signaling and vascular morphology in subgroup 2 and hormone synthesis and metabolism in subgroup 3. Yet all patients were clinically classified as severe preeclamptic. We infer that different molecular etiologies can elicit similar preeclampsia signs in humans as observed previously in animal models of this disease. Our findings further support this concept of divergent molecular etiologies causing similar disease phenotypes that within the enriched gene category of abnormal cardiovascular system morphology, common to all subgroups, there was little overlap of the specific genes affected in the three observed PE subgroups.
The combination of TC-enriched surface proteins with human PE gene expression data suggested GNA12 as a marker for PE subgroup 2. When we examined GNA12 protein expression in an independent human cohort, we found significantly increased expression at the protein level, specifically within a subset of PE patients with chronic hypertension (CHTN). Interestingly, we observed a higher percentage of PE patients with high GNA12 protein expression in our independent cohort than in the microarray study of Sitras et al. (49). This is likely because of our inclusion of chronic hypertension patients whereas the previous study had attempted to exclude them. However like the study of Sitras et al. (49), we found several PE cases with high levels of GNA12 but no reported CHTN in their records. Possibly, CHTN existed but was not indicated in the clinical record. Alternatively, factors other than CHTN may also elevate expression of this gene in the placenta in PE. The reduced incidence of GNA12 elevation in Sitras et al. (49) may also be because increased GNA12 protein may be caused by other means than increased mRNA expression. Future studies examining both protein and mRNA expression of GNA12 in PE placentas would be necessary to resolve this issue.
A role for GNA12 in the placenta, or its relationship to PE, has not been previously reported. GNA12 is a guanine nucleotide-binding protein (G protein), an important class of regulatory molecules that signal via transmembrane receptors (www.uniprot.org). GNA12 has been implicated in many biological processes including blood pressure regulation (54), as GNA12/GNA13 double knockout mice have a hypotensive phenotype (54). We infer that increased GNA12 expression may be associated with hypertensive phenotype but this remains to be shown. It is interesting that GNA12 expressed in fetal-derived trophoblast correlated with a maternal pre-pregnancy disorder. It remains to be seen if the increased level of GNA12 is a predictor or a result of CHTN in the mother. This maybe explained by two possibilities, one the increased placental GNA12 generates or is a result of a signal feedback between the maternal cardiovascular system and placenta trophoblast. A second possibility is that shedding of membrane vesicles from the trophoblast into the maternal blood may contain activated GNA12 signaling complexes that fuse with endothelial cells thus activating the pathway.
In future research it will be interesting to determine whether the 3 PE subgroups identified here on the basis of placental gene expression can be identified using blood biomarkers. Higher rates of trophoblast shedding are observed in PE as compared with normal pregnancies (11, 55). Trophoblast plasma membrane proteins shed into the maternal blood system may provide useful biomarkers. These vesicles can contain mRNA and miRNAs (11, 56), thus misexpressed genes identified here in placental tissue may be measured in vesicles isolated from maternal blood. Our observation of altered expression of four enzymes involved in steroid hormone biosynthesis (CYP11A1, HSD17B1, CYP11B2, and HSD11B1) in subgroup 3 would be interesting to study in more detail given that lower circulating levels of 17-beta-estradiol are observed in some preeclampsia patients (57). An important next step will be to devise methods to classify PE patients with different placental molecular pathologies using blood biomarkers or other clinically applicable approaches.
In summary, this study suggests different molecular pathology in the placenta can cause similar clinical phenotypes in the mother. This may explain why large linkage analysis studies have shown conflicting loci between studies and have not revealed many new biomarker candidates to date (50). Furthermore, diversity in PE etiology may explain the general failure of two large controlled clinical treatment trials (56, 58). We have also shown that different molecular pathologies can be used to classify patients with PE. We predict that, as in cancer, such classification will prove valuable for defining prognosis, and for individualizing diagnostic and treatment regimes. Our results show that there is a strong similarity between the mouse and human trophoblast surface proteome. This suggests that it may be possible to generate mouse models of the three identified human subgroups of PE. The mouse models may be useful in longitudinal studies to identify candidate blood biomarkers for human PE, similar to a recent strategy in prostate cancer biomarker discovery (59), as well as serving as models to study disease progression, understand mechanisms and test new therapies. Ultimately, knowledge of the unique proteins at the interface between maternal blood and fetal trophoblast reported here will help propel discovery of novel biomarkers to stratify disease by etiology, and may lead to novel targeted treatments based on a greater understanding of the molecular basis of placental pathology in this common and life-threatening disease of pregnancy.
Acknowledgments
The authors thank the donors, and the Research Centre for Women's and Infants' Health BioBank program of the CIHR Group in Development and Fetal Health (CIHR #MGC-13299), the Samuel Lunenfeld Research Institute and the Mount Sinai Hospital/University Healthy Network Department of Obstetrics & Gynaecology for the human specimens. We are grateful to Jorge Cabezas for animal breeding and mouse colony maintenance. The views expressed do not necessarily reflect those of the OMOHLTC. Contribution: P.S., B.C., A.I.E., K.W., V.I., A.I., D.B., M.C., S.K. and T.K. performed experiments and analyzed data; B.C., S.L.A. and T.K. designed the research; A.O.G., J.R., S.L.A., J.K. and T.K. supervised the project, and B.C., P.S., A.O.G., S.L.A., and T.K. wrote the paper. The authors declare no competing financial interests.
Footnotes
* This work was supported by the Heart and Stroke Foundation of Ontario to T.K., and A.O.G. (T-6281 and NS-6636). A.O.G. is a New Investigator of the Heart and Stroke Foundation of Canada and a Canada Research Chair in Cardiovascular Proteomics and Molecular Therapeutic. T.K. is a Canada Research Chair in Proteomics of Cancer Research. S.L.A. gratefully acknowledges support as the Anne and Max Tanenbaum Chair in Molecular Medicine at Mount Sinai Hospital. Operating funds from CIHR #MGC-13299 and CIHR#MOP-12772 to SLA also contributed to this work. This research was funded in part by the Ontario Ministry of Health and Long Term Care.
 This article contains supplemental Experimental Procedures, Figs. S1 to S3, and Tables S1 to S8.
This article contains supplemental Experimental Procedures, Figs. S1 to S3, and Tables S1 to S8.
1 The abbreviations used are:
- AEDF
- absence of umbilical end diastolic flow
- BIC
- Bayesian Information Criterion
- BP
- blood pressure
- BW
- birth weight (adjusted)
- CHTN
- chronic hypertension
- Comp
- component
- DM
- diabetes mellitus
- EC
- endothelial cell
- Gest HTN
- gestational hypertension
- GDM
- gestational DM
- HELLP
- Hemolytic anemia elevated liver enzymes low platelet count
- HTN
- hypertension
- IHC
- immunohistochemistry
- IUGR
- intrauterine growth restriction
- MGI
- Mouse Genome Informatics
- Mixed
- mixed IUGR and PE phenotype
- MudPIT
- multidimensional protein identification technology
- OMIM
- Online Mendelian Inheritance in Man
- PE
- preeclampsia
- REDF
- reversed umbilical end-diastolic flow
- SpC
- spectral count
- TC
- trophoblast cell.
REFERENCES
- 1. Roberts J. M., Cooper D. W. (2001) Pathogenesis and genetics of pre-eclampsia. Lancet 357, 53–56 [DOI] [PubMed] [Google Scholar]
- 2. van 't Veer L. J., Dai H., van de Vijver M. J., He Y. D., Hart A. A., Mao M., Peterse H. L., van der Kooy K., Marton M. J., Witteveen A. T., Schreiber G. J., Kerkhoven R. M., Roberts C., Linsley P. S., Bernards R., Friend S. H. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 [DOI] [PubMed] [Google Scholar]
- 3. Sibai B. M. (2003) Diagnosis and management of gestational hypertension and preeclampsia. Obstet. Gynecol. 102, 181–192 [DOI] [PubMed] [Google Scholar]
- 4. MacKay A. P., Berg C. J., Atrash H. K. (2001) Pregnancy-related mortality from preeclampsia and eclampsia. Obstet. Gynecol. 97, 533–538 [DOI] [PubMed] [Google Scholar]
- 5. Davisson R. L., Hoffmann D. S., Butz G. M., Aldape G., Schlager G., Merrill D. C., Sethi S., Weiss R. M., Bates J. N. (2002) Discovery of a spontaneous genetic mouse model of preeclampsia. Hypertension 39, 337–342 [DOI] [PubMed] [Google Scholar]
- 6. Furuya M., Ishida J., Inaba S., Kasuya Y., Kimura S., Nemori R., Fukamizu A. (2008) Impaired placental neovascularization in mice with pregnancy-associated hypertension. Lab. Invest. 88, 416–429 [DOI] [PubMed] [Google Scholar]
- 7. Kanayama N., Takahashi K., Matsuura T., Sugimura M., Kobayashi T., Moniwa N., Tomita M., Nakayama K. (2002) Deficiency in p57Kip2 expression induces preeclampsia-like symptoms in mice. Mol. Hum. Reprod. 8, 1129–1135 [DOI] [PubMed] [Google Scholar]
- 8. Levine R. J., Maynard S. E., Qian C., Lim K. H., England L. J., Yu K. F., Schisterman E. F., Thadhani R., Sachs B. P., Epstein F. H., Sibai B. M., Sukhatme V. P., Karumanchi S. A. (2004) Circulating angiogenic factors and the risk of preeclampsia. N. Engl. J. Med. 350, 672–683 [DOI] [PubMed] [Google Scholar]
- 9. Woods A. K., Hoffmann D. S., Weydert C. J., Butler S. D., Zhou Y., Sharma R. V., Davisson R. L. (2011) Adenoviral delivery of VEGF121 early in pregnancy prevents spontaneous development of preeclampsia in BPH/5 mice. Hypertension 57, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roberts J. M., Hubel C. A. (2009) The two stage model of preeclampsia: variations on the theme. Placenta 30 Suppl A, S32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang A., Rana S., Karumanchi S. A. (2009) Preeclampsia: the role of angiogenic factors in its pathogenesis. Physiology 24, 147–158 [DOI] [PubMed] [Google Scholar]
- 12. Huppertz B. (2008) Placental origins of preeclampsia: challenging the current hypothesis. Hypertension 51, 970–975 [DOI] [PubMed] [Google Scholar]
- 13. Pontén F., Gry M., Fagerberg L., Lundberg E., Asplund A., Berglund L., Oksvold P., Björling E., Hober S., Kampf C., Navani S., Nilsson P., Ottosson J., Persson A., Wernérus H., Wester K., Uhlén M. (2009) A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 5, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox B., Kotlyar M., Evangelou A. I., Ignatchenko V., Ignatchenko A., Whiteley K., Jurisica I., Adamson S. L., Rossant J., Kislinger T. (2009) Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol. Syst. Biol. 5, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durr E., Yu J., Krasinska K. M., Carver L. A., Yates J. R., Testa J. E., Oh P., Schnitzer J. E. (2004) Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol. 22, 985–992 [DOI] [PubMed] [Google Scholar]
- 16. Whiteley K. J., Pfarrer C. D., Adamson S. L. (2006) Vascular corrosion casting of the uteroplacental and fetoplacental vasculature in mice. Meth. Mol. Med. 121, 371–392 [DOI] [PubMed] [Google Scholar]
- 17. Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T., Rossant J., Hughes T. R., Frey B., Emili A. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173–186 [DOI] [PubMed] [Google Scholar]
- 18. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 19. Gortzak-Uzan L., Ignatchenko A., Evangelou A. I., Agochiya M., Brown K. A., St Onge P., Kireeva I., Schmitt-Ulms G., Brown T. J., Murphy J., Rosen B., Shaw P., Jurisica I., Kislinger T. (2008) A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J. Proteome Res. 7, 339–351 [DOI] [PubMed] [Google Scholar]
- 20. Sodek K. L., Evangelou A. I., Ignatchenko A., Agochiya M., Brown T. J., Ringuette M. J., Jurisica I., Kislinger T. (2008) Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT). Mol. Biosyst. 4, 762–773 [DOI] [PubMed] [Google Scholar]
- 21. Hall M., Frank E., Holmes G., Pfahringer B., Reutemann P., Witten I. H. (2009) The WEKA Data Mining Software: An Update. SIGKDD Explorations 11 [Google Scholar]
- 22. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 [Google Scholar]
- 23. Team R. D. C. (2010) R: A language and environment for statisitical computing. R Development Core Team (2010). R: A language and environment for statistical computing. (c) R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0, URL http://www.R-project.org [Google Scholar]
- 24. Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fraley C., Raftery A. E. (2002) Model-based clustering, discriminant analysis, and density estimation. J. Amer. Statistical. Assoc. 97, 611–631 [Google Scholar]
- 26. Culhane A. C., Thioulouse J., Perrière G., Higgins D. G. (2005) MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics 21, 2789–2790 [DOI] [PubMed] [Google Scholar]
- 27. Smyth G. K. (2005) Bioinformatics and Computational Biology Solutions using R and Bioconductor. Limma: Linear Models for Microarray Data. New York: Springer, 397–420 [Google Scholar]
- 28. Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 30. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Georgiades P., Ferguson-Smith A. C., Burton G. J. (2002) Comparative developmental anatomy of the murine and human definitive placentae. Placenta 23, 3–19 [DOI] [PubMed] [Google Scholar]
- 32. Mann S. E., Ricke E. A., Yang B. A., Verkman A. S., Taylor R. N. (2002) Expression and localization of aquaporin 1 and 3 in human fetal membranes. Am. J. Obstet. Gynecol. 187, 902–907 [DOI] [PubMed] [Google Scholar]
- 33. Nakajima A., Kataoka K., Takata Y., Huh N. H. (2000) Cathepsin-6, a novel cysteine proteinase showing homology with and co-localized expression with cathepsin J/P in the labyrinthine layer of mouse placenta. Biochem. J. 349, 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Challier J., Galtier M., Bintein T., Cortez A., Lepercq J., Hauguel-de Mouzon S. (2003) Placental leptin receptor isoforms in normal and pathological pregnancies. Placenta 24, 92–99 [DOI] [PubMed] [Google Scholar]
- 35. Reczek D., Bretscher A. (2001) Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J. Cell Biol. 153, 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. da Cunha J. P., Galante P. A., de Souza J. E., de Souza R. F., Carvalho P. M., Ohara D. T., Moura R. P., Oba-Shinja S. M., Marie S. K., Silva W. A., Jr., Perez R. O., Stransky B., Pieprzyk M., Moore J., Caballero O., Gama-Rodrigues J., Habr-Gama A., Kuo W. P., Simpson A. J., Camargo A. A., Old L. J., de Souza S. J. (2009) Bioinformatics construction of the human cell surfaceome. Proc. Natl. Acad. Sci. U.S.A. 106, 16752–16757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yan J., Li F., Ingram D. A., Quilliam L. A. (2008) Rap1a is a key regulator of fibroblast growth factor 2-induced angiogenesis and together with Rap1b controls human endothelial cell functions. Mol. Cell. Biol. 28, 5803–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murakami M., Simons M. (2008) Fibroblast growth factor regulation of neovascularization. Curr. Opin. Hematol. 15, 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mancini M. L., Terzic A., Conley B. A., Oxburgh L. H., Nicola T., Vary C. P. (2009) Endoglin plays distinct roles in vascular smooth muscle cell recruitment and regulation of arteriovenous identity during angiogenesis. Dev. Dyn. 238, 2479–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kruse A., Hallmann R., Butcher E. C. (1999) Specialized patterns of vascular differentiation antigens in the pregnant mouse uterus and the placenta. Biol. Reprod. 61, 1393–1401 [DOI] [PubMed] [Google Scholar]
- 41. Mohanty S., Anderson C. L., Robinson J. M. (2010) The expression of caveolin-1 and the distribution of caveolae in the murine placenta and yolk sac: parallels to the human placenta. Placenta 31, 144–150 [DOI] [PubMed] [Google Scholar]
- 42. Kouhara H., Hadari Y. R., Spivak-Kroizman T., Schilling J., Bar-Sagi D., Lax I., Schlessinger J. (1997) A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89, 693–702 [DOI] [PubMed] [Google Scholar]
- 43. Swift M. R., Weinstein B. M. (2009) Arterial-venous specification during development. Circ. Res. 104, 576–588 [DOI] [PubMed] [Google Scholar]
- 44. Benhayon D., Magdaleno S., Curran T. (2003) Binding of purified Reelin to ApoER2 and VLDLR mediates tyrosine phosphorylation of Disabled-1. Brain Res. Mol. Brain Res 112, 33–45 [DOI] [PubMed] [Google Scholar]
- 45. Trommsdorff M., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R. E., Richardson J. A., Herz J. (1999) Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97, 689–701 [DOI] [PubMed] [Google Scholar]
- 46. Yagyu H., Lutz E. P., Kako Y., Marks S., Hu Y., Choi S. Y., Bensadoun A., Goldberg I. J. (2002) Very low density lipoprotein (VLDL) receptor-deficient mice have reduced lipoprotein lipase activity. Possible causes of hypertriglyceridemia and reduced body mass with VLDL receptor deficiency. J. Biol. Chem. 277, 10037–10043 [DOI] [PubMed] [Google Scholar]
- 47. Kim D. H., Iijima H., Goto K., Sakai J., Ishii H., Kim H. J., Suzuki H., Kondo H., Saeki S., Yamamoto T. (1996) Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J. Biol. Chem. 271, 8373–8380 [DOI] [PubMed] [Google Scholar]
- 48. Montcouquiol M., Rachel R. A., Lanford P. J., Copeland N. G., Jenkins N. A., Kelley M. W. (2003) Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature 423, 173–177 [DOI] [PubMed] [Google Scholar]
- 49. Sitras V., Paulssen R. H., Gronaas H., Leirvik J., Hanssen T. A., Vårtun A., Acharya G. (2009) Differential placental gene expression in severe preeclampsia. Placenta 30, 424–433 [DOI] [PubMed] [Google Scholar]
- 50. Grill S., Rusterholz C., Zanetti-Dällenbach R., Tercanli S., Holzgreve W., Hahn S., Lapaire O. (2009) Potential markers of preeclampsia–a review. Reprod. Biol. Endocrinol. 7, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cox B., Kislinger T., Wigle D. A., Kannan A., Brown K., Okubo T., Hogan B., Jurisica I., Frey B., Rossant J., Emili A. (2007) Integrated proteomic and transcriptomic profiling of mouse lung development and Nmyc target genes. Mol. Syst. Biol. 3, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Washburn M. P., Koller A., Oshiro G., Ulaszek R. R., Plouffe D., Deciu C., Winzeler E., Yates J. R., 3rd (2003) Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 100, 3107–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adamson S. L., Lu Y., Whiteley K. J., Holmyard D., Hemberger M., Pfarrer C., Cross J. C. (2002) Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Develop. Biol. 250, 358–373 [DOI] [PubMed] [Google Scholar]
- 54. Wirth A., Benyó Z., Lukasova M., Leutgeb B., Wettschureck N., Gorbey S., Orsy P., Horvath B., Maser-Gluth C., Greiner E., Lemmer B., Schutz G., Gutkind J. S., Offermanns S. (2008) G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 14, 64–68 [DOI] [PubMed] [Google Scholar]
- 55. Orozco A. F., Lewis D. E. (2010) Flow cytometric analysis of circulating microparticles in plasma. Cytometry A 77, 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruano R., Fontes R. S., Zugaib M. (2005) Prevention of preeclampsia with low-dose aspirin – a systematic review and meta-analysis of the main randomized controlled trials. Clinics 60, 407–414 [DOI] [PubMed] [Google Scholar]
- 57. Hertig A., Liere P., Chabbert-Buffet N., Fort J., Pianos A., Eychenne B., Cambourg A., Schumacher M., Berkane N., Lefevre G., Uzan S., Rondeau E., Rozenberg P., Rafestin-Oblin M. E. (2010) Steroid profiling in preeclamptic women: evidence for aromatase deficiency. Am. J. Obstet. Gynecol. 203, 477, e471–479 [DOI] [PubMed] [Google Scholar]
- 58. Roberts J. M., Myatt L., Spong C. Y., Thom E. A., Hauth J. C., Leveno K. J., Pearson G. D., Wapner R. J., Varner M. W., Thorp J. M., Jr., Mercer B. M., Peaceman A. M., Ramin S. M., Carpenter M. W., Samuels P., Sciscione A., Harper M., Smith W. J., Saade G., Sorokin Y., Anderson G. B. (2010) Vitamins C and E to prevent complications of pregnancy-associated hypertension. N. Engl. J. Med. 362, 1282–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cima I., Schiess R., Wild P., Kaelin M., Schüffler P., Lange V., Picotti P., Ossola R., Templeton A., Schubert O., Fuchs T., Leippold T., Wyler S., Zehetner J., Jochum W., Buhmann J., Cerny T., Moch H., Gillessen S., Aebersold R., Krek W. (2011) Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]