Abstract
Type 1 diabetes genes within the interleukin (IL)-2, cytotoxic T-lymphocyte--associated protein 4 (CTLA-4), and natural resistance-associated macrophage protein (NRAMP1) pathways influence development of autoimmune diabetes in humans and NOD mice. In NOD mice, when present together, protective alleles encoding IL-2, Idd3 candidate gene, CTLA-4, NRAMP1, and acetyl-coenzyme A dehydrogenase, long-chain (ACADL) (candidate genes for the Idd5.1, Idd5.2, and Idd5.3 subregions) provide nearly complete diabetes protection. To define where the protective alleles of Idd3 and the Idd5 subregions must be present to protect from diabetes and tolerize islet-specific CD8+ T cells, SCID mice were reconstituted so that the host and lymphocytes expressed various combinations of protective and susceptibility alleles at Idd3 and Idd5. Although protective Idd3 alleles in the lymphocytes and protective Idd5 alleles in the SCID host contributed most significantly to CD8 tolerance, both were required together in both lymphocyte and nonlymphocyte cells to recapitulate the potent diabetes protection observed in intact Idd3/5 mice. We conclude that genetic regions involved in autoimmune disease are not restricted in their influence to individual cell types. Even a single protective gene product, such as IL-2, must be expressed in both the lymphocytes and dendritic cells to exert its full extent of disease protection. These studies highlight the pleiotropic effects of genes that determine autoimmune disease susceptibility.
Type 1 diabetes is an autoimmune disease that is caused by T cell–mediated destruction of the pancreatic islet β-cells. Multiple genetic and environmental factors contribute to disease progression in both humans and the NOD mouse model of type 1 diabetes (1). Considerable overlap exists between the mouse and human disease, exemplified by several common genetic risk pathways including genes encoding major histocompatibility complex (MHC)-II, insulin, interleukin (IL)-2/IL-2 receptor α (IL-2RA), cytotoxic T-lymphocyte--associated protein 4 (CTLA-4), and natural resistance-associated macrophage protein (NRAMP1) (2–5). In the mouse, type 1 diabetes susceptibility regions are termed insulin-dependent diabetes (Idd) regions and have been identified through the use of congenic strains in which genomic segments from diabetes-protected strains are introgressed onto the NOD background. Here we have examined the mechanisms of protection of four such regions: Idd3 on chromosome 3 and Idd5.1, Idd5.2, and Idd5.3 on chromosome 1.
Idd3 contains the candidate genes Il2 and Il21. The B6-derived Il2 allele produces twofold more IL-2 protein than the NOD-derived allele (6–8). Although the expression levels of both the Il2 (6,7,9) and Il21 (8) genes have been reported to differ between the NOD and B6 alleles, examination of other Idd3 haplotypes (CZECH, CAST, SWR, and A/J) introgressed onto the NOD background revealed that diabetes protection correlated with high expression levels of Il2 but no correlation was found with the Il21 expression level (6). Furthermore, the introduction of a single Il2 knockout allele onto NOD-Idd3 congenic mice, which reduces IL-2 production by 50%, abrogates protection despite the presence of two potentially protective Il21 alleles (6). This experiment proves that reduction of IL-2 causes increased diabetes susceptibility, verifying Il2 as Idd3.
Candidate genes for the Idd5 region include Ctla4 (Idd5.1) (10,11), Slc11a1 (formally Nramp1, Idd5.2) (12,13), and Acadl (Idd5.3) (13,14). CTLA-4 is a T cell–expressed inhibitory molecule, and enhanced expression of a ligand-independent isoform correlates with the protective allele (11). NRAMP1 is a phagosomal ion transporter expressed in dendritic cells (DCs) (15,16) and macrophages and a loss of function mutation is associated with disease protection (12). ACADL is a ubiquitously expressed enzyme involved in fatty-acid metabolism, and higher expression levels are associated with a protective phenotype (14,16). Despite the identification of candidate genes for these regions, it is not yet clear how they affect the disease process.
We demonstrated previously that protection from disease in mice expressing both Idd3 and Idd5 protective alleles (Idd3/5 mice) is highly correlated with restored CD8+ T-cell tolerance to islet antigens (16–18). Idd3/5 genes were found to function through both CD4+ T cells and DCs to restore proliferation and accumulation of islet-specific CD8+ T cells to a low level (16). Furthermore, in reconstituted SCID mice, in which the genotype of the donor T cells and host DCs could be manipulated separately, both host cells and donor lymphocytes needed to express protective Idd3/5 genes to completely protect from the development of insulitis. However, their contribution to diabetes was not explored; neither were the individual contributions of the Idd3 and Idd5 regions to donor T cells and host DCs explored. Because both Idd3 and Idd5 genes are expressed by both CD4+ T cells and DCs, we aimed to define where the individual Idd3 and Idd5 regions are required for the restoration of CD8+ T-cell tolerance, protection from insulitis, and diabetes onset.
RESEARCH DESIGN AND METHODS
Mice.
Experimental procedures were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Institutional Animal Care and Use Committee #09-0074). NOD/MrkTac and NOD-SCID mice were purchased from Taconic. Idd3/5, Idd3, Idd5, and Idd3/5-SCID congenic strains and NOD-8.3 Thy1.1+ T-cell receptor (TCR) transgenic mice have been described previously (6,16,19–21). The Idd3 (1098) strain contains a 2.7 Mb genetic interval derived from B6 bounded by the markers D3nds35 and D3nds76. The Idd3 region has been mapped to a 650 kb region that contains seven genes within line 1098 (6). The Idd5 (1094) strain contains a 28.3 Mb genetic interval derived from B10 and bounded by the markers D1Mit478 and D1mit134. The Idd5.1 region has been mapped to a 2.1 Mb region that contains four genes (20); the Idd5.2 region is 1.52 Mb and contains 45 genes (20) and the Idd5.3 region has been mapped to a 3.55 Mb region containing 11 genes (13). These strains can be visualized at http://dil.t1dbase.org/page/MouseStrainsHome and enter lines 1094 and 1098. Idd5-SCID mice were generated by intercrossing Idd5 and NOD-SCID mice and selecting mice homozygous for both alleles. Similarly, Idd3-SCID mice were made by intercrossing Idd3 and NOD-SCID mice.
Virus.
Recombinant vaccinia virus expressing the H-2Kd restricted epitope VYLKTNVFL, amino acid residues 206–214 of murine endogenous islet antigen, islet-specific glucose-6-phosphatase catalytic subunit related protein (IGRP; Vac-KdIGRP) was described previously (21). Mice were infected intraperitoneally with 1 × 107 plaque-forming units of virus, and CD8+ T-cell responses were measured in the spleen 7 days later.
Flow cytometry.
CD8+ T cells were stained with H-2Kd-IGRP206–214 –PE tetramers (National Institute of Allergy and Infectious Disease [NIAID] MHC Tetramer core facility) for 15 min RT followed by staining with anti-CD8-FITC at 4°C for 15 min. All mAbs were obtained from either eBioscience, BioLegend, or BD Pharmingen (all San Diego, CA). Cells were acquired with either a FACSCalibur or LSRII (Becton Dickinson, Mountain View, CA) and analyzed with FlowJo software (Tree Star, Inc., Ashland, OR).
Purification and adoptive transfer of T cells.
Naive CD8+Thy1.1+ 8.3 TCR transgenic cells were isolated from lymph nodes and spleen of NOD 8.3 Thy1.1+ TCR transgenic mice. CD8+ T cells were purified using a CD8+ T-cell enrichment kit (BD Biosciences), and carboxyfluorescein succinimidyl ester (CFSE) labeling was performed as described (22). Recipient mice were injected with 4 × 106 purified CD8+ T cells intravenously. Four days after the transfer, pancreatic lymph node (PcLN) cells were prepared for analysis.
Reconstitution of SCID mice.
SCID mice were reconstituted with total spleen and lymph node cells prepared from 3- to 5-week-old donor mice. DCs were depleted with Pan-DC microbeads (Miltenyi Biotec) according to the manufacturer’s instructions before transfer of 2 to 3 × 107 cells intravenously. Mice were rested for 6–8 weeks before adoptive transfer of CD8+Thy1.1+8.3 TCR cells or 10 weeks before infection with Vac-KdIGRP or harvesting pancreata.
Assessment of diabetes and insulitis.
Pancreata were fixed in 10% neutral buffered formalin and paraffin embedded. Hematoxylin and eosin–stained sections were scored for insulitis. At least 20 islets per mouse were scored blind as either 0, no infiltration; 1, peri-insulitis; 2, mild-invasive insulitis; or 3, severe invasive insulitis. Mice having elevated urinary glucose >500 mg/dL (detected using Diastix [Myles, Elkhart, IN]) were classified as diabetic. During the diabetes frequency study, some mice became sick or died without becoming diabetic. This was most likely from the development of thymomas, which commonly occur in SCID mice as they age. Nondiabetic mice were censored from the analysis either four weeks before death or when they became noticeably sick.
Statistical analysis.
Kaplan-Meier survival analysis of diabetes was performed for each group of reconstituted mice, and type 1 diabetes frequencies were compared using the log-rank test (GraphPad Prism software). Differences in the proportion or number of tetramer binding cells were compared between groups via the Mann-Whitney U test (GraphPad Prism software). Differences in the proportion of divided 8.3 cells were compared via Student t test (GraphPad Prism software).
RESULTS
Idd3 and Idd5 contribute to restored CD8+ T-cell tolerance to islet antigen IGRP.
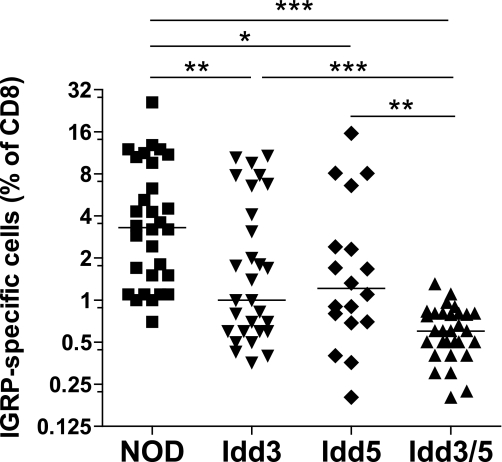
Protective alleles at Idd3 and Idd5 are associated with reduced CTL responses to IGRP. We chose to examine CD8+ T-cell tolerance to IGRP since these T cells are present at high frequency and their presence is predictive of diabetes progression in individual NOD mice (23). To assess the numbers of IGRP-specific CD8+ T cells, NOD mice congenic for the individual Idd3 and Idd5 regions were infected with Vac-KdIGRP and the frequency of IGRP-specific CD8+ T cells that expanded in the spleen were determined by tetramer staining (Fig. 1). Similar to our previous findings measuring CTL killing (18), Idd3/5 mice had a greatly reduced expansion of autoreactive IGRP-specific CD8+ T cells compared with NOD mice (P < 0.0001). Mice having diabetes-protective alleles at Idd3 alone (Idd3 mice) also had a strongly reduced expansion of IGRP-specific CD8+ T cells compared with NOD (P = 0.0037). Mice having diabetes-protective alleles at Idd5 (Idd5 mice) showed an intermediate level of CD8+ T-cell expansion compared with NOD (P = 0.0116). Total numbers of IGRP-specific cells were affected similarly (Supplementary Fig. 1). We conclude that diabetes-protective alleles at Idd3 and Idd5 both independently contribute to restored CD8+ T-cell tolerance to IGRP and that when diabetes-protective alleles at Idd3 and Idd5 are present together, the mechanisms by which tolerance is influenced by each of the regions are additive.
FIG. 1.
Restored IGRP-specific CD8+ T-cell tolerance in Idd3/5 subregion mice. Female, 10- to 14-week-old mice were infected with 1 × 107 pfu Vac-KdIGRP. On day 7 after infection, spleens were analyzed by fluorescence-activated cell sorter for CD8+ IGRP-tetramer+ cells. Results were compiled from five separate experiments with 4–8 mice per strain in each experiment. Horizontal lines depict median value. Pairs of strains were compared with the Mann-Whitney U test. *P < 0.05; **P < 0.01; ***P < 0.0001.
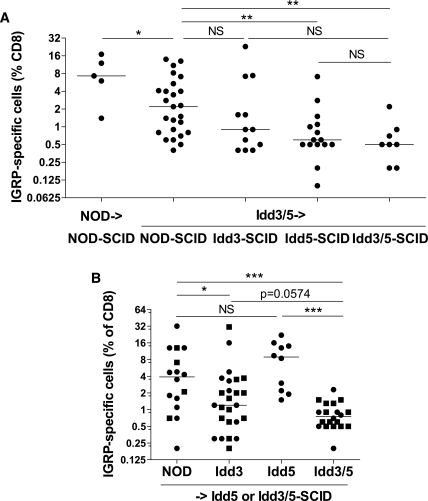
Although Idd3 (Il2) is expressed by DCs (24) in a genotype-dependent manner (16), IL-2 is primarily associated with T-cell functions. Therefore we predicted that Idd3 expression would be required on T cells. The Idd5.2 candidate gene product NRAMP1 is only expressed in phagocytic cell types such as DCs and macrophages; therefore, we predicted that Idd5 rather than Idd3 would be the primary region required on DCs to facilitate IGRP tolerance. To test these hypotheses, SCID mice expressing NOD, Idd3, Idd5, or Idd3/5 alleles were reconstituted with Idd3/5 or control NOD spleen and lymph node cells and after 10 weeks the mice were infected with Vac-KdIGRP and the expansion of IGRP-specific CD8+ T cells were assessed in the spleen (Fig. 2A). NOD->NOD-SCID and Idd3/5->NOD-SCID reconstituted mice had very high frequencies of IGRP-specific CD8+ T cells. Idd3-SCID mice reconstituted with Idd3/5 lymphocytes gave intermediate frequencies of IGRP-specific CD8+ T cells (Idd3/5->NOD-SCID vs. Idd3/5->Idd3-SCID, P = 0.2231; Idd3/5->Idd3-SCID vs. Idd3/5->Idd3/5-SCID, P = 0.2143). Expression of Idd5 genes on the SCID host mice prevented the expansion of IGRP-specific CD8+ T cells (Idd3/5->NOD-SCID vs. Idd3/5->Idd5-SCID, P = 0.0033). There was no significant difference in the level of IGRP-specific CD8+ T cells between Idd5-SCID and Idd3/5-SCID mice reconstituted with Idd3/5 lymphocytes (P = 0.4677, not significant [NS]). Thus the presence of diabetes-protective Idd5 alleles alone in the SCID host in the context of lymphocytes having protective alleles at both Idd3 and Idd5 was sufficient to restrain autoreactive, IGRP-specific CD8+ T cells.
FIG. 2.
Expression of Idd5 genes by nonlymphocytes and Idd3 genes by lymphocytes is required for restoration of CD8+ T-cell tolerance. A: NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice were reconstituted with spleen and lymph node cells depleted of DCs isolated from 3- to 4-week-old NOD or Idd3/5 donor mice. After 10 weeks, the mice were infected with Vac-KdIGRP, and 7 days later the spleens were analyzed for CD8+ IGRP-tetramer+ cells. Pooled results from four experiments are shown. Horizontal lines depict median value. Pairs of strains were compared with the Mann-Whitney U test. *P < 0.05; **P < 0.01. B: Idd5-SCID (■) and Idd3/5-SCID (●) mice were reconstituted with spleen and lymph node cells depleted of DCs isolated from 3- to 4-week-old NOD, Idd3, Idd5, or Idd3/5 donor mice. After 10 weeks, the mice were infected with Vac-KdIGRP, and 7 days later the spleens were analyzed for CD8+ IGRP-tetramer+ cells. Pooled results from four experiments are shown. Horizontal lines depict median value. Pairs of strains were compared with the Mann-Whitney U test. *P < 0.05; ***P < 0.001.
To assess the contribution of lymphocyte expression of protective genes, SCID hosts were reconstituted with either NOD, Idd3, Idd5, or Idd3/5 donor spleen and lymph node cells depleted of DCs. Both Idd5-SCID and Idd3/5-SCID hosts were used since they provided equivalent results (Fig. 2A), and no differences were seen between the two hosts (Fig. 2B, compare ● and ■). Idd3, but not Idd5, expressing donor cells were able to reduce the frequency of IGRP-specific CD8+ T cells (Fig. 2B, NOD vs. Idd3, P = 0.04; NOD vs. Idd5, P = 0.22, NS). However, Idd3 donor lymphocytes were almost significantly worse than Idd3/5 donor lymphocytes (Idd3 vs. Idd3/5, P = 0.0574, NS), and Idd3 had a significantly greater variance than Idd3/5 donor lymphocytes (P < 0.0001). This indicates that the presence of diabetes-protective Idd5 alleles in the donor lymphocytes also had a measurable effect on the frequency of IGRP-specific CD8+ T cells. Taken together these results suggest expression of Idd5 by DCs and Idd3 by lymphocytes contributes a highly significant level of tolerance.
The contribution of Idd3 and Idd5 to accumulation of IGRP-specific T cells in the PcLN.
The first encounter of naïve T cells in the periphery with islet antigens, such as IGRP, occurs in the PcLN where the cells undergo proliferation. In Idd3/5 mice this results in deletion of the autoreactive cells, whereas in NOD mice the cells accumulate and can later be found within the islets (16,17). We found previously that the degree of proliferation in the PcLN of transferred islet specific T cells is not influenced by the extent of pre-existing insulitis present in the pancreas (17). Similar to our previous findings (18), 8.3 cells, which are specific for IGRP (25), proliferated significantly more in NOD mice than Idd3/5 mice (P < 0.0001, Fig. 3). When 8.3 cells were transferred into Idd3 or Idd5 mice they proliferated at a slightly reduced level compared with 8.3 cells transferred to NOD mice (NOD vs. Idd3, P = 0.0363; NOD vs. Idd5, P = 0.0302). However, the presence of both diabetes-protective alleles together reduced the proliferation of 8.3 cells to a very low level (similar to what would be expected in a nondiabetes prone strain [16]). This demonstrates that diabetes-protective alleles together at Idd3 and Idd5 have an additive effect on reducing the ability of autoreactive CD8+ T cells to expand in response to islet-derived antigen presented in the regional lymph node, a very early check point in the autoimmune response.
FIG. 3.
Reduced proliferation of IGRP-specific 8.3 cells in Idd3/5 subregion mice. Mice were transferred with CFSE-labeled Thy1.1+ 8.3 CD8 T cells, and four days later the PcLN were analyzed by fluorescence-activated cell sorter. The proportion of Thy1.1+ CD8+ cells with diluted CFSE is plotted. Data are pooled from six independent experiments. Horizontal lines depict mean value. Pairs of strains were compared with the Student t test. *P < 0.05; **P < 0.01; ***P < 0.0001.
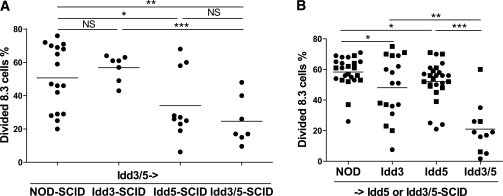
To assess the contribution of protective genes expressed by the donor lymphocytes or host, we repeated the SCID reconstitution experiments described in Fig. 2A and after 7 weeks transferred CFSE-labeled NOD 8.3 CD8+ T cells to the reconstituted mice. SCID host mice expressing protective Idd3 alleles and reconstituted with Idd3/5 lymphocytes were unable to reduce the proliferation of transferred 8.3 CD8+ T cells in the PcLN (Fig. 4A, NOD-SCID vs. Idd3-SCID, P = 0.429). However, host expression of protective Idd5 alleles reduced the accumulation of 8.3 CD8+ T cells (Fig. 4A, NOD-SCID vs. Idd5-SCID, P = 0.008). Idd3/5->Idd5-SCID mice had an equivalent low level of 8.3 T-cell proliferation as Idd3/5->Idd3/5-SCID mice (P = 0.241). We conclude that in the presence of Idd3/5 lymphocytes, expression of protective Idd5 alleles in the SCID host cells (including DCs) effectively allows deletion of autoreactive IGRP-specific CD8+ T cells.
FIG. 4.
Expression of Idd5 genes by nonlymphocytes and both Idd3 and Idd5 genes by lymphocytes is required for low proliferation of IGRP-specific T cells. A: NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice were reconstituted with spleen and lymph node cells depleted of DCs isolated from 3- to 4-week-old NOD or Idd3/5 donor mice. After 7 weeks, CFSE-labeled 8.3 Thy1.1+ CD8 T cells were transferred, and the PcLN was analyzed by fluorescence-activated cell sorter on day 4 for divided 8.3 cells. Pooled results from three experiments are shown. Horizontal lines depict mean value. Pairs of strains were compared by Student t test. *P < 0.05; **P < 0.01; ***P < 0.001. B: Idd5-SCID (■) and Idd3/5-SCID (●) mice were reconstituted with spleen and lymph node cells depleted of DCs isolated from 3- to 4-week-old NOD, Idd3, Idd5, or Idd3/5 donor mice. After 7 weeks, CFSE-labeled 8.3 Thy1.1+ CD8 T cells were transferred, and the PcLN was analyzed by fluorescence-activated cell sorter on day 4 for divided 8.3 cells. Pooled results from four experiments are shown. Horizontal lines depict mean value. Pairs of strains were compared by Student t test. *P < 0.05; **P < 0.01; ***P < 0.001.
To assess the contribution of lymphocyte expression of protective genes to tolerance in the PcLN, Idd5 or Idd3/5-SCID hosts reconstituted as described in Fig. 2B were given CFSE-labeled 8.3 cells. Neither Idd3 nor Idd5 donor cells were sufficient to fully restore low levels of accumulation of 8.3 cells in the PcLN (Fig. 4B). Diabetes-protective Idd3 alleles on the donor cells gave a partial reduction in 8.3 cell proliferation (NOD vs. Idd3, P = 0.019; Idd3 vs. Idd3/5, P = 0.0047), as did diabetes-protective Idd5 alleles (NOD vs. Idd5, P = 0.0187; Idd5 vs. Idd3/5, P = 0.0012). Only in Idd3/5 mice was low proliferation and accumulation of IGRP-specific 8.3 cells observed. Thus, tolerance in the PcLNs requires host expression of Idd5 and lymphocyte expression of both Idd3 and Idd5.
Protective alleles at both Idd3 and Idd5 must be present in lymphocytes and nonlymphocytes for protection from diabetes development.
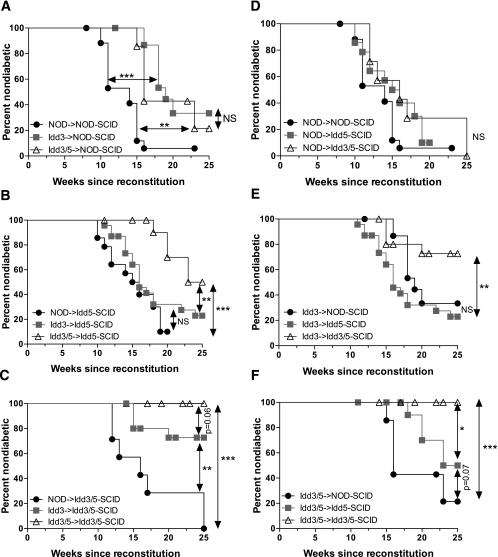
We now asked where the individual Idd3 and Idd5 regions must be expressed to prevent diabetes. Because Idd3-SCID mice had no or minor effects on CD8+ T-cell tolerance to IGRP (Figs. 2A and 4A), recipient SCID mice expressing protective Idd3 alleles only were not included in the diabetes study. Instead we compared the effect of Idd3 in the DCs for its effect on diabetes by comparing the diabetes results obtained with Idd3/5-SCID versus Idd5-SCID mice. NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice were reconstituted with donor spleen and lymph node cells (DC depleted) derived from young NOD, Idd3, or Idd3/5 mice. The frequency of diabetes was then monitored for 25 weeks (Fig. 5). When NOD-SCID mice were reconstituted with NOD lymphocytes, almost all mice became diabetic (Fig. 5A). Survival was significantly increased when Idd3 lymphocytes were used in NOD-SCID hosts (P < 0.0001), but Idd3/5 lymphocytes did not further enhance disease protection (Idd3->NOD-SCID vs. Idd3/5->NOD-SCID, P = 0.4467, NS; NOD->NOD-SCID vs. Idd3/5->NOD-SCID, P = 0.0065). When NOD, Idd3, and Idd3/5 lymphocytes were used to reconstitute Idd5-SCID hosts (Fig. 5B), a significant difference between Idd3 and Idd3/5 donor cells emerged (P = 0.0128), indicating that the presence of diabetes-protective Idd5 alleles in lymphocytes contributes to protection from type 1 diabetes. Idd5-SCID hosts receiving Idd3 donor lymphocytes were not protected from diabetes as compared with NOD->Idd5-SCID reconstituted mice (P = 0.2059), although Idd3->Idd5-SCID mice were significantly better protected than NOD->NOD-SCID mice (P = 0.0017). Finally, Idd3/5-SCID mice were reconstituted with NOD, Idd3, and Idd3/5 lymphocytes (Fig. 5C), resulting in greatly improved diabetes protection when Idd3 donor cells were used compared with NOD donor cells (P = 0.0023). Idd3/5 donor cells were strongly protective when transferred to Idd3/5-SCID recipients (NOD->Idd3/5-SCID vs. Idd3/5->Idd3/5-SCID, P < 0.0001) and further increased diabetes protection compared with Idd3 donor cells (P = 0.0610). In conclusion, protective alleles at both Idd3 and Idd5 contribute to diabetes protection when expressed by donor lymphocytes. However, the ability of protective alleles at Idd5 to add to the diabetes protection provided by Idd3 varies depending on the genotype of the SCID recipient.
FIG. 5.
Diabetes is dependent on both Idd3 and Idd5 region genes, expressed on both lymphocytes and nonlymphocytes. NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice were reconstituted with spleen and lymph node cells depleted of DCs isolated from 3- to 4-week-old NOD, Idd3, or Idd3/5 donor mice. NOD->NOD-SCID (n = 18), Idd3->NOD-SCID (n = 16), Idd3/5->NOD-SCID (n = 7), NOD->Idd5-SCID (n = 14), Idd3->Idd5-SCID (n = 23), Idd3/5->Idd5-SCID (n = 15), NOD->Idd3/5-SCID (n = 7), Idd3->Idd3/5-SCID (n = 16), and Idd3/5->Idd3/5-SCID (n = 13) mice were monitored for 25 weeks for elevated urine glucose. A: NOD-SCID hosts. B: Idd5-SCID hosts. C: Idd3/5-SCID hosts. D: NOD donor lymphocytes. E: Idd3 donor lymphocytes. F: Idd3/5 donor lymphocytes. Data are pooled from three experiments. Kaplan-Meier survival curves were compared with the log-rank test. *P < 0.05; **P < 0.01; ***P < 0.0001.
We also considered the same data presented in Fig. 5A-C to assess the effects of protective alleles at Idd3 and Idd5 on the host SCID cells. NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice reconstituted with each lymphocyte type are depicted in Fig. 5D (NOD lymphocytes), Fig. 5E (Idd3 lymphocytes), and Fig. 5F (Idd3/5 lymphocytes). Reconstitution of NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice with NOD lymphocytes did not result in significant differences in diabetes incidence (Fig. 5D), with relatively rapid diabetes occurring in almost all mice. These results underscore the conclusion that the presence of Idd3/5 diabetes-protective alleles in the SCID recipients only is not sufficient to provide any level of protection from diabetes. When Idd3 lymphocytes were used to reconstitute NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice, no increased diabetes protection resulted from expression of Idd5 on the SCID host (Fig. 5E; Idd3->NOD-SCID vs. Idd3-> Idd5-SCID, P = 0.1042, NS). However, Idd3->Idd3/5-SCID mice were highly significantly protected from diabetes compared with Idd3->Idd5-SCID mice (Fig. 5E; P = 0.0046). Finally, even though Idd3/5 donor lymphocytes transferred to NOD-SCID mice provide some diabetes protection as compared with NOD->NOD-SCID recipients (Fig. 5A) further decreases in disease were observed, revealing a dramatic effect of host genotype on diabetes incidence (Fig. 5F, Idd3/5->NOD-SCID vs. Idd3/5->Idd3/5-SCID, P = 0.0005). This effect was contributed to by both Idd5-SCID hosts (Idd3/5->NOD-SCID vs. Idd3/5->Idd5-SCID, P = 0.0754) and significantly when Idd3/5-SCID hosts were used (Idd3/5->Idd5-SCID vs. Idd3/5->Idd3/5-SCID, P = 0.0151). These combined data demonstrate that the presence of protective alleles at both Idd3 and Idd5 in the host SCID contributes to protection from diabetes.
Cellular requirements for protective Idd3 and Idd5 alleles to reduce insulitis development.
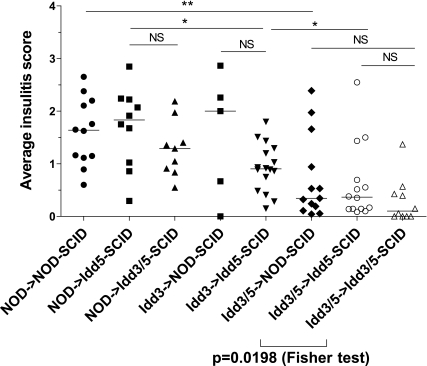
Disease progression is characterized by increasing insulitis. To assess the contribution of Idd3 and Idd5 on the lymphocyte and nonlymphocyte cell types to insulitis, NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice were reconstituted with NOD, Idd3, or Idd3/5 donor spleen and lymph node cells, and after 10 weeks, pancreata were harvested and assessed for insulitis development (Fig. 6). When the three SCID hosts were reconstituted with NOD lymphocytes and compared, little difference in insulitis was found (Fig. 6), a similar result to diabetes (Fig. 5D). Reconstitution of Idd5-SCID mice with Idd3 lymphocytes significantly decreased insulitis compared with NOD->Idd5-SCID mice (P = 0.0143), confirming a role for Idd3 on lymphocytes. However, Idd3->Idd5-SCID mice had significantly more insulitis than Idd3/5->Idd5-SCID mice (P = 0.0323), demonstrating a role for Idd5 on lymphocytes also. When Idd3/5 lymphocytes were used to reconstitute NOD-SCID and Idd5-SCID mice, no difference in the amount of insulitis was observed. However, when Idd3/5->Idd5-SCID and Idd3/5->Idd3/5-SCID mice were compared, although no significant difference in the median value was found using the Mann-Whitney U test, significantly more Idd3/5->Idd3/5-SCID mice had no insulitis (score 0, P = 0.0198, Fisher’s exact test). These data support our finding that the expression of protective Idd3 alleles by the host SCID mouse contributes to protection against type 1 diabetes.
FIG. 6.
Requirements for expression of protective Idd3 and Idd5 subregions for protection from insulitis development. NOD-SCID, Idd5-SCID, and Idd3/5-SCID mice were reconstituted with spleen and lymph node cells depleted of DCs isolated from 3- to 4-week-old NOD, Idd3, or Idd3/5 donor mice. After 10 weeks, pancreata were harvested and hematoxylin-eosin–stained sections were assessed for insulitis. Data are pooled from four experiments. Horizontal lines depict median value. Pairs of strains were compared with the Mann-Whitney U test. *P < 0.05; **P < 0.01.
DISCUSSION
Expression of protective alleles of Idd3 and Idd5 almost entirely abrogates diabetes development (19,26). In this study, we sought to define the cell type where each individual protective region acts alone as well as in combination. Because there is very strong evidence that the causative gene in the Idd3 locus is Il2 (6), which is most commonly associated with T-cell function, we hypothesized that Idd3 would function in the lymphocyte compartment. Because the Idd5 subregion with the strongest individual diabetes protective effect is Idd5.2 (13), and substantial evidence points to the DC and macrophage-expressed protein NRAMP1 (12), we hypothesized that the DC/host component of diabetes protection would be mediated by Idd5. The protective form of Slc11a1 (encoding NRAMP1) contains a loss of function mutation of this phagosomal ion transporter, which has been postulated to alter phagosome acidification and may influence antigen processing (15). Alternatively, NRAMP1 expression alters cytokine production in macrophages and DCs, which could alter differentiation or effector responses of phagocytic cells (27). We did find that Idd3->Idd5-SCID mice had significantly delayed diabetes as compared with NOD->NOD-SCID mice; however, it was not to the degree that we had expected since 80% of Idd3->Idd5-SCID mice still became diabetic. Therefore, significant Idd3/5 protective effects were missing in this combination.
In addition to the Idd3 candidate Il2, the Idd5.1 and Idd5.3 candidate genes Ctla4 and Acadl are also expressed by lymphocytes. The protective allele of Ctla4 produces increased amounts of a ligand-independent splice form (liCTLA-4). Expression of liCTLA4 inhibits TCR signaling by dephosphorylating the TCR-ζ chain, although it is not clear how this modulates disease (28); but one possibility is that liCTLA-4 and IL-2 both act to enhance Treg function. ACADL is a ubiquitously expressed enzyme involved in fatty acid metabolism. T cells have been shown to switch between primarily utilizing fatty acid metabolism to glycolytic metabolism upon proliferation, and regulation of this switch may be involved in the transition from effector to memory T cells (29). Therefore the study of ACADL in T-cell function is an area of interest, and we are currently testing protective alleles at individual Idd5 subregions together with Idd3 protective alleles for their effects on the diabetes protection mediated by donor cells. An additional complexity and caveat of these studies is that different subregion genes may act in different cell types. For example Idd5.1, Idd5.2, and Idd5.3 may act separately in lymphocytes and host cells and, furthermore, additional unidentified weak subloci may also contribute.
Our most surprising finding was that protective alleles at Idd3 as well as Idd5 contributed to diabetes protection when expressed by SCID host cells. Other than T cells, IL-2 can be produced by NK and NKT cells (30,31,32) as well as DCs after stimulation with microbial products (16,24), and DC-produced IL-2 has been shown to have a role in T-cell priming (24). Very recently, it has been shown that DCs provide IL-2 to T cells very early after activation, before the T cells produce their own IL-2, enhancing T-cell activation (33). However, an in vivo requirement for DC-produced IL-2 has not been shown previously in an animal model, and this is the first demonstration in which it may have a role in autoimmune disease. It is possible that DCs provide IL-2 to Tregs, enhancing their homeostatic proliferation or suppressor function.
The protective effect provided by the host cells was completely masked when NOD lymphocytes were present. It is noteworthy, however, that even when protective Idd3 alleles were expressed by T cells, a high frequency of diabetes occurred unless the host expressed Idd3 and Idd5 protective alleles. Thus the effects of both the lymphocytes and host are critical, and both must express protective alleles to dramatically reduce the incidence of diabetes.
To gain more information about what stage of the disease process the Idd3 and Idd5 genes function, we also examined the frequency of IGRP-specific CD8+ T cells in the reconstituted SCID mice as well as the proliferation of IGRP-specific 8.3 T cells in the PcLN. We found that host-expressed Idd5 was sufficient to tolerize IGRP-specific CD8+ T cells. This was in contrast with diabetes where host-expressed Idd5 was not able to provide as protective an environment as Idd3/5. This suggests that the effective CD8+ T-cell tolerance seen 7 and 10 weeks after reconstitution may not be maintained since the mice age to 17–25 weeks when they became diabetic.
On the lymphocytes, Idd3 alone had a strong effect on CD8+ T-cell tolerance, whereas Idd5 alone had no effect or only a slight effect (Fig. 2). Because of these minimal effects of diabetes-protective alleles at Idd5, we did not test whether protective alleles at Idd5 alone in the lymphocyte donor protect from diabetes. Similarly, because Idd5 hosts strongly promoted CD8+ T-cell tolerance, we did not include Idd3-SCID mice in the diabetes studies but compared Idd3 host effects in combination with Idd5.
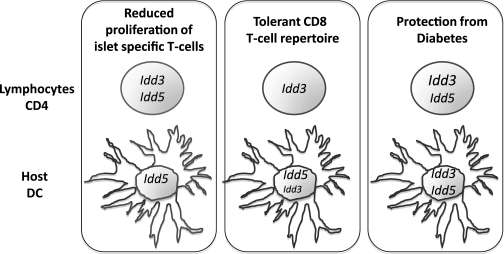
These studies demonstrate that the protective alleles of type 1 diabetes susceptibility genes act in multiple cell types and stages of disease to prevent diabetes. Our findings for each phenotype we have analyzed are summarized diagrammatically in Fig. 7. The important implication of these studies is that although Idd5 was sufficient on the host cells in the prediabetic period represented by early proliferation and tolerance of islet-specific CD8+ T cells, Idd3 on the host was critical to prevent conversion into clinical disease. This may have therapeutic consequences since, for example, treatments that enhance IL-2 production by DCs may be valuable at the time of disease onset.
FIG. 7.
Diagram of the phenotypic effects of the individual Idd3 and Idd5 regions. The individual protective alleles of the Idd3 and Idd5 regions have been identified to act in either the lymphocytes (including CD4+ T cells) or SCID host (including DCs) to restore a protective phenotype. Two subclinical phenotypes were examined: proliferation of transferred IGRP-specific CD8+ T cells and tolerance of the CD8+ T-cell repertoire to IGRP as well as the clinical phenotype of diabetes incidence.
ACKNOWLEDGMENTS
This work was supported by National Institute of Allergy and Infectious Diseases (NIAID) Grant AI 070351. E.E.H.-W. is supported by postdoctoral fellowships from the Juvenile Diabetes Research Foundation (JDRF). L.S.W. is supported by grants from the JDRF and the Wellcome Trust and is a Wellcome Trust Principal Research Fellow. The Cambridge Institute for Medical Research is the recipient of a Wellcome Trust Strategic Award (079895). The availability of NOD congenic mice through the Taconic Farms Emerging Models Program has been supported by grants from the Merck Genome Research Institute, NIAID, and the JDRF.
No potential conflicts of interest relevant to this article were reported.
E.E.H.-W. conceived and designed the study, performed experiments, analyzed data, and wrote the manuscript. J.C. performed experiments. E.E.H.-W., L.S.W., and L.A.S. are the guarantors for this article. D.B.R. and K.M.H. developed congenic mouse strains. L.S.W. developed congenic mouse strains, conceived the study, and edited and revised the manuscript. L.A.S. conceived the study and edited and revised the manuscript.
The authors thank all members of the Sherman laboratory (The Scripps Research Institute [TSRI]) for helpful discussion, Kristi Marquardt (TSRI) for technical assistance, and Michelle Gassert (TSRI) for administrative assistance. The authors also thank the employees of TSRI histology core facility, who prepared histology sections, and the employees of TSRI flow cytometry core facility, who sorted and helped with fluorescence-activated cell sorter analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0790/-/DC1.
REFERENCES
- 1.Kikutani H, Makino S. The murine autoimmune diabetes model: NOD and related strains. Adv Immunol 1992;51:285–322 [DOI] [PubMed] [Google Scholar]
- 2.Wicker LS, Clark J, Fraser HI, et al. Type 1 diabetes genes and pathways shared by humans and NOD mice. J Autoimmun 2005;25(Suppl.):29–33 [DOI] [PubMed] [Google Scholar]
- 3.Dendrou CA, Plagnol V, Fung E, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet 2009;41:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridgway WM, Peterson LB, Todd JA, et al. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol 2008;100:151–175 [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Downes K, Howson J, et al. Evidence of association with type 1 diabetes in the SLC11A1 gene region. BMC Med Genet 2011;12:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamanouchi J, Rainbow D, Serra P, et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 2007;39:329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sgouroudis E, Kornete M, Piccirillo CA. IL-2 production by dendritic cells promotes Foxp3+ regulatory T-cell expansion in autoimmune-resistant NOD congenic mice. Autoimmunity 2011;44:406–414 [DOI] [PubMed] [Google Scholar]
- 8.Goudy KS, Johnson MC, Garland A, et al. Reduced IL-2 expression in NOD mice leads to a temporal increase in CD62Llo FoxP3+ CD4+ T cells with limited suppressor activity. Eur J Immunol 2011;41:1480–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuire HM, Vogelzang A, Hill N, Flodström-Tullberg M, Sprent J, King C. Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus. Proc Natl Acad Sci USA 2009;106:19438–19443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueda H, Howson JM, Esposito L, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003;423:506–511 [DOI] [PubMed] [Google Scholar]
- 11.Araki M, Chung D, Liu S, et al. Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice. J Immunol 2009;183:5146–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS. In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat Genet 2006;38:479–483 [DOI] [PubMed] [Google Scholar]
- 13.Hunter K, Rainbow D, Plagnol V, Todd JA, Peterson LB, Wicker LS. Interactions between Idd5.1/Ctla4 and other type 1 diabetes genes. J Immunol 2007;179:8341–8349 [DOI] [PubMed] [Google Scholar]
- 14.Irie J, Reck B, Wu Y, et al. Genome-wide microarray expression analysis of CD4+ T cells from nonobese diabetic congenic mice identifies Cd55 (Daf1) and Acadl as candidate genes for type 1 diabetes. J Immunol 2008;180:1071–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai YD, Marrero IG, Gros P, Zaghouani H, Wicker LS, Sercarz EE. Slc11a1 enhances the autoimmune diabetogenic T-cell response by altering processing and presentation of pancreatic islet antigens. Diabetes 2009;58:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton-Williams EE, Martinez X, Clark J, et al. Expression of diabetes-associated genes by dendritic cells and CD4 T cells drives the loss of tolerance in nonobese diabetic mice. J Immunol 2009;183:1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez X, Kreuwel HT, Redmond WL, et al. CD8+ T cell tolerance in nonobese diabetic mice is restored by insulin-dependent diabetes resistance alleles. J Immunol 2005;175:1677–1685 [DOI] [PubMed] [Google Scholar]
- 18.Hamilton-Williams EE, Martinez X, Lyman M, Hunter K, Wicker LS, Sherman LA. The use of Idd congenic mice to identify checkpoints of peripheral tolerance to islet antigen. Ann N Y Acad Sci 2007;1103:118–127 [DOI] [PubMed] [Google Scholar]
- 19.Hill NJ, Lyons PA, Armitage N, Todd JA, Wicker LS, Peterson LB. NOD Idd5 locus controls insulitis and diabetes and overlaps the orthologous CTLA4/IDDM12 and NRAMP1 loci in humans. Diabetes 2000;49:1744–1747 [DOI] [PubMed] [Google Scholar]
- 20.Wicker LS, Chamberlain G, Hunter K, et al. Fine mapping, gene content, comparative sequencing, and expression analyses support Ctla4 and Nramp1 as candidates for Idd5.1 and Idd5.2 in the nonobese diabetic mouse. J Immunol 2004;173:164–173 [DOI] [PubMed] [Google Scholar]
- 21.Hamilton-Williams EE, Wong SB, Martinez X, et al. Idd9.2 and Idd9.3 protective alleles function in CD4+ T-cells and nonlymphoid cells to prevent expansion of pathogenic islet-specific CD8+ T-cells. Diabetes 2010;59:1478–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernández J, Aung S, Marquardt K, Sherman LA. Uncoupling of proliferative potential and gain of effector function by CD8+ T cells responding to self-antigens. J Exp Med 2002;196:323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trudeau JD, Kelly-Smith C, Verchere CB, et al. Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. J Clin Invest 2003;111:217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granucci F, Vizzardelli C, Pavelka N, et al. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat Immunol 2001;2:882–888 [DOI] [PubMed] [Google Scholar]
- 25.Lieberman SM, Evans AM, Han B, et al. Identification of the β-cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc Natl Acad Sci USA 2003;100:8384–8388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cordell HJ, Todd JA, Hill NJ, et al. Statistical modeling of interlocus interactions in a complex disease: rejection of the multiplicative model of epistasis in type 1 diabetes. Genetics 2001;158:357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stober CB, Brode S, White JK, Popoff JF, Blackwell JM. Slc11a1, formerly Nramp1, is expressed in dendritic cells and influences major histocompatibility complex class II expression and antigen-presenting cell function. Infect Immun 2007;75:5059–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vijayakrishnan L, Slavik JM, Illés Z, et al. An autoimmune disease-associated CTLA-4 splice variant lacking the B7 binding domain signals negatively in T cells. Immunity 2004;20:563–575 [DOI] [PubMed] [Google Scholar]
- 29.Pearce EL, Walsh MC, Cejas PJ, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 2009;460:103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3+ CD25+ CD4+ regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med 2005;201:723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang S, Game DS, Davies D, Lombardi G, Lechler RI. Activated CD1d-restricted natural killer T cells secrete IL-2: innate help for CD4+CD25+ regulatory T cells? Eur J Immunol 2005;35:1193–1200 [DOI] [PubMed] [Google Scholar]
- 32.Yui MA, Sharp LL, Havran WL, Rothenberg EV. Preferential activation of an IL-2 regulatory sequence transgene in TCR-γδ and NKT cells: subset-specific differences in IL-2 regulation. J Immunol 2004;172:4691–4699 [DOI] [PubMed] [Google Scholar]
- 33.Wuest SC, Edwan JH, Martin JF, et al. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat Med 2011;17:604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]