Abstract
Objective
Tacrolimus, an immunosuppressive drug widely prescribed in kidney transplantation, requires therapeutic drug monitoring due to its marked interindividual pharmacokinetic variability and narrow therapeutic index. Previous studies have established that CYP3A5 rs776746 is associated with tacrolimus clearance, blood concentration, and dose requirement. The importance of other drug absorption, distribution, metabolism, and elimination (ADME) gene variants has not been well characterized.
Methods
We used novel DNA biobank and electronic medical record resources to identify ADME variants associated with tacrolimus dose requirement. Broad ADME genotyping was performed on 446 kidney transplant recipients who had been dosed to steady state with tacrolimus. The cohort was obtained from Vanderbilt's DNA biobank, BioVU, which contains linked, de-identified electronic medical record data. Genotyping included Affymetrix DMET Plus (1936 polymorphisms), custom Sequenom MassARRAY iPLEX Gold assay (95 polymorphisms), and ancestry-informative markers. The primary outcome was tacrolimus dose requirement defined as blood concentration-to-dose ratio.
Results
In analyses that adjusted for race and other clinical factors, we replicated the association of tacrolimus blood concentration-to-dose ratio with CYP3A5 rs776746 (p = 7.15 × 10−29), and identified associations with nine variants in linkage disequilibrium with rs776746, including eight CYP3A4 variants. No NR1/2 variants were significantly associated. Age, weight, and hemoglobin were also significantly associated with the outcome. In final models, rs776746 explained 39% of variability in dose requirement, and 46% was explained by the model containing clinical covariates.
Conclusion
This study highlights the utility of DNA biobanks and electronic medical records for tacrolimus pharmacogenomic research.
Keywords: pharmacogenomics, pharmacokinetics, calcineurin inhibitor, tacrolimus, electronic medical records, kidney transplant, cytochrome P4503A5, genetic polymorphism, dosing
Introduction
The immunosuppressive calcineurin inhibitor, tacrolimus, is widely prescribed to kidney transplant recipients [1]. Its use, however, is complicated by marked interindividual pharmacokinetic variability and a narrow therapeutic index [2, 3]. To maximize efficacy and minimize toxicity, dosing is individualized based on monitoring of tacrolimus blood concentrations [4, 5]. Underdosing increases risk for transplant rejection, while overdosing increases risk for nephrotoxicity, hypertension, neurotoxicity, diabetes, and other metabolic derangements [6–9]. Clinical factors reported to affect tacrolimus pharmacokinetics include some concomitant medications, severe hepatic impairment, hematocrit, and serum albumin concentration but do not explain all the variability [10–14].
Previous analyses have demonstrated that tacrolimus disposition is affected by allelic variants in genes involved in drug absorption, distribution, metabolism and elimination (ADME) [15]. Tacrolimus is metabolized primarily by hepatic cytochrome P450 (CYP) 3A4 and 3A5 isoforms, and is a substrate for the multidrug efflux transporter P-glycoprotein (P-gp, now called ABCB1) which is expressed by hepatocytes, renal proximal tubular cells, enterocytes, and various other cells and tissues [16, 17]. The CYP3A isoforms and ABCB1 are polymorphic, [18, 19] and studies suggest that tacrolimus pharmacokinetic variability is affected by variable expression or function of CYP3A isoforms and ABCB1. Several studies have associated a CYP3A5 loss-of-function variant (rs776746, CYP3A5*3 haplotype) with lower tacrolimus clearance, higher blood concentrations, and lower dose requirements [20–22]. Studies of single nucleotide polymorphisms (SNPs) in CYP3A4 and ABCB1 have yielded conflicting results [24, 25].
The predictive value of ADME genes other than CYP3A5 for tacrolimus dose requirement in kidney transplant recipients has not been well characterized. Most published studies to date have considered few genes and SNPs, though recently a group reported results from a broad panel of variants, including almost 700 ADME variants, in a prospective multi-center cohort, again confirming the importance of CYP3A5*3 [26].
A clearer understanding of genetic and clinical factors that affect variability in tacrolimus concentration levels would improve individual tacrolimus dosing and possibly improve clinical outcomes. Previous studies have addressed this question through analysis of cohorts recruited from specific clinics and/or multicenter cohorts. Such an approach is relatively labor and resource-intensive. Here we present an alternative approach using a DNA repository linked to an electronic medical record at a large academic medical center. This approach has the potential advantages of the relatively rapid generation of a cohort and elimination of some biases that could be introduced through inclusion/exclusion criteria, patient willingness to participate, or patient co-morbidities. We assembled a retrospective cohort of kidney transplant patients who had been dosed to steady state with tacrolimus, and broadly assayed ADME gene variants in this cohort to identify variants associated with tacrolimus dose requirement.
Methods
Study Population
Samples were obtained from, BioVU, the DNA biobank for Vanderbilt University Medical Center. A full description of BioVU including its design, collection methods, and ethical considerations have been published previously [27]. Briefly, BioVU accrues DNA samples during routine clinical care from Vanderbilt patients who have not opted out of participation, using blood that would otherwise be discarded after clinical testing. Biological samples from BioVU are linked via anonymous research unique identifiers to the Synthetic Derivative, a de-identified version of Vanderbilt's electronic medical record. Using the Synthetic Derivative, we applied informatics methods to identify initial candidates who met screening criteria, including: 1) kidney transplant documented by ICD9 code V42.0 and CPT code 50360 or 50365; 2) assay results for tacrolimus blood concentrations; and 3) DNA in BioVU. This identified 776 potential candidates. Each subject's Synthetic Derivative chart was then manually reviewed for further inclusion criteria including: 4) documented receipt of tacrolimus-based immunosuppressive therapy; and 5) available tacrolimus dosing data. We excluded individuals with fewer than 3 tacrolimus blood assay results, or miscoded with heart, lung or liver transplant. After manual review, 527 subjects met all criteria and were used for genotyping.
BioVU accrues samples from approximately 800 additional patients per month, and by February 2011 comprised extracted DNA from more than 110,000 different patients. The samples are de-identified in accordance with provisions of Title 45 Code of Federal Regulations part 46 and considered “nonhuman subjects” research. Results of all genetic analysis of BioVU DNA are uploaded to BioVU and are available to investigators for future projects. BioVU has been reviewed at multiple levels, including the Vanderbilt Institutional Review Board, internal and external ethics committees, Community Advisory Board, legal departments, and the federal Office of Human Research Protection [28]. Developed by Vanderbilt's Department of Biomedical Informatics, the Synthetic Derivative is a research-enabled repository of nearly all available clinical data, with content changed by deleting or permutating all personal identifiers [27]. The Synthetic Derivative comprises records from approximately 1.8 million patients, with detailed longitudinal clinical data since the 1990s. Clinical data are updated regularly, to add patients new to Vanderbilt, and new data for existing patients. Data include diagnostic and procedure billing codes (ICD9 and CPT); basic demographics (age, sex, race); text from clinical care notes (e.g. inpatient and outpatient provide documentation, nursing notes, problem lists); laboratory values; inpatient and outpatient medication data; and other diagnostic reports. The ability to link genotype and phenotype through BioVU/Synthetic Derivative is a powerful platform for genetic association studies. Its strength for genetic discovery has been previously demonstrated in a study replicating known associations with five diseases and in a recent genome-wide analysis of atrioventricular conduction [29, 30].
Genotyping and Quality Control
In BioVU, DNA was extracted from whole blood using Autopure (Minneapolis, MN) systems in the Vanderbilt DNA Resources Core. Genotyping was performed primarily with Affymetrix drug-metabolizing enzymes and transporters panel (DMET™ Plus) chips (Affymetrix, Santa Clara, CA), which are designed to assay 1,936 polymorphisms across 225 genes, including 25 polymorphisms in CYP3A4, 14 in CYP3A5, and 40 in ABCB1 [31]. These comprise SNPs, insertion-deletions and copy number variants, and some highly polymorphic ADME genes not well interrogated by many other platforms (e.g. 31 variants in CYP2D6). To more thoroughly interrogate CYP3A4 and pregnane × receptor (P×R, now called NR1/2), which regulates expression of CYP3A4, CYP3A5 and other genes [32, 33], we used the MassARRAY® iPLEX Gold platform (Sequenom, Inc, San Diego, CA) to genotype an additional 95 polymorphisms including 51 across NR1/2 and 40 across CYP3A4 (final assay design available upon request). Our strategy was to use SeattleSNPs [34] to tag the entire NR1/2 and CYP3A4 gene, including 20 kB in each 5' and 3' untranslated region (UTR). We used a cosmopolitan approach across populations (Yoruba, Asian, African American, European-American, and Hispanic) with a 5% allelic frequency cutoff, a 0.80 threshold for r2, 85% data convergence for tagging polymorphisms, and 70% data convergence for clustering. The iPLEX Gold assay also included 3 SNPs in ABCB1 and 1 in CYP2C19. Eight variants shared between platforms were used to check for concordance (supplemental table of SNPs). For one tri-allelic SNP we could not model additive genetic effects, so we split it into three binary indicator variables; one variable indicated, for each chromosome, any variation from the major allele, while the other two indicated the particular minor allele variant in the individual. In addition to DMET Plus and iPLEX Gold, most subjects had ancestry informative markers (AIMs) available in BioVU from a separate project to examine population substructure among 1910 BioVU subjects. The AIMs were generated with the Illumina DNA Test Panel (Illumina, San Diego, CA) which assays 360 ancestry informative SNPs. Using AIMs data, principal components were generated using EIGENSTRAT [35].
Initially, 13 samples were excluded due to potential sex inconsistencies based on X chromosome variants, leaving 514 samples for ADME genotyping of 2,025 polymorphisms. The 2025 polymorphisms noted here include the 3 binary variables resulting from splitting the triallelic marker. We also excluded polymorphisms with less than 1% minor allele frequency or less than 95% genotyping efficiency, and subjects who were outliers in principal component analysis with AIMs.
Phenotyping
Clinical information was extracted from the Synthetic Derivative. The primary phenotype of interest was the tacrolimus dose requirement defined by concentration-to-dose ratio (C/D ratio). This was calculated as tacrolimus blood concentration in ng/mL divided by total daily dose in mg. Whole blood tacrolimus concentrations at trough were obtained during therapeutic drug monitoring at kidney transplant clinic visits, emergency department visits, and hospitalizations. Before February 2009 tacrolimus assays were by microparticulate enzyme immunoassay (MEIA, Abbott IMx Analyzer, Abbott Laboratories, Abbott Park, IL), and thereafter were by chemiluminescent microparticle assay (CMIA, Abbott ARCHITECT i2000, Abbott Laboratories, Abbott Park, IL). The correlation coefficient between MEIA and CMIA in the Vanderbilt Clinical Laboratory is 0.97, with 95% and 5% of the cohort tacrolimus levels measured by MEIA and CMIA, respectively. For each subject, the first 10 available tacrolimus blood concentration measurements beyond the first month of transplantation were obtained from a structured laboratory assay table in the Synthetic Derivative. For subjects with fewer than 10 results, all available measurements were used. Tacrolimus measurements during the first month were not used since that is an active time of dose titration to find the correct dose for target drug levels. Tacrolimus dosing information corresponding to each tacrolimus concentration date was collected from clinical notes using MedEx, a natural language processing tool that extracts drug names and dosing information such as dose and frequency from clinical notes [36]. An additional MedEx module was developed to calculate total daily tacrolimus dose [37]. For each tacrolimus concentration, MedEx dose data were verified and corrected as necessary by manual review of clinical data in the Synthetic Derivative by a trained nephrologist. Although we had access to every available tacrolimus measurement in every patient, to increase the quality of our primary outcome variable we focused on 10 measurements so that we could manually curate and validate every tacrolimus dose and confirm every trough time. Additional clinical information abstracted included sex, age, race (based on third party report), weight, days since transplant, and the laboratory measures hemoglobin and albumin. All covariates were selected a priori based on clinical considerations.
Statistical Analysis
Genotyping quality control (QC)
Polymorphisms with less than 1% minor allele frequency or less than 95% genotyping efficiency were excluded from the final analysis.
Population substructure adjustment
From 360 ancestry informative SNPs in the AIMs data, principal components were generated using a specialized software EIGENSTRAT which was developed to detect and adjust for population structure [35]. Without testing significance of principal components, a priori 10 principal components were selected to be included in the analyses to adjust for population substructure. Separate regression analysis was performed using the third party reported race indicator variables from the Synthetic Derivative, as an alternative method for population substructure adjustment. Sensitivity analysis was performed to evaluate different adjustment methods for population structure, and the number of principal components. The results were not sensitive to the choice of the methods or the number of principal components, and hence all 10 principal components were included in the final genetic association analysis models for adjustment of population substructure.
Statistical models for genetic association analysis
We used two approaches to examine genetic associations with tacrolimus C/D ratios; (1) multiple linear regression analysis using a summary measurement per subject as an outcome; (2) longitudinal data analyses (LDA) using all tacrolimus C/D ratios repeatedly measured within each subject. For the first approach, a summary outcome for each subject was derived based on the median of as many as 10 tacrolimus C/D ratio values. Medians of the corresponding continuous covariates (subject weight, days post transplant, albumin, hemoglobin, age) collected on the same date as the tacrolimus values were matched with the median of tacrolimus C/D ratio. Multiple linear regression analyses using the medians were performed for each genotype one-by-one after adjusting for the matched covariates as well as sex and population substructure described above. From this medians analysis we estimated percent variability in the outcome explained by the genetic variant alone, or the genetic variant together with all covariates. Sensitivity analysis was performed to evaluate different adjustment methods for population structure and the number of principal components using this analysis.
For LDA (which we consider the primary analysis), tacrolimus C/D ratios repeatedly measured over time within each subject were analyzed using mixed-effects models to take into account correlation due to repeated measurements in the same subject. The same covariates selected a priori (i.e. weight, days post transplant, sex, age, albumin, hemoglobin, and population substructure) were included as fixed effects. To adjust for nonlinear time trend for days post transplant and relationships with albumin and subject weight, restricted cubic spline functions with 3 knots were used [38]. To test additive effects of each genotype on the outcome, each SNP from the final set of SNPs was included one-by-one as the main fixed effect. This type of modeling was used to address the possibility of collinearity due to strong linkage disequilibrium, or of smaller effects being obscured by the large effect of a single polymorphism. Gene-gene interactions were not modeled in this analysis. The model included a random intercept and random slopes for albumin and hemoglobin, considering that each subject appeared to have their own slopes representing relationships between the outcome and albumin and hemoglobin. For correlation structure, an autoregressive-moving average model with 2 autoregressive and 1 moving average parameters was used for all SNPs except for 6 for which the model did not converge. For these 6 SNPs, a simpler autoregressive-moving average model with 1 autoregressive and 1 moving average parameters was used to insure the convergence of model fit.
To reduce skewness of the distribution, all outcomes were log-transformed in both approaches. Based on 1,000 independent tests of association, an unadjusted p value less than 1×10−5 was considered significant for each genetic association. Analyses were performed using the programming language R version 2.13.1 [39] and PLINK [40].
Results
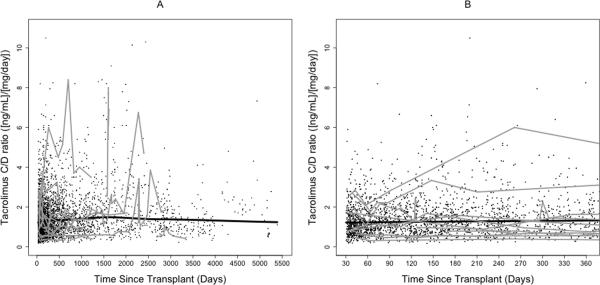
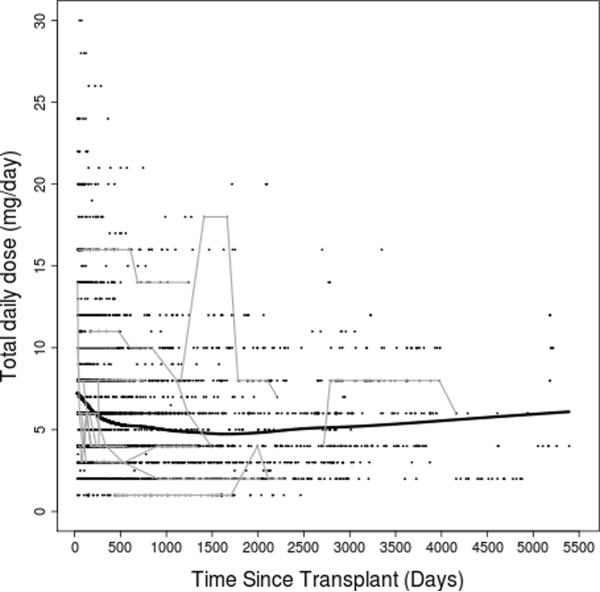
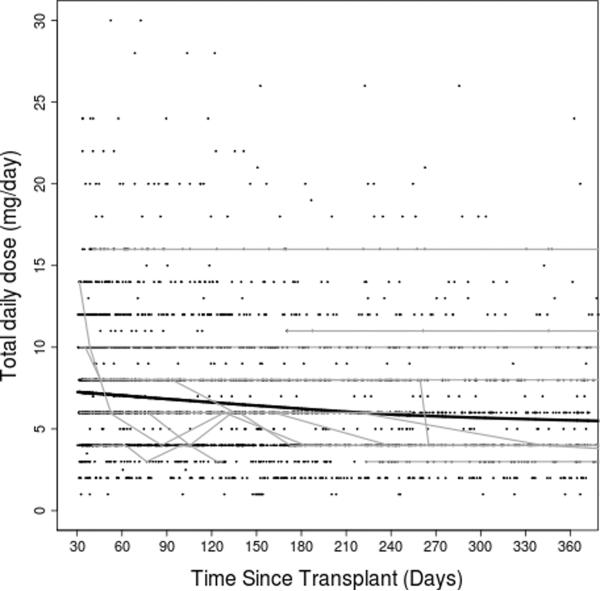
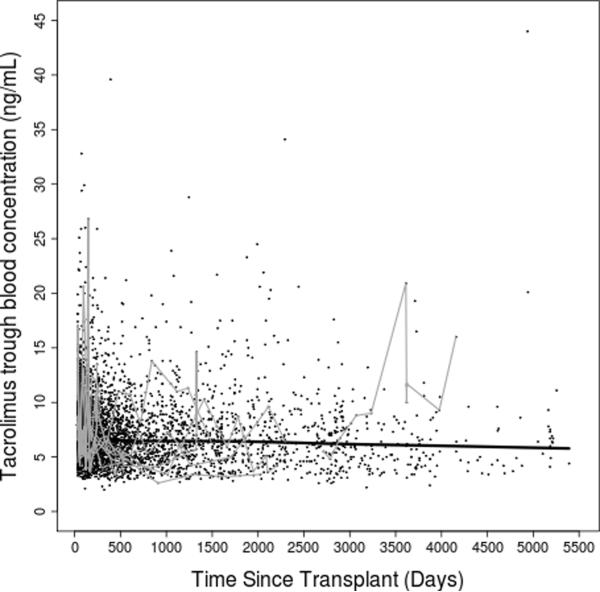
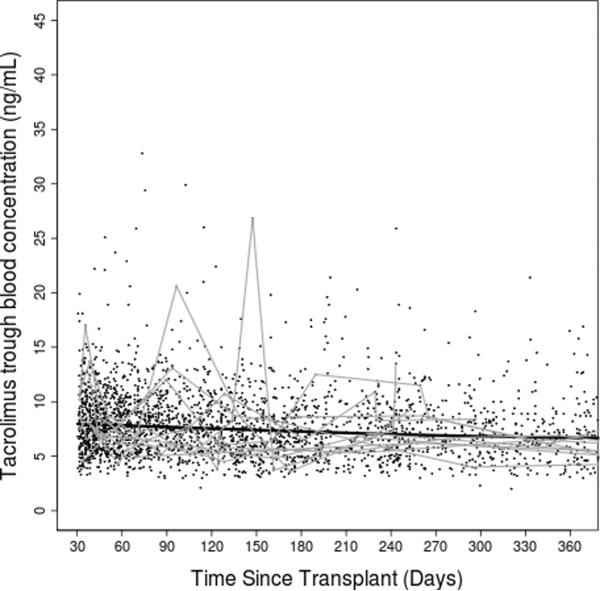
After clinical eligibility criteria validation and genotyping quality control (QC), a total of 446 individuals had evaluable results from both DMET Plus and iPlex Gold genotyping platforms. Of these 446 individuals, 399 also had available AIMs data (Figure 1). Of 2025 polymorphisms potentially assayed by DMET Plus and iPlex Gold, 936 remained after QC in the sample of 446 individuals. In the subset with AIMS data, 929 polymorphisms remained after QC (see Supplementary Table 1). The major reason for excluding polymorphisms was low minor allele frequency, with 1064 present at less than 1%, of which 855 were monomorphic. By pruning for r2 of ≥ 0.8 pair-wise for markers, 747 of the 929 polymorphisms remained. Of 360 AIMs assayed, 313 AIMs passed QC and were used to generate principal components. Following QC, sample genotyping rate exceeded 99%. For eight polymorphisms assayed both by DMET Plus and iPLEX Gold assay, results were 99.7% concordant. Demographic and clinical characteristics of the cohort are shown in Table 1. A total of 3731 tacrolimus levels for 399 subjects were obtained with a median of 10 per subject. The median tacrolimus C/D ratio was 1.3 (ng/mL)/mg, and the median of the medians (the individual summary measure) of time since transplant was 270 days (range: 34 to 5197; interquartile range: 118 to 834), with a highly skewed distribution due to some subjects with extremely long follow up time. Raw tacrolimus C/D ratios in the study cohort as a whole increased slightly from 30 days to 1 year post transplant, and were remarkably stable thereafter (Figure 2). By third party report, approximately three-fourths of subjects were white and one-fourth black.
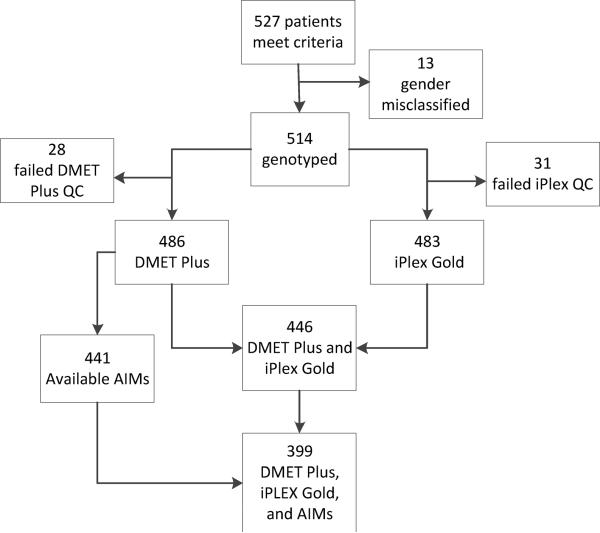
Figure 1. Derivation of the study cohort.
After applying inclusion criteria to the Synthetic Derivative, 527 subject samples were eligible for genotyping. Of these, 13 were removed for sex misclassification, 28 after DMET Plus quality control (QC), and 31 after iPlex Gold QC.
Table 1.
Selected Demographic and Clinical Characteristics of the Kidney Transplant Cohort
| Characteristic | N = 399 |
|---|---|
| Age (years) | 46 ± 13 |
| Sex: | |
| Male | 245 (61.4%) |
| Female | 154 (38.6%) |
| Race: | |
| White | 303 (75.9%) |
| Black | 84 (21.1%) |
| Other | 12 (3%) |
| Weight (kg)a | 83 ± 21 |
| Hemoglobin (g/dL)a | 12.6 ± 1.9 |
| Albumin (g/dL)a | 4.1 ± 0.4 |
| Time since transplant (days)b | 270 (118 – 834) |
| Tacrolimus C/D ratio ([ng/mL]/mg)a | 1.5 ± 0.9 |
| Tacrolimus trough blood concentration (ng/mL)a | 7.3 ± 2.1 |
| Total daily dose (mg/day)a | 6.5 ± 3.8 |
Note: All data are number (%) of subjects, mean ± standard deviation, or median (interquartile range). C/D ratio = normalized concentration-to-dose ratio.
For repeatedly measured variables, medians matched with tacrolimus C/D ratios were calculated for each patient, and then the means and SDs were estimated from all patients' median values.
For time since transplant, medians matched with tacrolimus C/D ratios were calculated for each patient, and then the median (interquartile range) was estimated from all patients' median values. The median and the interquartile range were provided to better represent the distribution of time since transplant due to high skewedness of the distribution.
Figure 2. Tacrolimus dose-adjusted trough concentrations over time.
(A) entire study period; (B) first year post transplant. Thick black lines represent smoothing lines fitted by the locally-weighted polynomial regression of the raw tacrolimus C/D ratio measurements (black dots) on time since transplant. Thin gray lines represent data from 15 randomly chosen subjects.
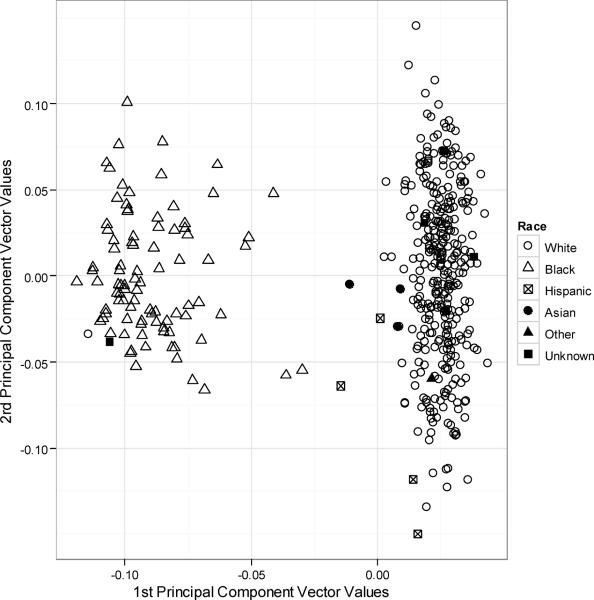
To assess for population stratification using genetic markers, AIMs data was used for principal components analysis without considering ADME genotypes. The principal components analysis identified two clusters corresponding to whites and blacks (Figure 3). The x and y values in Figure 2 correspond to values of the first and second principal component vectors, respectively. The first principal component describes variation on the axis from European to African descent while the second component vector describes variation within those of European descent. Due to admixture of European and African ancestry, the cluster comprising third-party report black individuals has more heterogeneity in principal component values reflecting greater genetic variability. In a separate analysis we used third party reporting of race, rather than principal components, to account for population differences. In sensitivity analyses, adjusting population substructure using 10 principal components, 1 principal component, or race indicator variables yielded similar results; we present the results from 10 principal components in 399 subjects.
Figure 3. Principal component analysis for race.
Using EIGENSTRAT to generate principal components, the cohort split into 2 clusters that correspond to black and white based on third party report. The x and y values correspond to values of the first and second principal component vectors, respectively, as derived by EIGENSTRAT's smartPCA program.
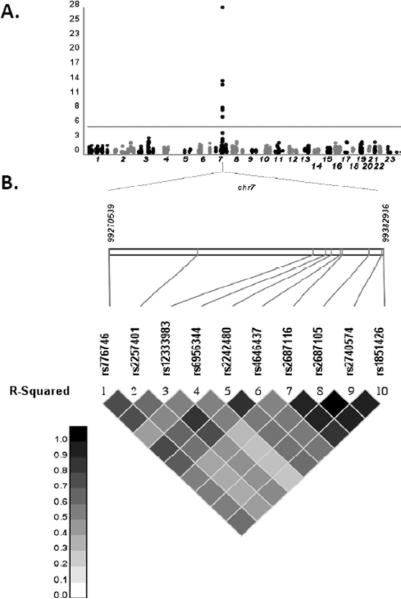
Results from the primary analysis (LDA) for association between genetic variants and tacrolimus C/D ratios are shown in Figure 4. (Medians analysis identified the same top 10 SNPs as LDA, data not shown). Only chromosome 7 variants were significantly associated with tacrolimus C/D ratios (Figure 3A). In analyses adjusted for age, sex, weight, hemoglobin, albumin, time since transplant, and first 10 principal components, the polymorphism most strongly associated with tacrolimus C/D ratio was the CYP3A5*3 loss-of-function SNP, rs776746 (p = 7.15 × 10−29, Table 2). The presence of CYP3A5*3 was associated with higher tacrolimus C/D ratios, with each G allele (*3) associated with an increase of 0.54 in log-transformed tacrolimus C/D ratio for subjects with the average of random effects. The allelic frequency of rs776746 (*3) was 0.92 among whites and 0.30 among blacks. Eight additional variants in CYP3A4 and one in CYP3A7 also exceeded the significance threshold for association with tacrolimus C/D ratios (Table 2). These other nine CYP3A4 and CYP3A7 variants were in linkage disequilibrium (LD) with rs776746 (Figure 3B).
Figure 4. Genotype association with tacrolimus dose-adjusted trough concentrations by multiple linear regression analysis.
(A) Manhattan plot of all evaluable ADME SNPs. The – log10 of the p value is shown for genotyped SNPs (circles); (B) Synthesis-View Plot [51]: Schematic of chromosome 7 region of interest. Linkage disequilibrium structure is shown.
Table 2.
Polymorphisms significantly associated with tacrolimus log-transformed, normalized concentration-dose ratio (adjusted analysis). All SNPs located on chromosome 7.
| SNP | Gene | Associated Allele | Reference Allele | Coefficienta | P value | Allelic Frequency in Whites | Allelic Frequency in Blacks |
|---|---|---|---|---|---|---|---|
| rs776746 | CYP3A5 | G | A | 0.54 | 7.15 × 10−29 | 0.92 | 0.30 |
| rs4646437 | CYP3A4 | C | T | 0.35 | 1.05 × 10−14 | 0.88 | 0.29 |
| rs2257401 | CYP3A7 | C | G | 0.37 | 1.08 × 10−14 | 0.91 | 0.48 |
| rs2242480 | CYP3A4 | C | T | 0.34 | 5.22 × 10−14 | 0.89 | 0.26 |
| rs12333983 | CYP3A4 | T | A | 0.32 | 7.35 × 10−14 | 0.88 | 0.32 |
| rs2687105 | CYP3A4 | A | T | 0.36 | 1.83 × 10−09 | 0.96 | 0.33 |
| rs6956344 | CYP3A4 | C | T | 0.29 | 3.34 × 10−09 | 0.91 | 0.67 |
| rs2740574 | CYP3A4 | A | G | 0.34 | 6.44 × 10−09 | 0.96 | 0.34 |
| rs1851426 | CYP3A4 | C | T | 0.33 | 9.35 × 10−08 | 0.96 | 0.27 |
| rs2687116 | CYP3A4 | T | G | 0.30 | 1.61 × 10−07 | 0.96 | 0.39 |
Adjusted for age, sex, weight, hemoglobin, albumin, time since transplant, and first 10 principal components from EIGENSTRAT.
Results for fixed effects in the final LDA model for CYP3A5*3 are presented in Table 3. Of the other pre-selected clinical covariates, overall effects of age, weight, hemoglobin albumin, and days post transplant on tacrolimus C/D ratio were each significant at p < 0.05. Increasing values of albumin and weight were each associated with a decrease in the log-transformed tacrolimus C/D ratio whereas age was associated with an increase. The medians analyses showed that the single predictor of CYP3A5*3 alone explained 39% of the variability and the final medians linear regression model with all the covariates including CYP3A5*3 explained 46% of the variability in log-transformed tacrolimus C/D ratio in the 399 subjects.
Table 3.
Results for fixed effects in the mixed-effects model for CYP3A5*3 in the longitudinal data analysis for log-transformed tacrolimus C/D ratio.
| Covariate | Coefficienta (S.E.) | P value |
|---|---|---|
| CYP3A5*3 | 0.54 (0.044) | 7.15 × 10−29 |
|
| ||
| Albuminb (x) | −0.17 (0.04) | 2.07 × 10−5 |
| (x') | 0.12 (0.04) | 0.005 |
|
| ||
| Age | 0.007 (0.002) | 1.80 × 10−5 |
|
| ||
| Weightb (x) | −0.007 (0.002) | 3.24 × 10−4 |
| (x') | 0.006 (0.002) | 0.004 |
|
| ||
| Hemoglobin | 0.015 (0.006) | 0.012 |
|
| ||
| Days post transplantb (x) | 0.0004 (6.2 × 10−5) | 2.97 × 10−9 |
| (x') | −0.0012 (2.5 × 10−4) | 2.44 × 10−6 |
|
| ||
| Sex (Male vs. Female) | 0.034 (0.043) | 0.431 |
|
| ||
| Principal components (PCs) | ||
| PC1 | −0.85 (0.59) | 0.155 |
| PC2 | 0.28 (0.40) | 0.487 |
| PC3 | −0.34 (0.40) | 0.404 |
| PC4 | −0.25 (0.40) | 0.535 |
| PC5 | −0.20 (0.40) | 0.616 |
| PC6 | −0.74 (0.40) | 0.064 |
| PC7 | 0.09 (0.40) | 0.816 |
| PC8 | −0.27 (0.40) | 0.504 |
| PC9 | 0.57 (0.40) | 0.157 |
| PC10 | 0.48 (0.40) | 0.228 |
For coefficients with a positive value, the covariate results in an increase in log-transformed tacrolimus C/D ratio, whereas a negative values results in a decrease in log-transformed tacrolimus C/D ratio.
Restricted cubic spline functions with 3 knots were used to model nonlinear time trend and relationships, and x and x' are the corresponding linear and nonlinear terms.
Discussion
The present study is the most comprehensive report to date of the ability of ADME variants to predict tacrolimus dose requirement. Using the novel genomics resource, BioVU, and its complementary informatics resource, the Synthetic Derivative, we readily replicated the previously described association between the rs776746 SNP in CYP3A5 and tacrolimus C/D ratio in a large cohort of “real-world” kidney transplant recipients. In an adjusted linear regression model, rs776746 explained 39% of interindividual variability in tacrolimus C/D ratio. Nine other chromosome 7 polymorphisms that were less strongly associated with tacrolimus C/D ratio were in LD with rs776746. Our broad ADME genotyping strategy with DMET Plus complemented by iPLEX Gold found no additional ADME variants significantly associated with tacrolimus C/D ratio. This included no significant associations with variants in CYP3A4 (other than those in LD with rs776746) or NR1/2 variants, which were particularly targeted for genotyping.
By replicating the known association between CYP3A5*3 and tacrolimus C/D ratio in our cohort, we highlight the ability of a DNA biobank linked to de-identified clinical electronic medical record data to reliably detect strong genotype-phenotype associations. The ready availability of well-documented longitudinal clinical information from the Synthetic Derivative should allow genetic association studies to be conceived and completed quickly and efficiently. These novel resources also provide a unique platform to execute broader GWAS.
Potential limitations of the Synthetic Derivative as a data source may include imprecision in classifying cases and controls, and erroneous or incomplete information for some phenotypes of interest. These limitations are largely offset by the availability of extensive longitudinal data entered by multiple care providers. In addition, the Synthetic Derivative is made robust by its data being derived from multiple complementary sources including clinic notes, admission and discharge summaries, inpatient daily and consult notes, medication orders, laboratory values, radiology studies, and procedure reports.
The CYP3A5 rs776746 variant (6986A→G, where A = *1 and G = *3) causes a splicing defect that yields a truncated, nonfunctional protein. Among subjects homozygous for this CYP3A5*3 allele, CYP3A5 comprises only 5% of hepatic CYP3A expression, compared to as much as 50% among individuals carrying at least one copy of the CYP3A5*1 reference allele [41]. Our study confirmed previous studies that showed a strong association between CYP3A5 and tacrolimus dose requirement, with CYP3A5*1 predicting more rapid tacrolimus clearance, lower blood concentrations, and higher total daily doses than CYP3A5*3/*3 [20–22, 24, 25]. A step toward translation into clinical practice has been undertaken, with a recent randomized trial in France comparing CYP3A5 genotype-based tacrolimus dosing to standard of care weight-based dosing in 280 new kidney transplant patients. Those with genotype-guided dosing were more likely to achieve target tacrolimus blood concentrations within 3 days after initiating tacrolimus [42]. Next steps include testing if dosing by genotype improves clinical outcomes that affect long-term graft survival, including acute rejection and calcineurin inhibitor toxicity.
Since CYP3A5 does not fully explain variability in tacrolimus dose-corrected blood concentrations, and overlap in these measurements occurs among the different CYP3A5 genotype groups, discovery of additional ADME genetic variants might better predict tacrolimus C/D ratio. In an effort to identify other relevant ADME variants, we used a broad approach to ADME genotyping complemented by more detailed genotyping of CYP3A4 and NR1/2, examining more ADME variants than have been considered in previous studies. These latter genes were targeted based on their putative role in tacrolimus metabolism, although results of previous studies to establish their importance in tacrolimus disposition have been inconsistent [24]. We also examined 16 variants of ABCB1 based on possible associations with tacrolimus disposition in previous studies [24, 25]. Other than nine variants in LD with rs776746, no additional ADME variants were significantly associated with C/D ratio. All nine variants were also on chromosome 7, one in CYP3A7 and the rest in CYP3A4. These variants included rs2257401 in CYP3A7, and rs2740574 (CYP3A4*1B), a CYP3A4 promoter variant whose previously reported associations with tacrolimus phenotypes were likely mediated by its LD with rs776746 [18, 25, 43]. The other seven variants in CYP3A4 have not been reported as clinically important in drug metabolism [44].
We had some limitations in obtaining our tacrolimus data. Unlike most previous studies that focused on tacrolimus blood concentrations obtained early in transplant (first few weeks to months), our unique resources allowed us to consider all concentrations over many years, summing to over 16,000 possible measurements. Although the laboratory data were excellent, the dosing data abstracted was less robust. In an effort to balance these electronic resources with high quality data, we decided to limit the tacrolimus blood concentrations to the first ten available after the first month post transplant for each patient. We then could manually confirm the dosing information for each concentration. This resulted in the median time post transplant for the dose-corrected blood concentrations as 9 months, somewhat later than in most previously studied cohorts. Although individual patient's tacrolimus C/D was from varying timepoints post transplant, which might be considered a limit, it also shows the ongoing prominence of rs776746 over time in a “real-world” kidney transplant population, making our results more generalizable
Our overall findings are consistent with a recent multi-center study by Jacobson et al., which also examined the association of genetic variants with tacrolimus dose requirement in a prospective study 695 kidney transplant patients [26]. In that study, tacrolimus trough concentrations were collected in the first 6 months post transplant. Both cohorts had a similar proportion of black participants. Our study focused exclusively on ADME variants with 2025 polymorphisms evaluated, whereas the Jacobson study examined variants in pathways associated with immunity, cell cycle, signaling, inflammation, and other pathways in addition to about 700 ADME variants. The clinical factors used in their final model were selected statistically, while in the present study we selected them a priori based on clinical relevance. They included covariates of diabetes and use of calcium channel blockers, which we did not abstract, though we included data on hemoglobin and albumin which was not part of their study, When considering all trough concentrations from the first 6 months, they also replicated the association of rs776746 with tacrolimus dose requirement. Like our study, they found several other variants in LD with rs776746 but no additional variants were significant when controlling for a false discovery rate of 20%. When considering only the first tacrolimus trough post transplant, they found eight other significant variants in addition to CYP3A5 rs776746, including CYP3A4 rs2687117. Our similar results underscore the use of a DNA biobank coupled with clinical information as a potential reliable alternative to more time and labor intensive cohort studies.
A strength of our study was inclusion of both whites and blacks in the cohort, with 21% of the cohort black, making our results generalizable to these two populations. Our results may not be as applicable to other populations (e.g. Asians). Previous non-genetic studies identified large racial differences in tacrolimus dosing requirement, with individuals of African descent requiring highest daily doses [45, 46]. This has been attributed to increased frequency of CYP3A5*1 among individuals of African descent.[23] We therefore rigorously accounted for ancestry, which could have confounded at least some previous genetic association studies. Population substructure was assessed using AIMs as well as third party report. By principal components analysis with AIMs, the black cluster was more heterogeneous than the white cluster, consistent with the greater genetic diversity among people of African descent [47]. After adjusting for race, rs776746 remained a highly significant predictor of tacrolimus C/D ratio, indicating a genetic effect independent of ancestry. Analyses that adjusted for race (based on third party report) yielded results consistent with those using AIMs data, indicating that BioVU third party reporting of race is sufficient for many association analyses. Indeed, a prior BioVU study using genetically determined ancestry from AIMs data showed that third-party reporting of race is nearly identical to that of self-report for European Americans and African Americans [48].
In our primary analysis adjusting for ancestry using principal components, we used the top 10 principal component vectors from AIMs. In univariable regression analysis with log-transformed tacrolimus C/D ratio, only the top vector was significant. However, because we predetermined that 10 principal components would be used, and because their significance was judged using the same data as for the ADME association analysis, it would have been statistically inappropriate to consider only the top vector in the final multiple regression model. This highlights the importance of determining a priori how many vectors to generate based upon knowledge of population substructure of the cohort (e.g. the number of race/ethnicity groups represented) as an alternative to using the study outcome to gauge significance of each vector and to include only a subset based on these results. We can see from Table 3 that ancestry as described through principal components ceases to be highly significant when rs776746 is included in the model. This is indicative of the relationship between race and CYP3A5 genotype where those of African descent have a much higher frequency of the CYP3A5*1 allele whereas those of European descent have a high frequency of the CYP3A5*3 allele.
In addition to genotype, the clinical variables age, weight, days post transplant, albumin, and hemoglobin were associated with tacrolimus C/D ratios. Since this was a retrospective cohort with tacrolimus levels obtained at different times post transplant, we attempted to account for this by adding time from transplant as a covariate in our model. This is important since tacrolimus dose-corrected exposure has been reported to increase during the first 12 months post transplant [49]. Indeed, our results showed tacrolimus C/D increases significantly initially post transplant and starts decreasing at later time. Thus, in our LDA analysis, we adjusted for linear and nonlinear time trend of tacrolimus C/D. Together, genetic and clinical variables explained approximately one-half of interindividual variability in tacrolimus C/D ratio. In a previous study, Wang et al. showed that among kidney transplant patients, a predictive model that included just age, race and concomitant use of medications that inhibit CYP3A explained approximately 30% of variability in dosing, which increased to 58% by including genotype in the model [50]. Both the present study and that of Wang et al. demonstrate increased predictive accuracy when including genetic variants.
Ongoing research in transplant pharmacogenomics may further delineate how ADME variants affect not only drug disposition but also clinical outcomes such as acute rejection and nephrotoxicity. If successful, pharmacogenomic approaches may ultimately improve long-term graft function. Future studies should also examine whether gene-gene interactions among frequent genetic variants, and/or main effects of rare variants affect tacrolimus dose requirement or outcomes.
Supplementary Material
Figure 5.
Figure 6.
Figure 7.
Figure 8.
Acknowledgements
The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center's BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant 1UL1RR024975-01 from NCRR/NIH. Genotyping experiments were performed in the Vanderbilt Center for Human Genetics Research DNA Resources Core and the Vanderbilt Functional Genomics Shared Resource. The Vanderbilt Functional Genomics Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Digestive Disease Center (P30 DK58404) and the Vanderbilt Vision Center (P30 EY08126). Statistical analysis was supported by the Vanderbilt CTSA and Vanderbilt Electronic Systems for Pharmacogenomic Assessment (VESPA) grant RC2 GM092618. Additional grant support included R01 AI077505 (Haas), the Vanderbilt Physician Scientist Development Program (Birdwell), and R21 AG034412 (Choi).
Dr. Marylyn Ritchie is now in the Department of Biochemistry and Molecular Biology at The Pennsylvania State University, but all work for this manuscript was performed while she was at Vanderbilt University.
Support: The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center's BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant 1UL1RR024975-01 from NCRR/NIH. Genotyping experiments were performed in the Vanderbilt Center for Human Genetics Research DNA Resources Core and the Vanderbilt Functional Genomics Shared Resource. The Vanderbilt Functional Genomics Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Digestive Disease Center (P30 DK58404) and the Vanderbilt Vision Center (P30 EY08126). Statistical analysis was supported by the Vanderbilt CTSA and Vanderbilt Electronic Systems for Pharmacogenomic Assessment (VESPA) grant RC2 GM092618. Additional grant support included R01 AI077505 (Haas), the Vanderbilt Physician Scientist Development Program (Birdwell), and R21 AG034412 (Choi).
Footnotes
Disclaimers: Dr. Haas has received research grants from Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead Sciences, and Merck. Drs. Haas and Ritchie have been consultants for Boehringer-Ingelheim.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Collins AJ, Foley RN, Herzog C, Chavers B, Gilbertson D, Ishani A, et al. United States Renal Data System 2008 Annual Data Report. Am J Kidney Dis. 2009;53:S1–374. doi: 10.1053/j.ajkd.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Staatz CE, Tett SE. Clinical pharmacokinetics and pharmacodynamics of tacrolimus in solid organ transplantation. Clin Pharmacokinet. 2004;43:623–653. doi: 10.2165/00003088-200443100-00001. [DOI] [PubMed] [Google Scholar]
- 3.Venkataramanan R, Swaminathan A, Prasad T, Jain A, Zuckerman S, Warty V, et al. Clinical pharmacokinetics of tacrolimus. Clin Pharmacokinet. 1995;29:404–430. doi: 10.2165/00003088-199529060-00003. [DOI] [PubMed] [Google Scholar]
- 4.Tsunoda SM, Aweeka FT. The use of therapeutic drug monitoring to optimise immunosuppressive therapy. Clin Pharmacokinet. 1996;30:107–140. doi: 10.2165/00003088-199630020-00003. [DOI] [PubMed] [Google Scholar]
- 5.Wallemacq P, Armstrong VW, Brunet M, Haufroid V, Holt DW, Johnston A, et al. Opportunities to optimize tacrolimus therapy in solid organ transplantation: report of the European consensus conference. Ther Drug Monit. 2009;31:139–152. doi: 10.1097/FTD.0b013e318198d092. [DOI] [PubMed] [Google Scholar]
- 6.Bechstein WO. Neurotoxicity of calcineurin inhibitors: impact and clinical management. Transpl Int. 2000;13:313–326. doi: 10.1007/s001470050708. [DOI] [PubMed] [Google Scholar]
- 7.Morales JM, Andres A, Rengel M, Rodicio JL. Influence of cyclosporin, tacrolimus and rapamycin on renal function and arterial hypertension after renal transplantation. Nephrol Dial Transplant. 2001;16(Suppl 1):121–124. doi: 10.1093/ndt/16.suppl_1.121. [DOI] [PubMed] [Google Scholar]
- 8.Kramer BK, Zulke C, Kammerl MC, Schmidt C, Hengstenberg C, Fischereder M, et al. Cardiovascular risk factors and estimated risk for CAD in a randomized trial comparing calcineurin inhibitors in renal transplantation. Am J Transplant. 2003;3:982–987. doi: 10.1034/j.1600-6143.2003.00156.x. [DOI] [PubMed] [Google Scholar]
- 9.Nankivell BJ, Borrows RJ, Fung CL, O'Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349:2326–2333. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 10.Undre NA, Schafer A. Factors affecting the pharmacokinetics of tacrolimus in the first year after renal transplantation. European Tacrolimus Multicentre Renal Study Group. Transplant Proc. 1998;30:1261–1263. doi: 10.1016/s0041-1345(98)00234-6. [DOI] [PubMed] [Google Scholar]
- 11.Armendariz Y, Garcia S, Lopez RM, Pou L. Hematocrit influences immunoassay performance for the measurement of tacrolimus in whole blood. Ther Drug Monit. 2005;27:766–769. doi: 10.1097/01.ftd.0000185769.36878.00. [DOI] [PubMed] [Google Scholar]
- 12.Brown NW, Gonde CE, Adams JE, Tredger JM. Low hematocrit and serum albumin concentrations underlie the overestimation of tacrolimus concentrations by microparticle enzyme immunoassay versus liquid chromatography-tandem mass spectrometry. Clin Chem. 2005;51:586–592. doi: 10.1373/clinchem.2004.043950. [DOI] [PubMed] [Google Scholar]
- 13.van Gelder T. Drug interactions with tacrolimus. Drug Saf. 2002;25:707–712. doi: 10.2165/00002018-200225100-00003. [DOI] [PubMed] [Google Scholar]
- 14.Christians U, Jacobsen W, Benet LZ, Lampen A. Mechanisms of clinically relevant drug interactions associated with tacrolimus. Clin Pharmacokinet. 2002;41:813–851. doi: 10.2165/00003088-200241110-00003. [DOI] [PubMed] [Google Scholar]
- 15.Moller A, Iwasaki K, Kawamura A, Teramura Y, Shiraga T, Hata T, et al. The disposition of 14C-labeled tacrolimus after intravenous and oral administration in healthy human subjects. Drug Metab Dispos. 1999;27:633–636. [PubMed] [Google Scholar]
- 16.de Jonge H, Kuypers DR. Pharmacogenetics in solid organ transplantation: current status and future directions. Transplant Rev (Orlando) 2008;22:6–20. doi: 10.1016/j.trre.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Vincent SH, Karanam BV, Painter SK, Chiu SH. In vitro metabolism of FK-506 in rat, rabbit, and human liver microsomes: identification of a major metabolite and of cytochrome P450 3A as the major enzymes responsible for its metabolism. Arch Biochem Biophys. 1992;294:454–460. doi: 10.1016/0003-9861(92)90711-5. [DOI] [PubMed] [Google Scholar]
- 18.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 19.Xie HG, Wood AJ, Kim RB, Stein CM, Wilkinson GR. Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics. 2004;5:243–272. doi: 10.1517/phgs.5.3.243.29833. [DOI] [PubMed] [Google Scholar]
- 20.de Jonge H, Naesens M, Kuypers DR. New insights into the pharmacokinetics and pharmacodynamics of the calcineurin inhibitors and mycophenolic acid: possible consequences for therapeutic drug monitoring in solid organ transplantation. Ther Drug Monit. 2009;31:416–435. doi: 10.1097/FTD.0b013e3181aa36cd. [DOI] [PubMed] [Google Scholar]
- 21.Haufroid V, Mourad M, Van Kerckhove V, Wawrzyniak J, De Meyer M, Eddour DC, et al. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics. 2004;14:147–154. doi: 10.1097/00008571-200403000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Thervet E, Anglicheau D, King B, Schlageter MH, Cassinat B, Beaune P, et al. Impact of cytochrome p450 3A5 genetic polymorphism on tacrolimus doses and concentration-to-dose ratio in renal transplant recipients. Transplantation. 2003;76:1233–1235. doi: 10.1097/01.TP.0000090753.99170.89. [DOI] [PubMed] [Google Scholar]
- 23.MacPhee IA, Holt DW. A pharmacogenetic strategy for immunosuppression based on the CYP3A5 genotype. Transplantation. 2008;85:163–165. doi: 10.1097/TP.0b013e3181609054. [DOI] [PubMed] [Google Scholar]
- 24.Hesselink DA, van Schaik RH, van der Heiden IP, van der Werf M, Gregoor PJ, Lindemans J, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74:245–254. doi: 10.1016/S0009-9236(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 25.Macphee IA, Fredericks S, Tai T, Syrris P, Carter ND, Johnston A, et al. Tacrolimus pharmacogenetics: polymorphisms associated with expression of cytochrome p4503A5 and P-glycoprotein correlate with dose requirement. Transplantation. 2002;74:1486–1489. doi: 10.1097/00007890-200212150-00002. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson PA, Oetting WS, Brearley AM, Leduc R, Guan W, Schladt D, et al. Novel Polymorphisms Associated With Tacrolimus Trough Concentrations: Results From a Multicenter Kidney Transplant Consortium. Transplantation. 2011 doi: 10.1097/TP.0b013e318200e991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci. 2010;3:42–48. doi: 10.1111/j.1752-8062.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deeken J. The Affymetrix DMET platform and pharmacogenetics in drug development. Curr Opin Mol Ther. 2009;11:260–268. [PubMed] [Google Scholar]
- 32.Zhang B, Xie W, Krasowski MD. PXR: a xenobiotic receptor of diverse function implicated in pharmacogenetics. Pharmacogenomics. 2008;9:1695–1709. doi: 10.2217/14622416.9.11.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burk O, Koch I, Raucy J, Hustert E, Eichelbaum M, Brockmoller J, et al. The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR) J Biol Chem. 2004;279:38379–38385. doi: 10.1074/jbc.M404949200. [DOI] [PubMed] [Google Scholar]
- 34.SeattleSNPs Variation Discovery Resource. 2008 http://pga.gs.washington.edu/
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H, Doan S, Birdwell KA, Cowan JD, Vincz AJ, Haas DW, et al. An automated approach to calculating the daily dose of tacrolimus in electronic health records. AMIA Summits Transl Sci Proc. 2010;2010:71–75. [PMC free article] [PubMed] [Google Scholar]
- 38.Harrell FE., Jr. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. Springer; New York: 2001. [Google Scholar]
- 39.Team RDC . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2005. [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 42.Thervet E, Loriot MA, Barbier S, Buchler M, Ficheux M, Choukroun G, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther. 2010;87:721–726. doi: 10.1038/clpt.2010.17. [DOI] [PubMed] [Google Scholar]
- 43.Macphee IA, Fredericks S, Mohamed M, Moreton M, Carter ND, Johnston A, et al. Tacrolimus pharmacogenetics: the CYP3A5*1 allele predicts low dose-normalized tacrolimus blood concentrations in whites and South Asians. Transplantation. 2005;79:499–502. doi: 10.1097/01.tp.0000151766.73249.12. [DOI] [PubMed] [Google Scholar]
- 44.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11:501–505. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mancinelli LM, Frassetto L, Floren LC, Dressler D, Carrier S, Bekersky I, et al. The pharmacokinetics and metabolic disposition of tacrolimus: a comparison across ethnic groups. Clin Pharmacol Ther. 2001;69:24–31. doi: 10.1067/mcp.2001.113183. [DOI] [PubMed] [Google Scholar]
- 46.Dirks NL, Huth B, Yates CR, Meibohm B. Pharmacokinetics of immunosuppressants: a perspective on ethnic differences. Int J Clin Pharmacol Ther. 2004;42:701–718. doi: 10.5414/cpp42701. [DOI] [PubMed] [Google Scholar]
- 47.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dumitrescu L, Ritchie MD, Brown-Gentry K, Pulley JM, Basford M, Denny JC, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med. 2010 doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuypers DR, Claes K, Evenepoel P, Maes B, Coosemans W, Pirenne J, et al. Time-related clinical determinants of long-term tacrolimus pharmacokinetics in combination therapy with mycophenolic acid and corticosteroids: a prospective study in one hundred de novo renal transplant recipients. Clin Pharmacokinet. 2004;43:741–762. doi: 10.2165/00003088-200443110-00005. [DOI] [PubMed] [Google Scholar]
- 50.Wang P, Mao Y, Razo J, Zhou X, Wong ST, Patel S, et al. Using genetic and clinical factors to predict tacrolimus dose in renal transplant recipients. Pharmacogenomics. 2010;11:1389–1402. doi: 10.2217/pgs.10.105. [DOI] [PubMed] [Google Scholar]
- 51.Pendergrass SA, Dudek SM, Crawford DC, Ritchie MD. Synthesis-View: visualization and interpretation of SNP association results for multi-cohort, multi-phenotype data and meta-analysis. BioData Min. 2010;3:10. doi: 10.1186/1756-0381-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.