Abstract
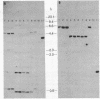
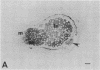
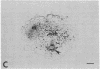
Symbiotically essential genes have been identified in Rhizobium meliloti that are structurally and functionally related to chromosomal virulence (chv) genes of Agrobacterium tumefaciens. Homologous sequences also exist in the genomes of other fast-growing rhizobia including Rhizobium trifolii, Rhizobium leguminosarum, and Rhizobium phaseoli. In Agrobacterium, the chvA and chvB loci are known to be essential for oncogenic transformation of dicotyledonous plants and for attachment to plant cells [Douglas, C. J., Staneloni, R. J., Rubin, R. A. & Nester, E. W. (1985) J. Bacteriol. 64, 102-106], and the chvB locus has been implicated in the production of (1→2)-β-glucan, a unique exopolysaccharide component [Puvanesarajah, V., Schell, F. M., Stacey, G., Douglas, C. J. & Nester, E. W. (1985) J. Bacteriol. 164, 102-106]. Site-directed transposon insertion mutants in the chvA and chvB-equivalent regions of R. meliloti are symbiotically defective. Mutants in the chvB-equivalent region have been examined in detail and have been found to induce the formation of nodule-like structures on alfalfa that are devoid of bacteroids, lack infection threads, and cannot fix nitrogen. Such mutants fluoresce normally in the presence of Calcofluor, a histochemical stain for β-linked polysaccharides, and produce normal amounts of total exopolysaccharide. The Rhizobium loci have been designated ndv because of their requirement for nodule development.
Keywords: symbiosis, fix- mutants, heterologous complementation
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyoshi D. E., Klee H., Amasino R. M., Nester E. W., Gordon M. P. T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5994–5998. doi: 10.1073/pnas.81.19.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Better M., Lewis B., Corbin D., Ditta G., Helinski D. R. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell. 1983 Dec;35(2 Pt 1):479–485. doi: 10.1016/0092-8674(83)90181-2. [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Douglas C. J., Peschon J. Flagella-specific bacteriophages of Agrobacterium tumefaciens: demonstration of virulence of nonmotile mutants. Can J Microbiol. 1984 May;30(5):676–681. doi: 10.1139/m84-101. [DOI] [PubMed] [Google Scholar]
- Corbin D., Barran L., Ditta G. Organization and expression of Rhizobium meliloti nitrogen fixation genes. Proc Natl Acad Sci U S A. 1983 May;80(10):3005–3009. doi: 10.1073/pnas.80.10.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C. J., Halperin W., Nester E. W. Agrobacterium tumefaciens mutants affected in attachment to plant cells. J Bacteriol. 1982 Dec;152(3):1265–1275. doi: 10.1128/jb.152.3.1265-1275.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C. J., Staneloni R. J., Rubin R. A., Nester E. W. Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol. 1985 Mar;161(3):850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Hirsch A. M., Leigh J. A., Johansen E., Kuldau G. A., Deegan S., Walker G. C., Signer E. R. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985 Apr;40(4):869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- Forrai T., Vincze E., Bánfalvi Z., Kiss G. B., Randhawa G. S., Kondorosi A. Localization of symbiotic mutations in Rhizobium meliloti. J Bacteriol. 1983 Feb;153(2):635–643. doi: 10.1128/jb.153.2.635-643.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., Simpson R. B., Ream L. W., White F. F., Gordon M. P., Nester E. W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981 Nov;27(1 Pt 2):143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- Klee H. J., White F. F., Iyer V. N., Gordon M. P., Nester E. W. Mutational analysis of the virulence region of an Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1983 Feb;153(2):878–883. doi: 10.1128/jb.153.2.878-883.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuykendall L. D., Elkan G. H. Rhizobium japonicum derivatives differing in nitrogen-fixing efficiency and carbohydrate utilization. Appl Environ Microbiol. 1976 Oct;32(4):511–519. doi: 10.1128/aem.32.4.511-519.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Signer E. R., Walker G. C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megias M., Caviedes M. A., Palomares A. J., Perezsilva J. Use of plasmid R68.45 for constructing a circular linkage map of the Rhizobium trifolii chromosome. J Bacteriol. 1982 Jan;149(1):59–64. doi: 10.1128/jb.149.1.59-64.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvanesarajah V., Schell F. M., Stacey G., Douglas C. J., Nester E. W. Role for 2-linked-beta-D-glucan in the virulence of Agrobacterium tumefaciens. J Bacteriol. 1985 Oct;164(1):102–106. doi: 10.1128/jb.164.1.102-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- SCHWINGHAMER E. A. Studies on induced variation in the rhizobia. I. Defined media and nodulation test techniques. Appl Microbiol. 1960 Nov;8:349–352. doi: 10.1128/am.8.6.349-352.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G., Waffenschmidt S., Weiler E. W., Schröder J. The T-region of Ti plasmids codes for an enzyme synthesizing indole-3-acetic acid. Eur J Biochem. 1984 Jan 16;138(2):387–391. doi: 10.1111/j.1432-1033.1984.tb07927.x. [DOI] [PubMed] [Google Scholar]
- Vandenbosch K. A., Noel K. D., Kaneko Y., Newcomb E. H. Nodule initiation elicited by noninfective mutants of Rhizobium phaseoli. J Bacteriol. 1985 Jun;162(3):950–959. doi: 10.1128/jb.162.3.950-959.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]