Abstract
We previously demonstrated that exonic selectivity for frameshift mutation (exon 10 over exon 3) of ACVR2 in mismatch repair (MMR)-deficient cells is partially determined by 6 nucleotides flanking 5’ and 3’ of each microsatellite. Substitution of flanking nucleotides surrounding the exon 10 microsatellite with those surrounding the exon 3 microsatellite greatly diminished heteroduplex (A7/T8) and full (A7/T7) mutation, while substitution of flanking nucleotides from exon 3 with those from exon 10 enhanced frameshift mutation. We hypothesized that specific individual nucleotide(s) within these flanking sequences control ACVR2 frameshift mutation rates. Only the 3rd nucleotide 5’ of the microsatellite, and 3rd, 4th, and 5th nucleotides 3’ of the microsatellite were altered from the native flanking sequences and these locations were individually altered (sites A, B, C, and D, respectively). Constructs were cloned +1 bp out-of-frame of EGFP, allowing a −1 bp frameshift to express EGFP. Plasmids were stably-transfected into MMR-deficient cells. Non-fluorescent cells were sorted, cultured for 35 days, and harvested for flow cytometry and DNA-sequencing. Site A (C to T) and B (G to C) in ACVR2 exon 10 decreased both heteroduplex and full mutant as much as the construct containing all 4 alterations. For ACVR2 exon 3, site A (T to C), C (A to G), and D (G to C) are responsible for increased heteroduplex formation, whereas site D is responsible for full mutant formation by ACVR2 exon 10 flanking sequences. Exonic selectivity for frameshift mutation within ACVR2’s sequence context is specifically controlled by individual nucleotides flanking each microsatellite.
Keywords: ACVR2, frameshift mutation, DNA mismatch repair, microsatellite instability, colorectal cancer
1. Introduction
Approximately 15% of colon caners display microsatellite instability (MSI) [1]. Microsatellites are nucleotide repeat sequences that are predominantly located in non-coding regions of the genome, but a minority are present in coding regions (exons) of critical growth regulatory genes such as transforming growth factor β receptor 2 (TGFBR2), activin receptor type 2 (ACVR2), and phosphatase and tensin homolog (PTEN) [2–7]. When the DNA mismatch repair (MMR) system, a key enzyme system that recognizes and coordinates repair of nucleotide base mismatches and slippage mistakes at microsatellite sequences, is defective, frameshift mutations within these genes are accumulated and their protein functions lost, resulting in deregulation of specific normal cellular processes that can drive the pathogenesis of colorectal cancer (CRC) and other tumors that manifest MSI [1, 7–14]. For example, an A8 microsatellite within ACVR2 exon 10 becomes − 1 bp frameshift mutated at both alleles in 70–90% of CRC with MSI [6, 15]. This frameshift mutation of ACVR2 causes loss of protein expression and its growth suppressive effect that is normally mediated by intracellular SMAD signaling [13]. In addition, ACVR2 shows exonic selectivity for frameshift mutation. ACVR2 contains identical A8 microsatellite sequences in exon 3 and 10, but only its exon 10 microsatellite is mutated in CRC with MSI [6, 15]. We replicated these observations in a cell model. We have measured frameshift mutation rate at the A8 microsatellite of ACVR2 exon 10 in real time and showed that it accumulates −1 bp frameshift mutation in human colorectal cancer cells with DNA MMR defects (frameshifts with hMLH1 and hMSH6 deficiencies) after forming heteroduplexes at a constant rate [16, 17]. We also demonstrated frameshift mutation selectivity for the ACVR2 exon 10 over the ACVR2 exon 3 microsatellite in DNA MMR defective cells in real time [17]. Furthermore, we showed that this selectivity for frameshift mutation within ACVR2 lies in part with the immediate flanking nucleotides surrounding each coding microsatellite [17]. Briefly, we substituted a grouping of 6 nucleotides immediately the flanking 5’ and 3’ ends of each ACVR2 exon 3 and ACVR2 exon 10 microsatellite. Substitution of the flanking nucleotides surrounding the exon 10 microsatellite with those surrounding the exon 3 microsatellite greatly diminished heteroduplex (A7 paired with T8) and full mutant (A7 paired with T7) formation in hMLH1−/− cells, while substitution of the flanking nucleotides from exon 3 with those from the exon 10 enhanced frameshift mutation [17]. These observations demonstrated a critical role of the flanking DNA sequence context for microsatellite frameshift mutation when DNA MMR is defective, possibly explaining why certain genes containing coding microsatellites become frameshift mutated at various frequencies.
In this study, we tested further if specific individual nucleotide(s) within these flanking sequences, versus all flanking nucleotides, control frameshift mutation rates of ACVR2. We performed single nucleotide substitutions within the six flanking nucleotides surrounding the exon 3 and 10 microsatellites of ACVR2, and measured mutation rates in MMR-deficient cells to assess their relative effect on frameshift mutation. We demonstrate that single nucleotide changes at the 3rd nucleotide 5’ of the microsatellite and the 3rd nucleotide 3’ of the microsatellite of ACVR2 exon 10 are responsible for a decreased −1 bp frameshift mutation rate. In addition, we observed that single nucleotide changes at the 5th nucleotide 3’ of the microsatellite of ACVR2 exon 3 are responsible for the appearance of full mutants. These findings suggest that exonic selectivity for frameshift mutation within ACVR2’s sequence context is specifically regulated by individual nucleotides flanking each coding microsatellite of ACVR2.
2. Materials and Methods
2.1. Cloning of pIREShyg2-ACVR2 exon 10-EGFP and pIREShyg2-ACVR2 exon 3-EGFP plasmids containing single nucleotide substitutions
Plasmids pIREShyg2-ACVR2 exon 10-EGFP and pIREShyg2-ACVR2 exon 3-EGFP, containing portions of exon 10 and exon 3 of human ACVR2 were constructed previously [16, 17]. Plasmids pIREShyg2-ACVR2 exon 10-exon 3 flanking (F) OF and pIREShyg2-ACVR2 exon 3-exon 10 F OF were also constructed previously in which six nucleotides flanking ACVR2 exons 10 or 3 microsatellites at the 5’ and 3’ ends were swapped with the 6 nucleotides flanking ACVR2 exons 3 or 10 microsatellites, respectively [17]. Single nucleotide changes within the 6 nucleotides flanking the 5’ and 3’ ends of each ACVR2 exon 10 and 3 microsatellite were performed using a Quikchange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) (Table 1). Details of plasmid cloning were previously described [16, 17]. Briefly, portions of native or modified sequences of exon 10 or 3 of ACVR2 (Table 1) were inserted immediately after the start codon of the EGFP gene, either in-frame of EGFP (IF) or +1 bp out of frame of EGFP (OF) in pIREShyg2-EGFP. For experimental plasmids, various ACVR2 sequences were cloned +1 bp OF in pIREShyg2-EGFP and thus a −1 bp frameshift mutation at the coding microsatellite would allow expression of EGFP. Mutation resistant (MR) counterpart plasmids were constructed by interrupting the microsatellites (A8 to A3GA4) using a Quikchange II site-directed mutagenesis kit (Stratagene). The MR ACVR2 plasmids were placed OF (+1bp) and IF to be used as negative and positive controls for EGFP expression, respectively.
Table 1.
Sequences of exon 10 and exon 3 of ACVR2 used for cloning into pIREShyg2-EGFP plasmid.
| ACVR2 plasmids | ACVR2 sequences |
|---|---|
| MR ACVR2 exon 10 OF | -----TGTGCATAAAGAAAAGAGGCC----- |
| ACVR2 exon 10 OF | -----TGTGCATAAAAAAAAGAGGCC----- |
| ACVR2 exon 10-exon 3 F OF | -----TGTGTATAAAAAAAAGACAGC----- |
| ACVR2 exon 10-A mut OF | -----TGTGTATAAAAAAAAGAGGCC----- |
| ACVR2 exon 10-B mut OF | -----TGTGCATAAAAAAAAGACGCC----- |
| ACVR2 exon 10-C mut OF | -----TGTGCATAAAAAAAAGAGACC----- |
| ACVR2 exon 10-D mut OF | -----TGTGCATAAAAAAAAGAGGGC----- |
| MR ACVR2 exon 3 OF | -----TGTGTAGAAAGAAAAGACAGC----- |
| ACVR2 exon 3 OF | -----TGTGTAGAAAAAAAAGACAGC----- |
| ACVR2 exon 3-exon 10 F OF | -----TGTGCAGAAAAAAAAGAGGCC----- |
| ACVR2 exon 3-A mut OF | -----TGTGCAGAAAAAAAAGACAGC----- |
| ACVR2 exon 3-B mut OF | -----TGTGTAGAAAAAAAAGAGAGC----- |
| ACVR2 exon 3-C mut OF | -----TGTGTAGAAAAAAAAGACGGC----- |
| ACVR2 exon 3-D mut OF | -----TGTGTAGAAAAAAAAGACACC----- |
Various exon 10 or exon 3 ACVR2 sequences were inserted immediately after the translation initiation codon of EGFP gene +1 bp out of reading frame of the EGFP gene (OF) in the pIREShyg2-EGFP plasmid. Mutation resistant (MR) plasmids were constructed by interrupting microsatellite sequences (A8 to A3GA4). Mutation resistant (MR) OF plasmids served as negative controls for EGFP expression. Six nucleotides (in italic) flanking ACVR2 exons 10 or 3 microsatellites at the 5’ and 3’ ends were replaced with the 6 nucleotides (in italic) flanking exons 3 or 10 microsatellites to construct ACVR2 exon 10-exon 3 flanking (F) and ACVR2 exon 3-exon 10 F plasmids, respectively. Single nucleotide substitutions within these flanking sequences were performed. Only the 3rd nucleotide 5’ of the microsatellite, and 3rd, 4th and 5th nucleotides 3’ of the microsatellite were altered compared to the wild type (WT) sequence upon swapping the exonic surrounding sequences, and these locations were individually altered to assess their relative effect (sites A, B, C and D, respectively).
2.2 Cell lines, transfection, and selection
The human colon cancer cell lines, HCT116 (hMLH1−/−, hMSH3−/−) were cultured as described previously [16]. Cells were transfected with various pIREShyg2-ACVR2-EGFP plasmids (described in Table 1) by using Nucleofector V kit (Amaxa, Cologne, Germany) and stable cell lines were generated by hygromycin B (Invitrogen Corp, Carlsbad, CA) selection. After selection, colonies from each cell line were initially pooled and cultured for mutation analysis. All stable cell lines were confirmed by DNA sequencing.
2.3. Mutation Analysis by flow cytometry
Details of analysis of mutant cells by flow cytometry were previously described [16]. Briefly, ten thousand non-fluorescent cells expressing MR ACVR2 exon 10 OF, ACVR2 exon 10 OF, ACVR2 exon 10-exon 3 F OF, ACVR2 exon 10-A mut OF, ACVR2 exon 10-B mut OF, ACVR2 exon 10-C mut OF, ACVR2 exon 10-D mut OF, MR ACVR2 exon 3 OF, ACVR2 exon 3 OF, ACVR2 exon 3-exon 10 F OF, ACVR2 exon 3-A mut OF, ACVR2 exon 3-B mut OF, ACVR2 exon 3-C mut OF, or ACVR2 exon 3-D mut OF were sorted into 24-well plates by FACS ARIA (Becton Dickinson, San Jose, CA). During the analysis period, cultures were expanded as required to keep cells in exponential growth. Cells were trypsinized, washed in PBS, and resuspended in a total volume of 200 µl of PBS/ 0.5 µg/ml of propidium iodide (PI) and 3% BSA. At specified time points, cell suspensions from two cultures were analyzed in parallel on a FACSCalibur (Becton Dickinson). The population with dim EGFP fluorescence was designated M1, and the population with bright EGFP fluorescence was designated M2. The counts of M1 and M2 cells were expressed as percentages of total live cell number.
2.4. PCR and DNA sequencing
Total cellular DNA from stable cell lines and the M1 cell populations were PCR-amplified by specific primers using a cloned Pfu DNA polymerase (Stratagene) as described previously [17]. The PCR products were used for DNA sequencing to identify stable cell lines and frameshift mutations at the microsatellites of various ACVR2 exon 10 and 3 sequences.
2.5. Determination of −1 bp frameshift mutation rates of ACVR2 sequences in MMR-deficient cells
Mutation rates were calculated by the “method of the mean” developed by Luria and Delbruck [18] as described previously [16].
2.6. Statistical analysis
Mutation frequencies and rates of cell lines were compared by T-test or one-way ANOVA.
3. Results
3.1. Exonic selectivity for frameshift mutation rates of ACVR2 in the hMLH1−/− background is determined by individual nucleotides that flank the coding microsatellites of ACVR2
We hypothesized that specific individual nucleotide(s) within the flanking sequences surrounding the microsatellites of ACVR2 exon 10 and exon 3 are responsible for frameshift mutation selectivity (exon 10 over exon 3) and tested this hypothesis using the same cell model system we previously established [16, 17]. We constructed EGFP plasmids in which a −1 bp frameshift mutation at the coding microsatellites of ACVR2 exons 10 and 3 could be measured in the context of native or altered flanking micorsatellite sequences, and be detected by EGFP expression in DNA MMR-deficient cells once frameshift mutation occurs [16, 17].
To test our hypothesis, we additionally constructed various ACVR2 plasmids in which individual nucleotide substitution within the flanking sequences of ACVR2 exon 10 and 3 microsatellites were performed. Only the 3rd nucleotide 5’ to the microsatellite, and 3rd, 4th and 5th nucleotides to 3’ to the microsatellite of both exon 10 and exon 3 were altered compared to the native ACVR2 sequence upon swapping the exonic flanking sequences [17]. These four locations were individually altered (sites designated as A, B, C and D, respectively, Table 1) within the native ACVR2 sequences, and stable cell lines containing each single nucleotide substituted sequence were generated in hMLH1-deficient (completely DNA MMR defective) cells to assess their relative effect on frameshift mutation. These cells were compared to stable cell lines containing the native and swapped ACVR2 exon 10 or 3 sequences in the hMLH1−/− background. Additionally, hMLH1−/− MR ACVR2 exon 10 OF and hMLH1−/− MR ACVR2 exon 3 OF cells were used as negative controls for EGFP expression.
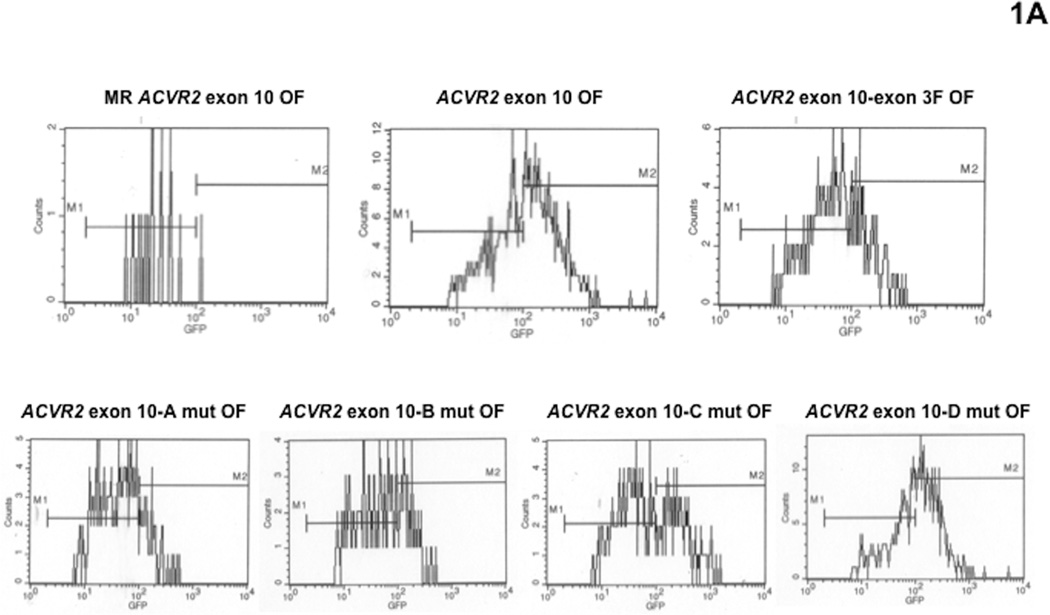
To compare mutation frequencies and rates of cells containing the native ACVR2 sequences with ones of cells containing modified ACVR2 sequences in the hMLH1−/− background, non-fluorescent cells containing either MR ACVR2 exon 10 OF, ACVR2 exon 10 OF, ACVR2 exon 10-exon 3 F OF, ACVR2 exon 10-A mut OF, ACVR2 exon 10-B mut OF, ACVR2 exon 10-C mut OF, ACVR2 exon 10-D mut OF, MR ACVR2 exon 3 OF, ACVR2 exon 3 OF, ACVR2 exon 3-exon 10 F OF, ACVR2 exon 3-A mut OF, ACVR2 exon 3-B mut OF, ACVR2 exon 3-C mut OF or ACVR2 exon 3-D mut OF were sorted and exponentially grown for 7 to 35 days. At specific time points (day 7, 14, 21, 28, and 35), a −1 bp frameshift mutation was detected by EGFP expression using flow cytometry. EGFP flow cytometric histograms at day 35 were shown as representatives of mutation analysis in Fig. 1. As observed previously [17], cells containing the native ACVR2 exon 10 or modified ACVR2 exon 10 sequences revealed both M1 and M2 populations (Fig. 1A), whereas cells containing the native ACVR2 exon 3 sequence revealed only the M1 population (Fig. 1B), re-confirming the exonic selectivity of ACVR2 for frameshift mutation. Previously we demonstrated that the M1 population represents a heteroduplex (A7/T8) population expressing dim fluorescence, whereas the M2 population represents a full mutant (A7/T7) population expressing bright fluorescence [16, 17]. In particular, ACVR2 exon 3-D mut OF cells revealed a small proportion of full mutant (M2) population as observed in ACVR2 exon 3-exon 10 F OF cells (Fig. 1B).
Fig. 1. Mutation analysis of hMLH1−/− cells containing various ACVR2 exon 10 or exon 3 sequences by flow cytometry.
Non-fluorescent cells were sorted and exponentially grown for 7 to 35 days. At specific time points, cells were harvested, and 200,000 cells were analyzed for EGFP expression (identifying a −1bp frameshift mutation) by flow cytometry. Gated live cells were analyzed on an EGFP histogram and two distinct EGFP populations were plotted. The population with dim EGFP expression was designated M1 and the population with bright EGFP expression was designated M2. EGFP histograms of MR ACVR2 exon 10 OF, ACVR2 exon 10 OF, ACVR2 exon 10-exon 3 F OF, ACVR2 exon 10-A mut OF, ACVR2 exon 10-B mut OF, ACVR2 exon 10-C mut OF, and ACVR2 exon 10-D mut OF cells (A) as well as MR ACVR2 exon 3 OF, ACVR2 exon 3 OF, ACVR2 exon 3-exon 10 F OF, ACVR2 exon 3-A mut OF, ACVR2 exon 3-B mut OF, ACVR2 exon 3-C mut OF, and ACVR2 exon 3-D mut OF cells (B) at day 35 were shown as representatives of mutation analysis. Note that scaling of cell counts for each EGFP histogram is different.
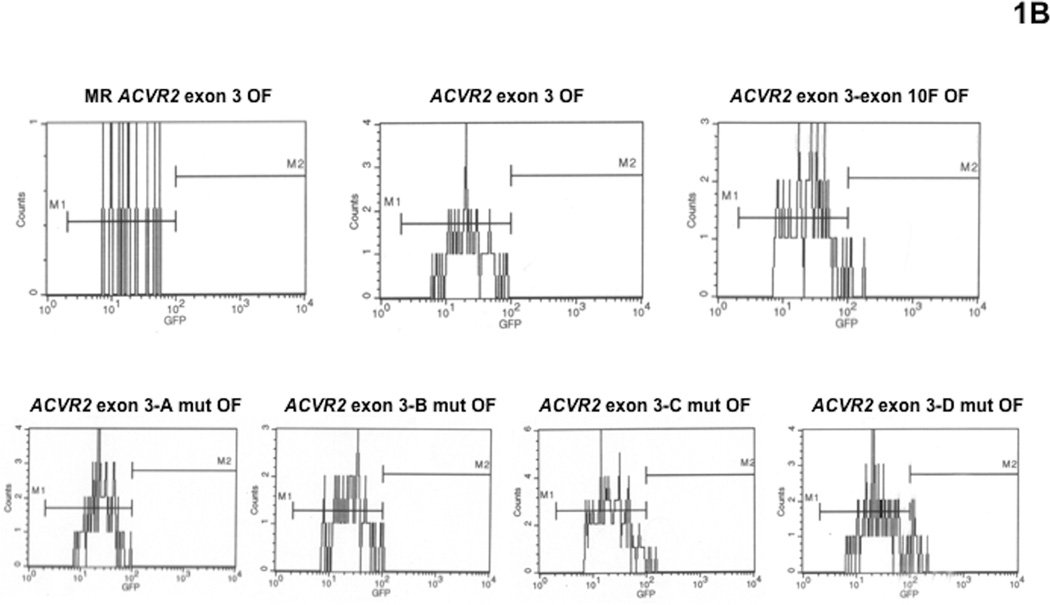
The −1 bp frameshift mutation frequency at each time point was expressed as a fold change using the following formula: (EGFP positive cells /total live cells from ACVR2 OF cells)/(EGFP positive cells /total live cells from MR ACVR2 OF cells) (Fig. 2). As we observed previously [16, 17], the M2 population from various ACVR2 exon 10 sequences in hMLH1−/− cells dramatically accumulated over time, whereas the M1 population showed relatively little change over time (Fig. 2A and 2B), again indicating that the M1 and M2 are distinct populations.
Fig. 2. Mutation frequencies of the native and modified ACVR2 exons 10 and 3 sequences in the hMLH1−/− background.
Non-fluorescent cells were analyzed for EGFP expression by flow cytometry at 7, 14, 21, 28, and 35 days after being sorted and cultured, and EGFP analysis was performed at each day as shown in Fig. 1. Mutation frequency at each time point was expressed as a fold change using the following formula: (EGFP positive cells/total live cells from ACVR2 OF cells)/(EGFP positive cells/total live cells from MR ACVR2 OF cells). In various ACVR2 exon 10 sequences, the mutation frequencies of the M1 populations (A) were much lower and showed relatively little change compared to ones of the M2 populations (B) which dramatically accumulated over time. (A): At day 28 and 35, the native ACVR2 exon 10 sequence showed the highest M1 mutation frequency but its modified sequences such as ACVR2 exon 10-exon 3F, ACVR2 exon 10-A mut, ACVR2 exon 10-B mut, and ACVR2 exon 10-C mut sequences lower its mutation frequencies. In particular, ACVR2 exon 10-B mut sequence showed significantly less M1 population compared to ACVR2 exon 10-exon 3 F sequence at all time points (P < 0.05). (B): The mutation frequency of the native ACVR2 exon 10 sequence in the M2 population was significantly higher than ones of ACVR2 exon 10-exon 3F, ACVR2 exon 10-A mut, ACVR2 exon 10-B mut sequences at all time points (P < 0.05). Note that ACVR2 exon 10-A mut and ACVR2 exon 10-B mut sequence (except day 21) showed less M2 population compared to ACVR2 exon 10-exon 3 F sequence at all time points. Sequences showing lower mutation frequencies (less than 150-fold change) were separately plotted in the right panel using a smaller y-axis scale. (C): The native ACVR2 exon 3 sequence showed the least M1 population but its modified sequences (except ACVR2 exon 3-B mut sequence) showed significantly higher M1 populations over time (P < 0.05). Overall, these observations indicate that not only the flanking sequences of ACVR2 exon 10 and exon 3 microsatellites but also specific single nucleotides within these flanking sequences altered frameshift mutation at each microsatellite. Data are means from two independent experiments at each time point.
There was no dramatic increase in the frequencies in the M1 population from any cell lines containing various ACVR2 exon 10 sequences (Fig. 2A). At day 35, ACVR2 exon 10-exon 3 F OF cells showed ~2 fold less M1 population compared to ACVR2 exon 10 OF cells as we observed previously [17]. In addition, compared to ACVR2 exon 10 OF cells, ACVR2 exon 10-A mut, ACVR2 exon 10-B mut, ACVR2 exon 10-C mut, and ACVR2 exon 10-D mut OF cells showed ~2-, 3-, 2-, and 1.3-fold less M1 population, respectively (Fig. 2A), indicating all single nucleotide substitutions of ACVR2 exon 10 sequence decreased the M1 population compared to the native ACVR2 exon 10 sequence at day 35. However, considering all time points, only ACVR2 exon 10-A mut OF (except day 7) and ACVR2 exon 10-B mut OF cells showed comparable or significant less M1 population compared to ACVR2 exon 10-exon 3 F OF cells, respectively (P < 0.05). In the M2 population, ACVR2 exon 10 OF and ACVR2 exon 10 D-mut OF cells revealed prominent M2 populations compared to other cell lines (Fig. 2B). In particular, ACVR2 exon 10 D-mut OF cells showed the highest mutation frequency over time (except day 35) and was comparable to the ACVR2 exon 10 WT sequence. ACVR2 exon 10 C-mut OF cells showed significantly less M2 population at day 28 and 35 compared to ACVR2 exon 10 OF cells but showed a greater M2 population than ACVR2 exon 10-exon3 F OF cells (P < 0.05) at all time points (Fig. 2B). On the other hand, compared to ACVR2 exon 10-exon3 F OF cells, ACVR2 exon 10 A-mut OF cells and ACVR2 exon 10 B-mut OF cells (except day 21) showed less M2 population at all time points. Overall, these data suggest that single nucleotide substitution at sites A and B in the native ACVR2 exon 10 sequence are responsible for the decreased M1 and M2 populations.
In the M1 population of cells containing the various ACVR2 exon 3 sequences, all cell lines except ACVR2 exon 3-B mut OF cells showed significantly higher mutation frequencies than ACVR2 exon 3 OF cells at all time points (P < 0.05) (Fig. 2C). Compared to ACVR2 exon 3-exon 10 F OF cells, ACVR2 exon 3-C mut OF cells showed a higher mutation frequency, and ACVR2 exon 3-A mut OF and ACVR2 exon 3-D mut OF cells showed comparable mutation frequencies over time (Fig. 2C), suggesting that single nucleotide mutation at A, C, and D sites in the native ACVR2 exon 3 sequence are responsible for the increased M1 populations by swapping its flanking sequences. It should be noted that the M2 frequencies for the various ACVR2 exon 3 sequences were too low to make comparisons.
3.2. Flanking nucleotides alter heteroduplex formation in ACVR2 microsatellites
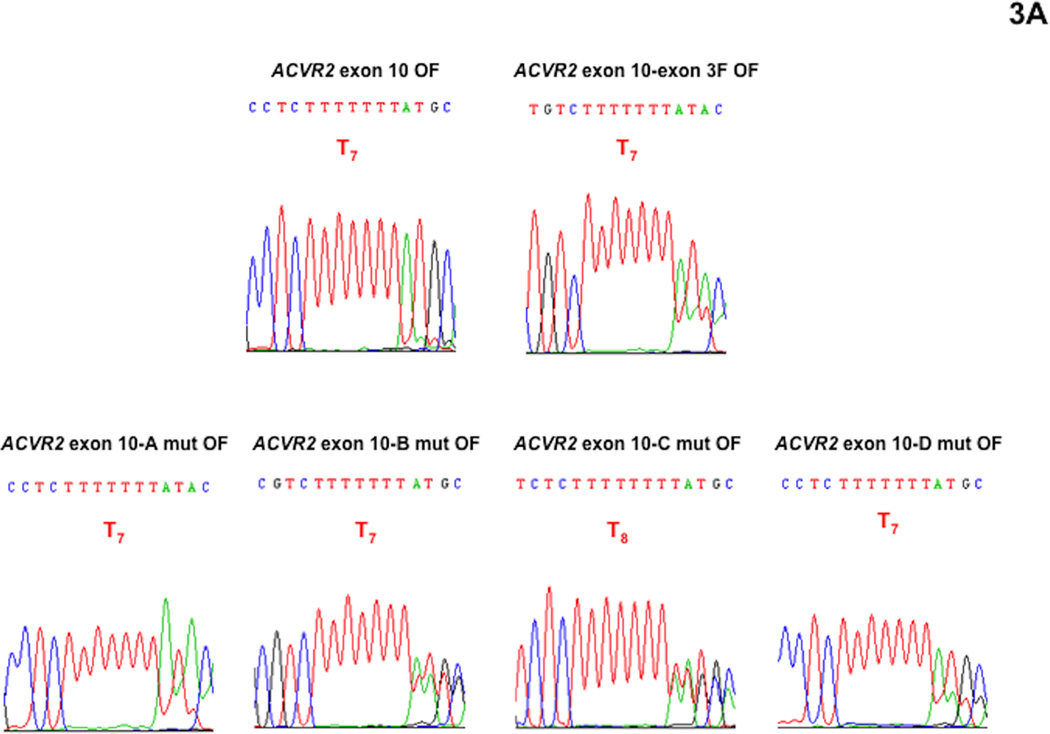
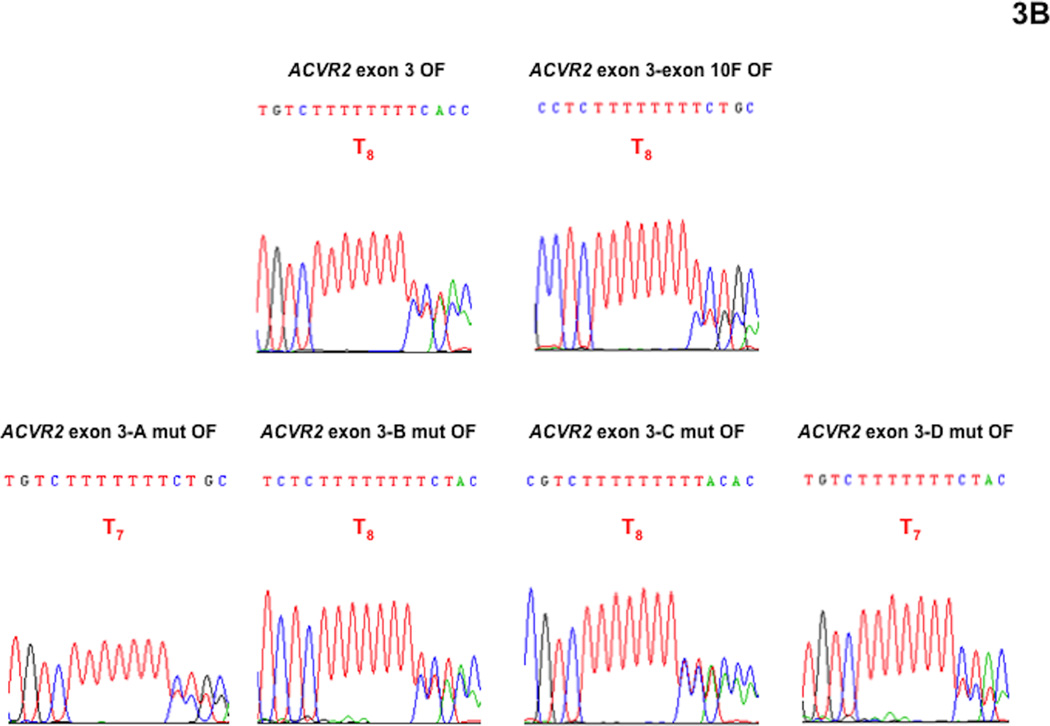
We performed DNA sequencing analysis to confirm that fluorescence from the M1 population was driven by heteroduplex formation (A8/T7) and to dissect its overlapped sequences (Fig. 3). We did not performed DNA sequencing analysis with the M2 population in this study because we had demonstrated that the M2 population is a “full mutant” population showing A7/T7 sequence (−1 bp frameshifted) previously [16, 17, 19]. At day 21 after being plated as non-fluorescent cells, the M1 populations of ACVR2 exon 10 OF, ACVR2 exon 10-exon 3 F OF, ACVR2 exon 10-A mut OF, ACVR2 exon 10-B mut OF, ACVR2 exon 10-C mut OF, ACVR2 exon 10-D mut OF, ACVR2 exon 3 OF, ACVR2 exon 3-exon 10 F OF, ACVR2 exon 3-A mut OF, ACVR2 exon 3-B mut OF, ACVR2 exon 3-C mut OF or ACVR2 exon 3-D mut OF cells were sorted and expanded for DNA sequencing analysis which was done at day 35.
Fig. 3. DNA sequencing analysis showing effect of single nucleotide substitution within the flanking sequences of ACVR2 exon 10 and exon 3 on heteroduplex formation at their microsatellites in the hMLH1−/− background.
The M1 cell populations from various ACVR2 exon 10 and exon 3 OF cells were sorted and cultured for DNA sequencing analysis. DNA from each cell line was amplified by PCR and the anti-sense strand was sequenced to assess for frameshift mutation at the microsatellites of ACVR2 exon 10 and exon 3. All cell lines revealed overlapped sequences, indicating heteroduplex formation in the M1 population. (A): All various ACVR2 exon 10 OF cell lines except ACVR2 exon 10-C mut OF cells (showing a T8 sequence as dominant) revealed −1 bp frameshifts (T7) as a dominant sequence. Compared to ACVR2 exon 10 OF cells, all cells containing modified ACVR2 exon 10 sequences showed decreased −1 bp frameshifts. In particular, ACVR2 exon 10-B mut OF and ACVR2 exon 10-C mut OF cells decreased −1bp frameshifts greater than ACVR2 exon 10-exon 3 F OF cells. (B): ACVR2 exon 3 OF, ACVR2 exon 3-exon 10 F, ACVR2 exon 3-B mut OF, and ACVR2 exon 3-C mut OF cells revealed a WT sequence (T8) as a dominant sequence. ACVR2 exon 3-A mut OF and ACVR2 exon 3-D mut OF cell lines showed a mutant sequence (T7) as dominant, indicating that sites A and D in the native ACVR2 exon 3 sequence are critical nucleotides to increase frameshift mutation in the M1 population. Overall, these data suggest that specific single nucleotide within the flanking sequences of ACVR2 regulate heteroduplex formation.
As shown in Fig. 3A, ACVR2 exon 10 OF cells revealed ~ 95% of mutant sequence (T7) overlapped with ~5% WT sequence (T8) (based on relative heights of peaks in the chromatograms) and ACVR2 exon 10-exon 3 F OF cells showed less mutant sequence (~75%) compared to ACVR2 exon 10 OF cells, confirming our previous finding that substitution of flanking sequences surrounding exon 10 microsatellite with ones surrounding exon 3 microsatellite decreases the frameshift mutation (heteroduplex and full mutant formation) of the native ACVR2 exon 10 [17]. Heteroduplex (A8/T7) formation was observed in all ACVR2 exon 10 cell lines containing single nucleotide substitutions. ACVR2 exon 10-A mut OF cells showed ~80% mutant sequence overlapped with ~20% WT sequence. ACVR2 exon 10-B mut OF cells and ACVR2 exon 10-C mut OF cells showed about 60 and 50% mutant sequence overlapped with WT sequence, respectively, congruent with that these single nucleotide substituted cell lines had less mutation frequencies in the M1 population at day 35 compared to ACVR2 exon 10-exon 3 F OF cells (Fig. 2A). ACVR2 exon 10-D mut OF cells showed about 75% mutant sequence overlapped with WT sequence (Fig. 3A). Thus, DNA sequencing analysis indicates that all 4 single nucleotide substitutions of ACVR2 exon 10 sequence altered heteroduplex formation and decreased the mutant sequence similarly (A and D mutation) or more (B and C mutation) compared to ACVR2 exon 10-exon 3F sequence at day 35.
We also observed heteroduplex formation from the M1 population of various ACVR2 exon 3 cell lines (Fig. 3B). ACVR2 exon 3 OF cells showed about 40% mutant sequence overlapped with ~60% WT sequence as observed previously [17]. Compared to ACVR2 exon 3 OF cells, we did not observe an increase of −1bp frameshifs in ACVR2 exon 3-exon 10 F OF cells (~25% mutant) for the M1 population (Fig. 3B) although a 50% higher mutation frequency was obtained (Fig. 2C). However, DNA sequencing analysis indicated that single nucleotide substitutions within flanking sequences of ACVR2 exon 3 alter heteroduplex formation (Fig. 3B). ACVR2 exon 3-A mut OF, ACVR2 exon 3-B mut OF, ACVR2 exon 3-C mut OF, and ACVR2 exon 3-D mut OF cells revealed ~60%, 40%, 50% and 70% mutant sequence overlapped with WT sequence, respectively, indicating that A and D sites (showed T7 as the main sequence) within the ACVR2 exon 3 sequence are critical nucleotides to increase frameshift mutation in the M1 population. Overall, This DNA analysis confirms that the M1 populations of various ACVR2 cell lines form heteroduplexes, and that specific single nucleotide within the flanking sequences of ACVR2 regulate heteroduplex formation.
3.3 Full mutant mutation rates are determined by flanking nucleotides in ACVR2 microsatellites
The mutation rates for various hMLH1−/− ACVR2 exon 10 cell lines were shown in Table 2. The native ACVR2 exon 10 sequence showed the highest mutation rate: 5.17 × 10−4 ± 1.47 × 10−4. The mutation rate for ACVR2 exon 10-exon 3 F sequence is ~ 4 times lower compared to the native ACVR2 exon 10 sequence (P < 0.05), confirming that the exon 3 flanking sequences make a microsatellite less prone to full mutation with DNA MMR deficiency. The mutation rates for ACVR2 exon 10-A mut OF and ACVR2 exon 10-B mut OF sequences are ~6 and ~4.5 times lower compared to the native ACVR2 exon 10 sequence (P < 0.05) and these are comparable to the mutation rate of ACVR2 exon 10-exon 3 F sequence. The mutation rate of ACVR2 exon 10-C mut OF sequence is ~2 times less than the native ACVR2 exon 10 sequence but it is higher than ACVR2 exon 10-exon 3 F sequence. There is no significant difference in mutation rate between ACVR2 exon 10-D mut OF sequence and native ACVR2 exon 10 sequence. On the other hand, as shown Fig. 1B, we observed the appearance of the M2 population from ACVR2 exon 3-exon 10 F OF and ACVR2 exon 3-D mut OF cells but their mutation frequencies were not high enough for statistical comparisons. Calculated mutation rates for ACVR2 exon 3-exon 10 F OF and ACVR2 exon 3-D mut OF cells are 0.11 × 10−4 ± 0.02 × 10−4 and 0.17 × 10−4 ± 0.03 × 10−4, respectively.. These data indicate that A and B sites in the native ACVR2 exon 10 sequence and D site in the native ACVR2 exon 3 sequence are responsible for altered −1 bp frameshift mutation rates.
Table 2.
Calculated −1bp frameshift mutation rates at the microsatellite within various ACVR2 exon 10 sequences in the hMLH1−/− background.
| Gene | Rate for mutation |
|---|---|
| ACVR2 exon 10 OF | 5.17 × 10−4 ± 1.47 × 10−4a |
| ACVR2 exon 10-exon 3 F OF | 1.30 × 10−4 ± 0.30 × 10−4b |
| ACVR2 exon 10-A mut OF | 0.89 × 10−4 ± 0.22 × 10−4b |
| ACVR2 exon 10-B mut OF | 1.13 × 10−4 ± 0.09 × 10−4b |
| ACVR2 exon 10-C mut OF | 2.68 × 10−4 ± 0.27 × 10−4c |
| ACVR2 exon 10-D mut OF | 5.76 × 10−4 ± 1.84 × 10−4a |
Data from the M2 population at each time point between day 21 and day 35 were used for −1 bp frameshift mutation rate analysis. Single mutation rates were calculated by combining and averaging time-specific mutation rates. Rates are expressed as mutations at microsatellite sequence per cell per generation. Data shown are mean ± SEM. a, b, and c represent significant difference in the mutation rate (P < 0.05).
ACVR2 exon 10 OF versus ACVR2 exon 10-exon 3 F OF, ACVR2 exon 10-A mut OF, ACVR2 exon 10-B mut OF, and ACVR2 exon 10-C mut OF;
ACVR2 exon 10-exon 3 F OF versus ACVR2 exon 10 OF, ACVR2 exon 10-C mut OF and ACVR2 exon 10-D mut OF;
ACVR2 exon 10-C mut OF versus ACVR2 exon 10 OF, ACVR2 exon 10-exon 3 F OF, ACVR2 exon 10-A mut OF, ACVR2 exon 10-B mut OF, and ACVR2 exon 10-D mut OF.
4. Discussion
We previously demonstrated that six nucleotides that flank the microsatellties of ACVR2 exon 10 and exon 3 are partly responsible for exonic selectivity of ACVR2 for frameshift mutation in colorectal cancer cells with DNA MMR defect [17]. In this study, we hypothesized that specific individual nucleotide(s) within these flanking sequences regulate frameshift mutation rates of ACVR2. In the sequence context of ACVR2 exon 10 and exon 3, only 4 nucleotides within the 5’ and 3’ immediate flanking sequences were altered compared to the native ACVR2 sequences (Table 1). Specifically, the 3rd nucleotide 5’ to the microsatellite, and 3rd, 4th and 5th nucleotides 3’ of the microsatellite were altered and these sites were individually substituted to elucidate their relative effect (sites A, B, C and D, respectively) on frameshift mutation. To test the hypothesis, we utilized stably transfected hMLH1−/− cell lines containing various ACVR2 sequences (Table 1) linked 1 bp OF to EGFP and these cell lines allowed us to observe −1 bp frameshift mutations occurring in real time with the onset of EGFP fluorescence.
We observed the following: i) in the context of both of ACVR2’s two coding microsatellites, the 3rd flanking nucleotide 5’ and 3’ to the exon 10 microsatellite are the primary drivers for influencing frameshift mutation, and the 5th flanking nucleotide 3’ to the exon 3 microsatellite is the key nucleotide driving frameshift mutation, ii) individual flanking nucleotides appear to at best modestly alter DNA heteroduplex formation (a DNA MMR independent event), but more dominantly drive heteroduplexes toward fully mutant frameshift mutation, and iii) the primary DNA sequences surrounding these coding microsatellites influence frameshift mutation. These findings highlight a poorly understood process where neighboring DNA nucleotides affect mutation of microsatellites. In addition to these flanking nucleotides influencing frameshifts, our findings further lend credence to the notion that the primary surrounding DNA sequence helps determine which coding microsatellite is altered, and thus loss of function of a gene’s protein, in microsatellite unstable cancers.
The formation of heteroduplexes is incompletely understood in the context of DNA MMR. We have previously observed heteroduplex formation at microsatellite sequences with a fully intact and functioning MMR system, indicating that heterodulpex formation is not dependent on loss of DNA MMR [16]. Heteroduplex DNA formation could be the result of a natural tendency for slippage at microsatellites during DNA replication, and easily repaired before mutation by the DNA MMR system. Certainly, the more slippage occurrences would require more response by competent MMR proteins to prevent full mutation. Our observation in this study suggests the individual flanking nucleotides modestly influence the natural rate for heteroduplex formation. In our data for ACVR2 exon 3, certain nucleotides do not alter, whereas others change the frequency for microsatellite slippage. Thus, the natural tendency for heteroduplex formation at these sites appears to be directed by the surrounding primary DNA sequence.
Since we observed that immediate flanking sequences as well as specific single nucleotides within these flanking sequences surrounding ACVR2 microsatellites altered frameshift mutation, we speculated the existence of consensus sequence(s) that regulate frameshift mutation at coding microsatellites. Therefore, we examined some human target genes that have shown frameshift mutations at their coding microsatellites in CRC with MSI: TGFBR2 exon 3 (A10), Caspase-5 exon 2 (A10), MBD4 exon 2 (A10), TCF4 exon 17 (A9), ACVR2 exon 3 and 10 (A8), MSH3 exon 7 (A8), MSH6 exon 5 (C8), BAX exon 3 (G8), IGF2R exon 22 (G8), and PTEN exon 7 and 8 (A6) sequences [3, 4, 6, 7, 20–25]. We aligned 10 nucleotides surrounding these coding microsatellites at both 5’ and 3’ ends to find any potential consensus sequence(s) (Table 3). We could not identify any consensus sequence among these target genes as shown in a notation: NN{C}NNN{C}NNN-microsatellite-{C}NNNNNN{T}NN (N represents any nucleotides and {X} represents any nucleotides except X) but ~67 (8/12)% of target genes including ACVR2 exons 3 and 10 revealed “G” at the 1st nucleotide 3’ to the microsatellite (Table 3).
Table 3.
There is no consensus sequence for frameshift mutation within immediate flanking sequences surrounding coding microsatellites of target genes.
| Genes | Sequence |
|---|---|
| TGFR2 | -----ATTATGAAGG (A10) GCCTGGTGAG----- |
| Caspase-5 | -----AGACAGTGGC (A10) GGCGTAAGAA----- |
| MBD4 | -----ACAGCCTTGT (A10) GAAAGATCAT----- |
| TCF4 | -----CTTGCAGGAG (A9) GTGCGTTCGC----- |
| ACVR2 exon 3 | -----GATTGTGTAG (A8) GACAGCCCTG----- |
| ACVR2 exon 10 | -----TGTTGTGCAT (A8) GAGGCCTGTT----- |
| MSH3 | -----TGTTAGGGAC (A8) GGGCAACATT----- |
| MSH6 | -----CCGGAAGATA (C8) TTCTTAGAGC----- |
| BAX | -----CAGGGCGAAT (G8) AGGCACCCGA----- |
| IGF2R | -----TGAACTTCAC (G8) ACACTTGCCA----- |
| PTEN exon 7 | -----ACAAGATGCT (A6) GGTTTGTACT----- |
| PTEN exon 8 | -----TTACTTTAAC (A6) TGATCTTGAC----- |
TGFBR2, Caspase-5 and MBD4 genes containing an A10 microsatellite, TCF4 gene containing an A9 microsatellite, ACVR2 exon 3, ACVR2 exon 10, and MSH3 exon 7 genes containing an A8 microsatellite, MSH6 gene containing a C8 microsatellite, BAX and IGF2R genes containing a G8 microsatellite, and PTEN exon 7 and PTEN exon 8 genes containing an A6 microsatellite were examined. Ten nucleotides surrounding the coding microsatellites were aligned to identify consensus sequence(s) for frameshift mutation at both 5’ and 3 ends. No consensus sequence was identified among these target genes.
In conclusion, our data suggest that exonic selectivity for frameshift mutation within ACVR2 is specifically controlled by individual nucleotides flanking each coding ACVR2 microsatellite, even out of the context of the tertiary DNA structure. Our study has implications for the frequency of heteroduplex formation and frameshift mutation at other microsatellite sequences and for how MMR deficiency drives frameshift mutation.
Highlights.
The primary sequence surrounding DNA microsatellites help determine microsatellite mutability
Flanking nucleotides around coding ACVR2 microsatellites alter heteroduplex DNA formation
Frameshift mutation is determined by individual nucleotides surrounding ACVR2 microsatellites
Acknowledgements
Supported by the U.S. Public Health Service (DK067287), the UCSD Digestive Diseases Research Development Center (DK080506), and the SDSU/UCSD Comprehensive Cancer Center Partnership (CA132379 and CA132384).
Abbreviations
- OF
out of frame
- IF
in frame
- MMR
mismatch repair
- MSI
microsatellite instability
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributions: HC and JMC designed the project; HC, JC, JFL, and DJY performed all experiments; HC and JMC wrote the manuscript.
Conflict of Interest: All authors declare no conflict of interest.
References
- 1.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 3.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 4.Guanti G, Resta N, Simone C, Cariola F, Demma II, Fiorente P P, Gentile M. Involvement of PTEN mutations in the genetic pathways of colorectal cancerogenesis. Hum. Mol. Genet. 2000;9:283–287. doi: 10.1093/hmg/9.2.283. [DOI] [PubMed] [Google Scholar]
- 5.Mori Y, Yin J, Rashid A, Leggett BA, Young J, Simms L, Kuehl PM, Langenberg P, Meltzer SJ, Stine OC. Instabilotyping: comprehensive identification of frameshift mutations caused by coding region microsatellite instability. Cancer Res. 2001;61:6046–6049. [PubMed] [Google Scholar]
- 6.Jung B, Doctolero RT, Tajima A, Nguyen AK, Keku T, Sandler RS, Carethers JM. Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology. 2004;126:654–659. doi: 10.1053/j.gastro.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Woerner SM, Kloor M, Mueller A, Rueschoff J, Friedrichs N, Buettner R, Buzello M, Kienle P, Knaebel HP, Kunstmann E, Pagenstecher C, Schackert HK, Moslein G, Vogelsang H, von Knebel Doeberitz M, Gebert JF. Microsatellite instability of selective target genes in HNPCC-associated colon adenomas. Oncogene. 2005;24:2525–2535. doi: 10.1038/sj.onc.1208456. [DOI] [PubMed] [Google Scholar]
- 8.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 9.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD MD. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 10.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 12.Fishel R. Mismatch repair, molecular switches, and signal transduction. Genes Dev. 1998;12:2096–2101. doi: 10.1101/gad.12.14.2096. [DOI] [PubMed] [Google Scholar]
- 13.Jung B, Smith EJ, Doctolero RT, Gervaz P, Alonso JC, Miyai K K, Keku T, Sandler RS, Carethers JM. Influence of target gene mutations on survival, stage and histology in sporadic microsatellite unstable colon cancers. Int. J. Cancer. 2006;118:2509–2513. doi: 10.1002/ijc.21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, Yeo CJ, Hruban RH, Kern SE. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- 16.Chung H, Young DJ, Lopez CG, Le TA, Lee JK, Ream-Robinson D, Huang SC, Carethers JM. Mutation rates of TGFBR2 and ACVR2 coding microsatellites in human cells with defective DNA mismatch repair. PLoS ONE. 2008;3:e3463. doi: 10.1371/journal.pone.0003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung H, Lopez CG, Young DJ, Lai JF, Holmstrom J, Ream-Robinson D, Cabrera BL, Carethers JM. Flanking sequence specificity determines coding microsatellite heteroduplex and mutation rates with defective DNA mismatch repair (MMR) Oncogene. 2010a;29:2172–2180. doi: 10.1038/onc.2009.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luria SE, Delbruck M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung H, Lopez CG, Holmstrom J, Young DJ, Lai JF JF, Ream-Robinson D, Carethers JM. Both microsatellite length and sequence context determine frameshift mutation rates in defective DNA mismatch repair. Hum. Mol. Genet. 2010b;19:2638–2647. doi: 10.1093/hmg/ddq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader S, Walker M, Hendrich B, Bird A, Bird C, Hooper M, Wyllie A. Somatic frameshift mutations in the MBD4 gene of sporadic colon cancers with mismatch repair deficiency. Oncogene. 1999;18:8044–8047. doi: 10.1038/sj.onc.1203229. [DOI] [PubMed] [Google Scholar]
- 21.Barnetson R, Eckstein R, Robinson B, Schnitzler M. There is no increase in frequency of somatic mutations in metastases compared with primary colorectal carcinomas with microsatellite instability. Genes Chromosomes Cancer. 2003;38:149–156. doi: 10.1002/gcc.10262. [DOI] [PubMed] [Google Scholar]
- 22.Malkhosyan S, Rampino N, Yamamoto H, Perucho M M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 23.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz S, Jr, Yamamoto H, Navarro M, Maestro M, Reventos J, Perucho M. Frameshift mutations at mononucleotide repeats in caspase-5 and other target genes in endometrial and gastrointestinal cancer of the microsatellite mutator phenotype. Cancer Res. 1999;59:2995–3002. [PubMed] [Google Scholar]
- 25.Souza RF, Appel R, Yin J, Wang S, Smolinski KN, Abraham JM, Zou TT, Shi YQ, Lei J, Cottrell J, Cymes K, Biden K, Simms L, Leggett B, Lynch PM, Frazier M, Powell SM, Harpaz N, Sugimura H, Young J, Meltzer SJ. Microsatellite instability in the insulin-like growth factor II receptor gene in gastrointestinal tumors. Nat. Genet. 1996;14:255–257. doi: 10.1038/ng1196-255. [DOI] [PubMed] [Google Scholar]