Abstract
DNA mutations are the source of genetic variation within populations. The majority of mutations with observable effects are deleterious. In humans mutations in the germ line can cause genetic disease. In somatic cells multiple rounds of mutations and selection lead to cancer. The study of genetic variation has progressed rapidly since the completion of the draft sequence of the human genome. Recent advances in sequencing technology, most importantly the introduction of massively parallel sequencing (MPS), have resulted in more than a hundred-fold reduction in the time and cost required for sequencing nucleic acids. These improvements have greatly expanded the use of sequencing as a practical tool for mutation analysis. While in the past the high cost of sequencing limited mutation analysis to selectable markers or small forward mutation targets assumed to be representative for the genome overall, current platforms allow whole genome sequencing for less than $5,000. This has already given rise to direct estimates of germline mutation rates in multiple organisms including humans by comparing whole genome sequences between parents and offspring. Here we present a brief history of the field of mutation research, with a focus on classical tools for the measurement of mutation rates. We then review MPS, how it is currently applied and the new insight into human and animal mutation frequencies and spectra that has been obtained from whole genome sequencing. While great progress has been made, we note that the single most important limitation of current MPS approaches for mutation analysis is the inability to address low-abundance mutations that turn somatic tissues into mosaics of cells. Such mutations are at the basis of intra-tumor heterogeneity, with important implications for clinical diagnosis, and could also contribute to somatic diseases other than cancer, including aging. Some possible approaches to gain access to low-abundance mutations are discussed, with a brief overview of new sequencing platforms that are currently waiting in the wings to advance this exploding field even further.
Keywords: Massively parallel sequencing, Somatic mutation, Germ line mutation, Low-abundance mutations, Whole genome sequencing, Mosaicism, Aging, Cancer
1. Introduction
DNA mutations are a double-edged sword. On the one hand they provide a template for natural selection in creating the bewildering diversity of life on Earth, while on the other hand, their random occurrence can disturb highly conserved and interconnected gene regulatory networks resulting in altered genes or gene expression, defective cell functioning and disease. To maintain germ line mutation rate at an optimal level, allowing organisms to adapt to new environments while avoiding deleterious effects on fitness, an efficient system of genome maintenance has evolved over the millennia. This has led to a mutation rate that is surprisingly well conserved among unicellular and multicellular organisms with rates between 1×10−9 and 1×10−10 mutations per base per cell division (Table 1)[1]. The majority of deleterious mutations that spontaneously arise in a population are quickly removed from the gene pool through selection. However, mutations with mildly deleterious effects can remain in the population and lead to disease-related phenotypes[2].
Table 1.
Organismal germline basepair mutation rates per cell division
| Species | Assay | Cell divisions per generationa | Mutation rateb (×10−9) |
|---|---|---|---|
| H. sapiens | MPS - Family trio | 225 | 0.05 |
| D. melanogaster | MPS - Accumulation lines | 36 | 0.13 |
| C. elegans | MPS - Accumulation lines | 8.5 | 0.32 |
| A. thaliana | MPS - Accumulation lines | 30 | 0.16 |
| S. cerivisiae | MPS - Accumulation lines | 1 | 0.33 |
| E. coli | Reporter genes | 1 | 0.26 |
For multicellular organisms, it is necessary to differentiate between mutational processes that occur within the germline and those that arise in the soma. Although selective pressures in the germline and soma differ, they share a common set of repair enzymes that likely evolved prior to the emergence of multicellular organisms[3]. Mutations in the germline, so-called de novo mutations, can be passed on to offspring and may have adverse phenotypic consequences. Mutations can also arise in the soma, contributing to the development of both neoplastic and non-neoplastic syndromes.
In spite of the importance of DNA mutation as the substrate of evolution and a major cause of human disease, there is very little direct information about mutation frequencies and spectra in metazoans. This is entirely due to the lack of methods for quantifying and characterizing germline and somatic mutations. Especially low-abundance, somatic mutations are currently beyond the reach of most molecular analysis methods. With the emergence of massively parallel sequencing (MPS) methods, the direct measurement of genetic mutations is now possible and has already led to new data on germline mutation frequencies in invertebrate organisms. Here, we give a short historical background of the field of mutation research with the technology platforms it has used to estimate mutation rates and study mutation spectra in different cells and organisms. We then review MPS technologies and their applications in mutation research with a focus on mutation detection in mammalian systems. Finally, we briefly discuss new approaches to capture low-abundance mutations and experimentally address cell-to-cell variation in mutation loads, including the impact of new, experimental systems for single molecule sequencing.
1.1. Germline mutations
Some of the earliest attempts to define the rate of germline mutation were described at the beginning of the 20th century[4, 5]. The first demonstration of an induced mutation load was provided by Muller’s X-ray experiments on Drosophila[6]. This work formed the basis of forward genetics and expanded the tools available to estimate germline mutation rates. Muller used a phenotypic scoring approach, counting mutant lethals in order to measure the effect of irradiation on mutation frequencies. Building upon Muller’s work, Stadler irradiated maize and scored mutants using qualitative traits at eight loci to estimate the mutation frequency of each gene (between 10−4 and 10−6). Haldane provided the first estimate (indirect) for human mutation frequency using the principle of selection balance[7]. Based on demographic data on the fitness and frequency of hemophilia-affected males, he was able to estimate the frequency of new mutations arising at the haemophilia locus in the general population[8].
Due to a lack of knowledge on the size of the genome, the size of the locus, and the fraction of the locus that, when mutated, led to a dysfunctional protein product, extrapolations of the mutation frequency to the entire genome were not possible until very recently. The availability of fully annotated genomes has produced accurate estimations of the fraction of functional sites per locus and has allowed for the quantification of genome-wide per generation base-pair mutation rates in many organisms (Table 1)[9–12].
In principle, non-synonymous mutations can be detected at the protein level. Neel and co-workers examined children whose parents had been exposed to radiation at the time of the atomic bombings of Hiroshima and Nagasaki for the occurrence of mutations altering the electrophoretic mobility or activity of a series of proteins[13]. The mutation rate observed in the children of exposed individuals was 0.60 × 10−5/locus/generation compared to 0.64 × 10−5/locus/generation in the control children, whose parents had not been exposed to radiation. Apart from the fact that the mutant frequency in this case appeared not to depend on parental exposure, these results reveal a spontaneous mutation frequency that is very similar to the estimates made 50 years earlier by Haldane[8].
A major limitation of approaches based on phenotypic scoring is the high number of individuals that must be screened in order to detect spontaneous mutations. One way to circumvent this problem is to look at hundreds or thousands of loci simultaneously, e.g. using two-dimensional protein gel electrophoresis-based methods. Classical phenotypic scoring of visible markers or mutant lethals, however, can often be extremely labor intensive, requiring millions of screened cells or samples, e.g., 2.8 million mice, from multiple labs across the US, were screened for seven visible markers in the specific locus test[14].
The large sample size needed in phenotype-based assays can be circumvented through mutation accumulation experiments[15]. These assays tested for a reduction in fitness, or for a visible phenotypic change, after an inbred population accumulated mutations over many generations. By measuring the fitness of Drosophila at different generational time points, Mukai was able to provide an estimate for the deleterious (but non-lethal) mutation rate[16]. The validity of data obtained using this method has recently been questioned due to consistent overestimation of the genome-wide rate when compared with other assays[17]. A dependence on deleterious mutations of large effect has limited its usefulness and applications. Also, variation in the selective effect of deleterious mutations is not accounted for and could be the reason for overestimation of the mutation rate.
Prior to the introduction of DNA sequencing, mutation research was limited to estimating mutation rates and mapping new mutations through linkage analysis, i.e., by tracking the co-segregation of phenotypic marker loci. The emergence of assays to directly analyze DNA sequence variation enabled investigators to identify mutations at the molecular level and explore their mechanisms of action[18]. The first DNA-based assays screened for mutations at restriction sites that resulted in a restriction fragment length polymorphism (RFLP)[19]. The development of nucleotide sequencing methods subsequently permitted the analysis of small sections of genomic DNA for allelic variants after cloning[20, 21]. However, the high cost of sequencing led to the development of alternative assays to scan DNA fragments for sequence variants. In the 1980s multiple techniques were developed that screened samples for single base variants at specific, PCR-amplified loci[22, 23]. Examples include denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE), which are based on the exquisite sensitivity of DNA denaturation for sequence variants; two 500-bp fragments of similar size, differing in only one base pair melt at different temperatures and can be separated by gel electrophoresis in a gradient of chemical denaturants or temperature[24–26]. The sensitivity of these assays can be greatly increased by first allowing a mixture of mutant and wildtype fragments to denature and then slowly reanneal. The subsequent heteroduplex fragments are then easily distinguished by denaturing gradient gel separation. To increase efficiency, denaturing gradient assays can be run in a two-dimensional format allowing the detection of all possible sequence variants in the PCR-amplified coding and regulatory regions of large genes[27]. Other frequently used formats of DNA melting assays for mutation detection are denaturing high performance liquid chromatography (DHPLC)[28] and high-resolution melting curve analysis [29]. These approaches are used as a preliminary screen for mutants, which can then be characterized using Sanger sequencing. The arrival of shotgun sequencing enabled labs to screen larger fractions of the genome for mutations. Several groups have sequenced specified regions from their mutation accumulation lines to identify germline mutations and estimate locus-specific base-pair substitution rates[30, 31].
Meanwhile, array-based methods, such as comparative genomic hybridization (aCGH), allowed the identification of large germline variation, such as copy number variation[32]. While the resolution of these assays has recently been improved to 500bp with Nimblegen’s 4.2M arrays, their sensitivity and accuracy remain poor for events smaller than 10kb[33]. Still, they are much more sensitive than cytogenetic assays, based on fluorescence in situ hybridization (FISH), which have a resolution of no more than 10Mb[34], but offer the advantage that low-abundant chromosomal mutations can be detected in single cells. More recently, single cell assays have also emerged for aCGH based on whole genome amplification[35–37]. However, while useful in their own right, aCGH and FISH are incapable of detecting small mutations, which comprise the majority of the mutational landscape.
1.2 Somatic mutations
A connection between somatic mutations and human disease was first proposed by Carl Nordling who proposed that cancer is the result of accumulated mutations to a cell’s DNA[38, 39]. Around the same time, Leo Szilard attempted to explain the nature of aging through a somatic mutation framework. Szilard postulated that aging is caused by the accumulation of damaged chromosomes or genes in somatic tissue, realizing that ‘when a chromosome suffers an aging hit, the cell will cease to be functional if the homologous chromosome has either previously suffered an aging hit or if it carries a fault’ [40]. Almost two decades later, Knudson, based on these ideas, proposed his two-hit hypothesis, i.e., hereditary cancer is caused by the inheritance of a mutant allele in the germinal cells and acquisition of a mutation in the normal allele in a somatic cell; in nonhereditary cancers both mutations occur in somatic cells. As Szilard predicted, the frequency of somatic mutations has now been demonstrated to increase with age, but their relationship with aging and disease remains unclear[41]. However, somatic mutations are now considered to be the driving force behind the majority of cancers as well as a causal factor in some non-neoplastic human diseases, such as neurofibromatosis 1 and 2[42].
The discovery of the role played by somatic mutations in cancer initiation led to the emergence of assays to measure somatic mutation rates and to test the mutagenicity of different chemical agents and compounds in human cell lines and mice. For this purpose, cytogenetic methods were initially used, which proved capable of detecting the mutagenic effect of clastogens[43]. For smaller mutations bacterial systems, such as the Ames test, and endogenous and transgenic reporter-gene based mutation assays in mammals have dominated the field[44].
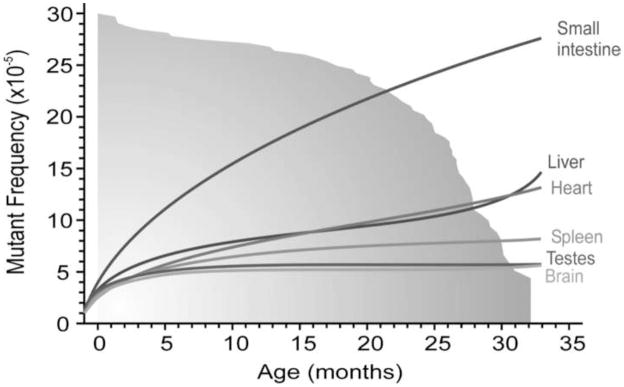
The endogenous, X-linked hypoxanthine-guanine phosphoribosyltransferase (HPRT) assay was one of the first reporter systems available and remains the most widely used assay for the analysis of somatic mutation frequencies in humans[45, 46]. A similar system in mice, based on a heterozygous mutation in the (autosomal) Aprt gene, also allows mutation analysis in vivo by screening for loss of the remaining wild-type allele in somatic cells[47]. These assays, however, can only be performed on cells that can actively proliferate in culture. The need for an in vivo assay that can be applied to all organs and tissues led to the development of transgenic reporter based methods. Mice harboring a LacZ or LacI reporter transgene emerged in the late 80s [48–50]. These reporter genes are part of a lambda or plasmid construct that can be recovered from genomic DNA and tested in E. coli for mutational inactivation of the reporter gene. Since then, a plethora of data has been published on somatic mutation frequencies and spectra in these animals, both induced mutations after exposure to a variety of mutagenic agents and spontaneous mutations accumulating during the normal aging process[51–53]. Using transgenic reporter genes, it has been shown that mutation frequencies in most tissues, especially actively proliferating tissues, such as the epithelia in the small intestine, consistently show an age-dependent increase (Fig. 1)[54].
Figure 1.
Somatic mutation frequencies in the aging mouse. Spontaneous lacZ mutant frequencies increase at different rates during aging in the brain, testis, spleen, liver, heart and small intestine of lacZ transgenic mice. The lines represent the mean mutant frequencies in different age groups. The gray fading area represents the survival curve of the mice, with 50% survival at 26.5 months. (Taken from Jan Vijg and Martijn E. T. Dollé. Large genome rearrangements as a primary cause of aging. Mechanisms of Ageing and Development 123, 907–915, 2002.)
While reporter-based assays have provided us with information about the frequency and spectra of mutations in human lymphocytes and varioust organs and tissues of aging animals, they are limited to a single locus and so might not be representative of genome-wide events. Additionally, their reliance on a robust phenotypic change, i.e. a dysfunctional protein product, ignores slightly deleterious mutations and therefore leads to a gross underestimation of the actual mutation frequency at the reporter locus. But the most significant drawback of transgenic reporter gene-based methods is the fact that they are restricted to model organisms and therefore unable to replace the HPRT assay for studying mutational processes in humans.
2. Massively parallel sequencing
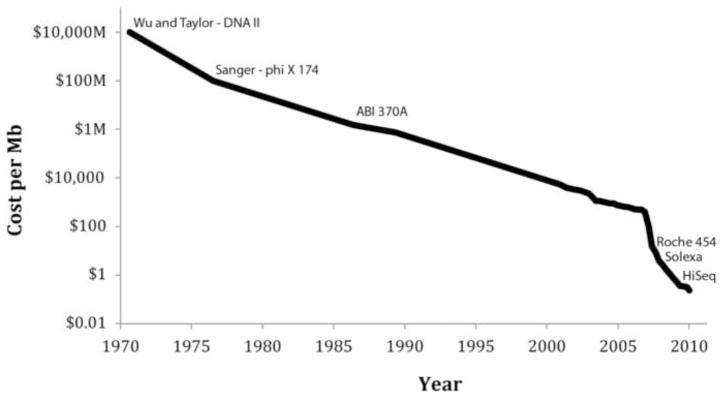
The main difference between massively parallel sequencing (MPS) and the classical Sanger sequencing is that in the former each target is sequenced multiple times as a series of overlapping short sequencing ‘reads’. To do this a DNA sample is first randomly fragmented, for example, by sonication, into fragments that can vary from 200 to 500 basepairs (bp). These fragments are then sequenced, millions at a time, in a random fashion. The resulting short sequence reads are aligned to reference sequences (when available), and consensus base calls are made. The emergence of MPS, which allows for the sequencing of 500Gb (500 billion) nucleotides of DNA sequence in a week on a single machine, has opened the door for projects previously thought unfeasible. This is reflected in the dramatically lower cost of sequencing. Figure 2 shows the cost per basepair since 1971, when the first DNA molecule was sequenced[55]. Initially, improvements in Sanger sequencing drove this development but with the introduction of the first ‘next-generation’ sequencing machine, the Roche 454 in 2005, the costs have come down by orders of magnitude. During the last 10 years, Genbank has grown from 5Gb to over 285Gb[56]. More impressively, a new database dedicated to the open access of short-read sequencing data, the Short Read Archive (SRA), has grown to contain over 60 terabases(1012) of sequence data[57].
Figure 2.
Sequencing cost per megabase since 1971. The sequencing cost per megabase has decreased rapidly since the first 12 bp were sequenced in 1971. The cost is displayed using a logarithmic scale, with key events in the history of sequencing plotted on the curve. Of note, the cost of mutation discovery is dependent on the platform error rate and therefore reductions in cost may be less significant then they appear.
The importance of this development for the future of genetics and medicine is well recognized. The new technologies have been used to sequence hundreds of human genomes, novel organisms, and metagenomes (genomes recovered from environmental samples). Multiple human cancer genomes have been interrogated leading to the discovery of new oncogenes and tumor suppressor genes. With most of the focus on applications directed to cancer genomics and personalized medicine, the potential for MPS (or next-generation sequencing, NGS, as it is often called) to revolutionize mutation research and lead to more sensitive assays to test for mutagen exposure and genome instability has received less attention.
Here, we will address the impact of this new technology on mutation research (Table 2) per se, i.e., the study of the natural tendency of genomes to be unstable and its consequences with respect to evolution and as a cause of disease, aging and death.
Table 2.
Advantages of using high-throughput sequencing for mutation discovery
| Advantages | Disadvantages |
|---|---|
| Direct measurement (as opposed to indirect phenotypic change) | Difficulty in assaying low- abundance mutations |
| Analysis of whole genomes | Sequencing error rates |
| Analysis of transcription-coupled repair | |
| Analysis of mutation localization | |
| More accurate data on mutational spectra |
2.1. Platforms
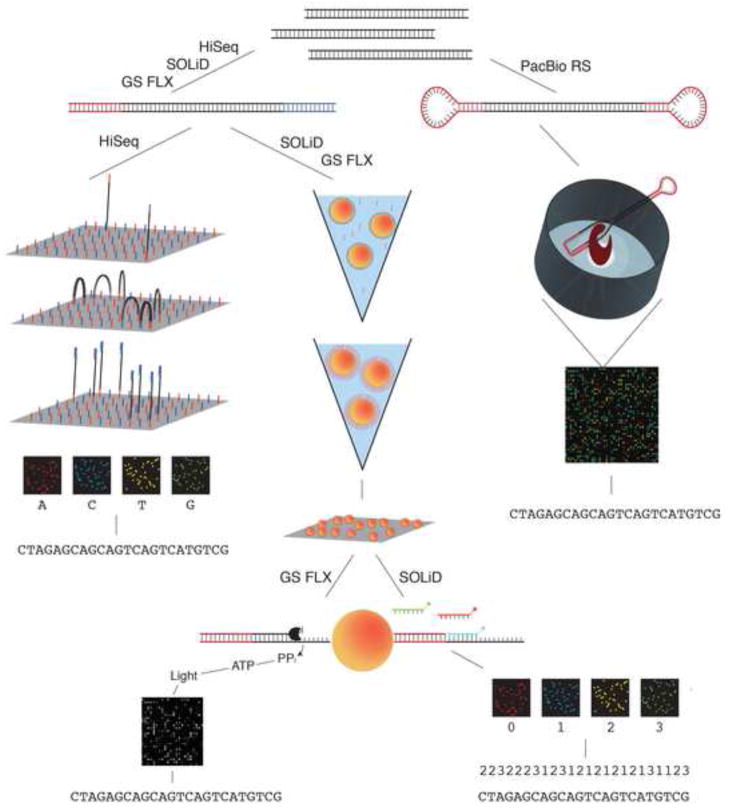
There are currently three routinely used platforms for MPS: the Roche 454 system[58], the Illumina HiSeq[59], and the SOLiD system of Applied Biosystems[60]. A fourth, the PacBio RS single molecule sequencing system of Pacific Biosciences[61], was introduced fairly recently and will be briefly discussed later. The principles behind these four systems are schematically depicted in Figure 3. Additional platforms exist, such as the Polonator[62], the Helicos Single Molecule Sequencer[63], and an in-house only nanoarray-based sequencing-by-ligation technology developed by Complete Genomics[64]. These latter systems remain quite limited in their use and will not be discussed.
Figure 3.
Massively parallel sequencing technologies. A schematic showing sample preparation and sequencing technologies for the four major commercially available sequencers: the GS FLX Titanium by 454 (Roche), the HiSeq by Illumina, the SOLiD system by Life Technologies and the PacBio RS by Pacific Biosciences.
The Roche 454 system employs a “sequencing-by-synthesis” strategy based on pyrosequencing, which detects pyrophosphate molecules as they are cleaved during nucleotide incorporation. Although the system was the first massively parallel technology released, a high error rate at homopolymer sites and a low throughput compared to the two newer platforms has narrowed its applications. It is now primarily used for sequencing metagenomes, e.g., human microbial communities, and for gap filling in de novo sequencing projects, both of which benefit from its long read length (500bp).
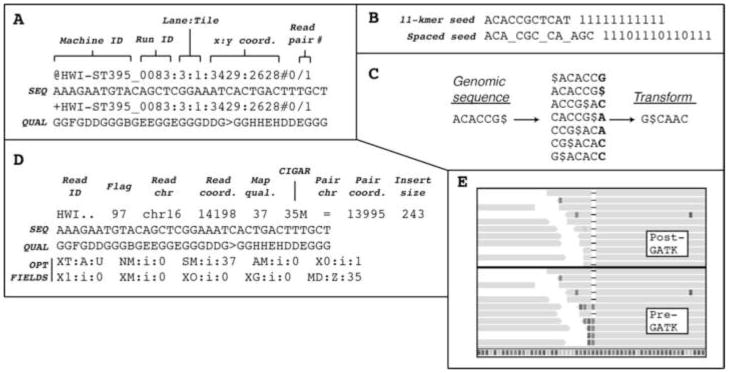
The SOLiD sequencer and the Illumina HiSeq, an updated version of Illumina’s earlier Genome Analyzer, are competing technologies that use different sequencing strategies. In the HiSeq, a library of double-stranded adapter-ligated template molecules between 300 and 600bp in size, constructed from fragmented nucleic acids, is flowed across a hollow glass slide coated on the inside with polyacrylamide to which forward and reverse primers are attached. The adapter-ligated template DNA hybridizes to the primers and is copied onto the flow-cell surface by extension of the flow-cell primer to which it is hybridized. These newly synthesized strands serve as templates for an isothermal amplification reaction, resulting in clusters of amplified strands. One strand is selectively removed before a sequencing primer is hybridized. Sequencing begins using reversible fluorescent terminator dNTPs. Each DNA strand within a cluster incorporates the same, single nucleotide during each chemistry cycle. At the end of every cycle, the clusters are imaged, before the blocking groups and fluorophores on the newly incorporated nucleotides are removed by chemical cleavage and the next round of nucleotide incorporation begins. Four images are the output, one for each fluorophore. A base and associated base quality score (which is estimated using the background fluorophore levels at each cluster on logarithmically linked to error probabilities, similar to Phred quality scores used in Sanger sequencing) are called for each cluster using built-in analysis software. By sequentially sequencing from both ends of a DNA fragment, it is possible to obtain so-called paired end sequences of up to 150 bases each. This entire process (Fig. 3) is described in detail on the Illumina website and in some excellent reviews[65, 66]. The output format for the sequencing files is FASTQ, which contains information on the cluster location, the sequence of called bases, and the associated base-quality scores (Fig. 4A).
Figure 4.
Bioinformatics formats and tools. a. FASTQ format, which uses four lines per read, is the preferred output format for MPS data. The example shown is Illumina FASTQ format, with the read identifier occupying the first and third lines, and the sequence and associated base qualities occupying the second and fourth lines, respectively. b. Spaced seeds are used by modern alignment algorithms in order to improve alignment performance around variants and sequencing errors. c. The Burrows-Wheeler transform, used by the aligners BWA and bowtie, rearranges the order of a sequence in a programmed fashion in order to cluster similar sequence patterns and thereby improve data compression. d. The SAM format is the alignment output for BWA, as well as other programs. It is the preferred format for downstream variant analysis tools. The flag field is a decimal number that has to be interpreted as a 16-bit binary number. It contains information on the read and its alignment that can be used to filter or select for a subset of reads. The CIGAR field gives the location of insertions/deletions as well as the location of clipped bases. e. Reads aligning across a 1-bp deletion are shown before (bottom half) and after (top half) the local realignment step performed by GATK. The realignment helps to reduce false positive SNP calls.
In the SOLiD machine, adapter-ligated template molecules are individually captured on a bead and subjected to PCR. This produces an emulsion of beads each containing thousands of copies of an identical template molecule. The beads flow across a glass slide and are chemically cross-linked to its surface. Instead of sequencing by synthesis, the SOLiD platform exploits so-called 2-base encoding, utilizing a ligation-mediated sequencing reaction. Basically, hexamer probes, which contain a 5′ di-base specific label, are added to the slide during each cycle and are ligated to the 3′ end of a sequencing primer hybridized to the template molecule. Each di-base sequence is matched to a fluorophore that is detected at each subsequent ligation step. Following fluorescence detection, the 3′ base of the incorporated hexamer is removed and the entire process is repeated with a net extension of five bases per cycle. After 10 cycles, the entire synthesized strand is removed and the process is repeated beginning with an “n-1” sequencing primer, which is offset by a single base. The SOLiD machine currently has read lengths of 50bp and has kits designed for single-end reads and paired-end reads for fragments between 600 bp and 10 kbp (the so called mate-pair approach). The SOLiD machine outputs color space data, where a nucleotide is called based on the sequence of two emitted flurophores instead of a single fluorophore. The output format is similar to the FASTQ format in that it contains associated quality scores. However, instead of presenting a sequence of called bases, a string of the numbers 0, 1, 2, and 3 is used, which represent the four different emitted fluorophores. The string of numbers can then be converted into a nucleotide sequence (Fig. 3).
2.2. Alignment
The shift from Sanger sequencing to high-throughput technologies has required new computational approaches to deal with the considerably larger data sets[67]. The first challenge is to match the sequence reads to a reference sequence of the species under study. This so-called sequence alignment profits from the many consensus genome sequences that are now available for a large number of animal and plant species. To process the millions of reads produced by the SOLiD and Illumina machines, alignment algorithms (Table 3) have been optimized for speed and memory usage. Additionally, because the major application of these platforms lies in genome re-sequencing rather than de novo sequencing, the alignment algorithms have been designed for low divergence rates. Expectations for the average number of mismatches between the short read and the reference sequence are driven by the species polymorphism rate and the platform error rate instead of evolutionary divergence. These assumptions have allowed for faster alignments of considerably larger datasets without a large increase in the required computational resources.
Table 3.
MPS data analysis toolkit for mutation research
| Software | Website |
|---|---|
|
Alignment
|
|
| Bowtie | http://bowtie-bio.sourceforge.net/ |
| BWA | http://bio-bwa.sourceforge.net/ |
| SOAP | http://soap.genomics.org.cn/ |
| Novoalign | http://www.novocraft.com/main/index.php |
|
Post-alignment processing
|
|
| SAMtools | http://samtools.sourceforge.net/ |
| GATK | http://www.broadinstitute.org/gsa/wiki/ |
| Picard | http://picard.sourceforge.net |
|
SNP calling
|
|
| SAMtools | http://samtools.sourceforge.net/mpileup.shtml |
| GATK | http://www.broadinstitute.org/gsa/wiki/ |
| SomaticSniper | http://genome.wustl.edu/software/somaticsniper |
| VarScan | http://varscan.sourceforge.net/ |
|
Structural variation calling
|
|
| Breakdancer | http://sourceforge.net/projects/breakdancer/ |
| Breakway | http://breakway.sf.net |
| GASV | http://code.google.com/p/gasv/ |
| SVMerge | http://svmerge.sourceforge.net/ |
| TigraSV | http://sourceforge.net/projects/tigrasv/ |
The two major alignment algorithms used are hash table–based algorithms (BLAST, MAQ[68], Eland[59]) and Burrows Wheeler transform (BWT)-based methods (SOAP2[69], BWA[70], Bowtie[71]). Hash table-based algorithms use an indexing scheme that enables ultra-fast searches for short sequences of a defined length k (k-mer matches or seeds) with up to m mismatches. These seeds are then extended to find the optimal hit. In other words, a small piece of the target sequence or read (k-mer) is aligned to the reference and if a match is found the small piece is extended to see if the whole read matches. Unlike BLAST, which seeds alignments for consecutive matches, the algorithms used by Eland and MAQ utilize spaced seeds (Fig. 4B) to improve sensitivity around polymorphisms and single-base errors.
BWT-based methods also use a hash table, but apply a Burrows Wheeler transform (Fig. 4C), which helps build a more efficient index of either the reference sequence or the sequencing reads, leading to a reduction in the memory-footprint. Currently the gold standard for short-read alignments is BWA, a BWT-based method that is both accurate and fast. BWA produces aligned sequences in the SAM/BAM format, which contains all of the information needed by downstream variant calling programs (Fig. 4D).
2.3. Single nucleotide variants/InDel calling
Ideally, following an alignment one could easily determine mismatches between the genome or genomic region under study and the reference sequence by ‘calling’ the correct base or set of bases at each position in the genomic target. This would allow one to detect single nucleotide variants and small deletions or insertions (InDels). Unfortunately, locus specific differences in sequencing depth, mapping quality scores (a measure of the probability that the read is correctly aligned) and allelic imbalance (where one allele makes up a greater fraction of reads than the second allele), as well as a high frequency of sequencing errors (i.e., in the range of 0.1–1%), necessitate variant calling algorithms that normalize the sequencing data and eliminate sources of error and bias. Although there are many homebrew programs available, developed in bioinformatics groups all over the world, two SNP calling pipelines have dominated modern genotyping (Table 3): the recently published Genome Analysis Tool Kit (GATK)[72], which was used to analyze most of the data from the 1000 Genomes Project, and SAMtools[73]. GATK is a comprehensive package that contains tools for working with aligned BAM files. Its main advantages over other software tools lie in its ability to recalibrate base quality scores, which helps to lower the false positive rate of SNP calling by lowering the base quality scores of specific dinucleotides (and other variables such as homopolymer tracts) that are associated with higher error rates, and its ability to identify small intra-read insertions and deletions (responsible for misaligned reads) and realign the reads at these loci using indel-friendly parameters (the alignment algorithm’s penalties for insertions and deletions will be reduced) leading to cleaner consensus calls (Fig. 4E).
2.4. Structural variation calling
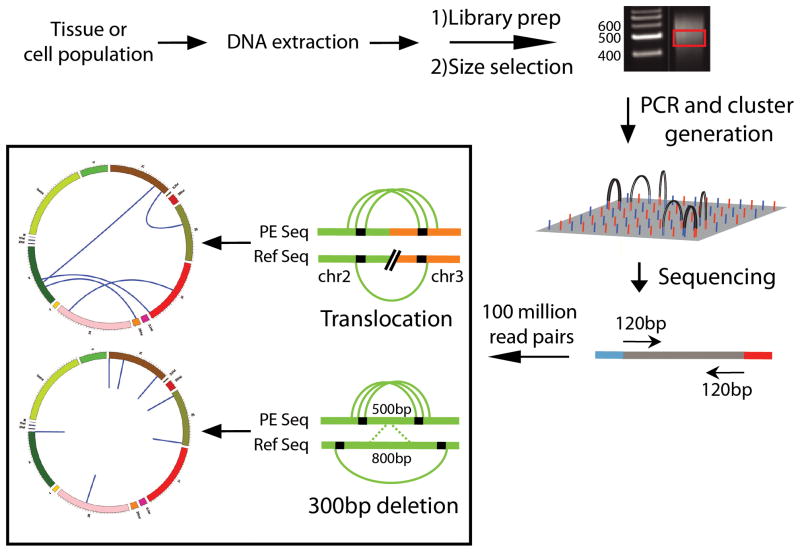
Analysis of structural variation can be performed using paired-end sequencing data from either the Illumina or SOLiD platform. Basically, from the sequencing library of randomly fragmented DNA a particular size class is selected, for example, all fragments of 500 bp ±10bp, and sequenced from both ends. The two sequenced ends should now align to the reference sequence within the 500 bp range. But when, for example, one of the paired reads maps to another chromosome this is taken as evidence for an interchromosomal rearrangement (Fig. 5). Similarly, when the two end sequences align too far out this is evidence for a deletion (Fig. 5) with the opposite situation indicative for an insertion. The read lengths vary between 50 and 150bp depending on the platform and kit used. Alternatively, a mate-paired approach can be used where larger fragments (between 2kb and 10kb) are circularized after their ends have been labeled with biotinylated nucleotides. Following fragmentation of the circularized molecules, the fragments containing biotin groups, representing the circularized ends of the original fragments, are captured on streptavidin magnetic beads. The eluted fragments are sequenced using the standard paired-end module producing paired reads with an inverted orientation. The mate-pair approach produces a considerably higher mapping coverage of the genome from the same number of reads and provides a cost-effective approach for identifying large structural variants genome-wide. Recently two groups used this approach to identify rearrangements in tumor samples[74, 75].
Figure 5.
Schematic depiction of the Illumina protocol for structural variation detection. DNA is extracted from a tissue or cell population and randomly fragmented and gel size-selected to approximately 500bp. Adapters are ligated to both ends of the fragments and an enrichment PCR is used to select for fragments with adapters on both ends. The completed library is then diluted and applied to the Illumina flow cell for cluster generation and sequencing. Both ends of the fragments are sequenced in succession producing paired sequencing reads. The paired reads are compared to a reference sequence to identify genome loci where clusters of read pairs provide evidence of a deletion or a translocation.
Similar to calling single nucleotide variants and InDels, analyzing structural variations is prone to artifacts. A common artifact involves chimeras between two fragments in the library that can be miscalled as genome rearrangements. Chimeras are thought to originate from the ligation reaction during the library preparation that attaches sequencing adapters to all DNA fragments. While the nature of the adapters should prevent such self-ligation, it apparently happens at low frequency. To address this problem one approach[76] uses a series of stringent gel-based size-selected fragments both before and after the ligation reaction. Any chimeras that are produced will be double the size of the selected fragment range and thus will not be retained after the second size selection. Another source of false positive variant calls are incorrectly mapped paired-end reads resulting from multiple single nucleotide errors in a single read. To filter out these artifacts and produce high-quality variant calls, most programs require a cluster of read-pairs supporting any aberration.
A comprehensive and proven package for the detection and characterization of structural variations is yet to be released. The spectrum of structural variation has made it difficult to design a single program that can accurately call all types of structural variation. Instead, three distinct types of calling algorithms have been developed (Table 3) that together, are able to capture the full spectrum of these events.
The most basic algorithm is that applied by Breakdancer[77], Breakway[78], SVDetect[79], BreakSeq[80] and GASV[81]. These programs use mapping distance data provided through the paired-end alignment statistics to estimate the average fragment size of the library. Clusters of aligned reads that appear to be mapping at a distance that is more than three standard deviations away from the average are identified and called as structural variants. The stringency of this algorithm depends on a number of key parameters: the minimum number of reads supporting a cluster (normally >4), the frequency of the event at the locus (commonly an underestimate due to reads that span the breakpoint and thus are not aligned), and the map quality score, which is a measure of the probability that the paired read is correctly aligned. By narrowing the size distribution of fragments that are selected during the library preparation, it is possible to call smaller insertions and deletions. Events that are missed using these algorithms include deletions and insertions smaller than 100bp, insertions larger than 400bp, and some segmental duplications (CNVs). To correctly identify these events, two additional algorithms must be implemented. The first uses an approach[82] similar to that used for the analysis of array-based Comparative Genomic Hybridizatin (aCGH) data. Bins of a width defined by the user are constructed to span the genome, and the number of aligned reads in each bin is recorded. Bins that have an average read depth that is significantly different from the norm can be investigated manually using a genome visualizer and can be validated using aCGH or qPCR. This approach can be used to look for both insertions and deletions and can be used in parallel with a program like Breakdancer to provide additional support for these events.
For the investigation of large insertions, so called orphan reads (only one of the two read pairs map) are extracted from the alignment data and used as input in a local de novo assembly using the assembly software ABySS or Velvet[83]. The assembled fragments are aligned to the reference genome and the alignments are parsed for contigs that provide evidence of insertional breakpoints. An open-source pipeline called SVMerge[84] was recently released that uses the output from the various classes of variant calling algorithms in order to filter and classify a complete set of structural variants. The pipeline runs a de novo local assembly using reads that align at the genome coordinates associated with each identified structural variant. This provides an additional validation step to filter out false positives and identify exact breakpoints.
3. MPS Applications in mutation analysis
3.1. Genome-wide germline mutations
Until recently, our knowledge of the rate and spectra of de novo mutations was limited to studies in a number of reporter genes. The reduced cost and higher throughput of MPS has led to the first genome-wide estimates for germline mutation rates and spectra in yeast[85], worms[86], plants[87], and flies[88]. The experiments, run on both 454 and Illumina machines, used accumulation lines, where inbred populations accumulated mutations over many generations. A wide range of generational time points were used, with an average of 4800 generations from the founder for S. cerevisae, 300 generations for C. elegans, 30 generations for A. thaliana, and 262 generations for D. melanogaster. The criterion for calling germline mutations in any one of the accumulation lines was that the variant must not be present in the composite control, representing the consensus sequence for all other accumulation lines and/or the founder line. Using data on the number of base-substitutions in each line and the number of generations from the founder, a mutation rate was calculated for each species (Table 1). Missing from all four experiments was comprehensive data on structural variants. Although two of the four groups presented data on InDels, the absence of paired-end sequencing data prevented the groups from analyzing large deletions, insertions and segmental-duplications. Decreasing sequencing costs should allow future studies to be performed using shorter generational time points.
3.2. 1000 human genomes
With the completion of the draft sequence of the human genome, the human genetics community has turned to analyzing human genetic variation. The HapMap project, which was started in 2003[89], has genotyped four million Single Nucleotide Polymorphisms (SNPs) in 1301 individuals from eleven populations distributed across the world[90]. The data has provided an abundance of information on common SNPs and their association with human disease, but many variants, including disease-causing variants, that occur at a low allele frequency in the population have been missed. The emergence of next-generation sequencing technologies led to a proposal to sequence the entire genomes of 1000 individuals of different ethnicity. This would provide investigators conducting genome-wide association studies (GWAS) with all variants of at least 1% minor allele frequency in the disease-associated regions. In addition, it allows imputation of many millions of variants identified in the 1000 genomes in the GWAS studies, based on shared haplotype stretches[91, 92]. The project data is available through the NIH and European Bioinformatics Institute (EBI) websites and is updated in real time. So far, data for more than 1100 individuals has been released, with an additional 1400 genomes planned for 2011 (http://www.1000genomes.org/).
Two family trios were sequenced as part of the project. Working with a combination of calling algorithms, de novo mutations arising in the parental gametes were identified. Based on an initial validation using targeted resequencing, 1001 and 669 germline de novo mutations were scored in the offspring from the two families. The majority of these “de novo” mutations represented lymphoblastoid cell-line specific somatic mutations resulting from clonal selection during passaging. Therefore, a second validation was performed using primary DNA sources and by testing for segregation to offspring. This resulted in 35 and 45 “de novo” mutations for the two trios, respectively. Mutation rates were estimated by correcting for the false negative rate of mutation discovery from the three calling algorithms and the false negative rate in the validation experiments (Table 1)[91]. Structural variant analysis did not reveal any confident calls in either trio; only deletions were identified with high sensitivity[92]. It is likely that with improved calling algorithms and higher sequencing coverage, the full spectrum of events will be uncovered.
3.3. Cancer genomics
During the last two decades of the 20th century, advances in cancer genetics were hindered by the limited resolution of available techniques and by an incomplete map of the human genome. With both limitations now overcome, the full spectrum of somatic mutations in cancer genomes has begun to emerge[93]. Since 2002, the number of recognized cancer genes has grown from 115 to 457[94]. Multiple international efforts, including the Cancer Genomes Project[95], the Cancer Genome Atlas[96] and the International Cancer Genome Consortium[97], have set out to uncover new driver mutations and survey the association between mutated genes and drug efficacy.
Traditionally, genome-wide discovery efforts were performed using FISH or array-based methods, leading to the identification of translocations, for example, those common to Chronic Myelogenous Leukemia (CML) and Burkitt’s lymphoma[98, 99]. These assays were limited to analyzing large translocations or copy number changes. In an attempt to identify both somatic and germline mutations in protein coding genes, and thereby discover new oncogenes and tumor suppressor genes, genome resequencing projects have targeted the exome[100], the transcriptome[101] and even the complete genomes of tumor and matched normal tissues[102]. Although some of the early data was generated using capillary sequencing[95], the majority of cancer sequencing projects have relied on MPS[102].
The analysis of these MPS data sets has posed unique challenges. A cancer is a mass of cells that has gone through multiple rounds of selection and clonal expansion[103, 104] and has both heterogeneous and homogeneous components to its mutation profile[105]. Arising from a single cell, with its own spectrum of unique somatic mutations that accumulated over the more than 50 rounds of cell division since fertilization of the egg, a million or more cells arise that share the same set of clonally amplified somatic mutations (both drivers and passengers). In parallel, many low-abundance mutations arise as a consequence of what has been termed a mutator phenotype, resulting from mutations in genes involved in genome maintenance[106]. This creates a large degree of mutational heterogeneity within the tumor. Identification of the clonally amplified mutations is akin to looking for germline events, but investigating the low-abundant random somatic mutations requires new strategies and approaches. Due to this heterogeneity, and the frequent contamination of tumor samples with adjacent normal tissue, allele frequencies do not always fall within standard genotyping windows, i.e. 0.0, 0.5, 1.0. Therefore, standard SNP-calling algorithms have been modified to allow for the full spectrum of genotyping calls. SomaticSniper[107] and VarScan[108] use aligned data from both the tumor sample and matched control sample as input to identify low frequency events specific to the tumor. The combination of these stringent algorithms and the higher coverage available through high-throughput technologies has enabled the identification of mutant alleles present in tumor samples in proportions as low as 0.2% [109]. Translocations and copy-number changes have also been investigated using paired-end sequencing coupled with the aforementioned structural variation callers, e.g., Breakdancer, GASV, BreakSeq. Following the identification of somatic variants in genome-wide studies, investigators have performed large scale resequencing projects to determine whether the gene is mutated at a statistically significant frequency in the cancer subtype, and if so, what the underlying biological mechanism might be [110–113].
The falling costs of high-throughput sequencing, and improvements in statistical methods and data analysis tools, have opened the door to additional medical applications. The Cancer Translation Project was created within the Cancer Genomes Project to investigate the genomics of drug sensitivity in cancer. The first results from the project were released in 2010 and provide evidence of strong correlations between gene-specific mutations and drug responses[114].
High-throughput sequencing is also being used as a tool to monitor disease progression through the identification of cancer-specific rearrangements[74, 75]. After discovering these rearrangements through whole-genome sequencing, simple PCR assays were designed to target the rearrangement signatures in circulating tumor cells in plasma (CTCs) and thereby track disease progression and response to treatments. A mate-paired sequencing approach was used in one of these studies, with a cost of only $2000 per patient[74]. The ability to track the disease in real-time and monitor the effectiveness of different treatments represents a potentially invaluable tool for clinicians that could reduce both the cost and treatment time associated with ineffective chemotherapy drugs.
3.4. Low-abundant, somatic mutations
Heterogeneity among cells in their mutation spectra is not limited to tumors. Due to the inevitability of errors in the processing of DNA damage, mutations accumulate from the zygote through development, adulthood and aging. Life style factors, environmental exposure and the quality of one’s genome maintenance systems determine the rate and severity of this process of somatic mutation accumulation[41]. The stochasticity of mutagenesis with its many low-abundance or even unique mutations has necessitated the use of reporter assays in mutation research. As we have seen, MPS has now essentially broken through the high-cost barrier that thus far essentially constrained whole genome sequencing to identify mutational differences between tissues. However, is it also capable of accurately determining how individual cells in a cell population genetically diverge over time or during cell culture?
Using MPS it is now possible to sequence an entire mammalian genome to find mutational variants present in all or most of the cells of a tissue. This readily allows the determination of mutation rates and spectra in germlines and clonally derived tissues, such as tumors. However, low-abundant, somatic mutations remain a major challenge for two reasons. First, they require a significantly high coverage to identify them. And, second, the nature of MPS, which is based on a consensus model, tends to discard low-abundant variants as potential artifacts.
Due to sequencing errors produced by all contemporary technologies, the identification of somatic variants that are present in a single copy, or a few copies (if clonally amplified), poses many problems. Illumina sequencing runs consistently display a base-pair error rate of 0.05 to 1%[115]. The di-base encoding used by the SOLiD machine is able to lower this error rate to about 0.075% (SOLiD application note). These error rates constitute a background that is several orders of magnitude higher than the recorded somatic base-pair mutation rate and therefore low-abundance somatic events are effectively masked.
Similarly, the detection of random somatic rearrangements by paired-end sequencing is limited by the generation of chimeric sequences, i.e., ligation of two genomic sequences to each other, during the library preparation[76]. Normally, the DNA sample is subjected to random fragmentation, after which the DNA fragments are end-polished and appended with an A-overhang, which promotes preferential annealing with the T-overhang-containing sequencing adapters and precludes cross-ligation. However, as already discussed above, cross-ligation does occur at a very low rate. Such artifacts are not a problem in consensus-based procedures, since unique rearrangement events are discarded, but for somatic mutation research they will lead to high false positive rates.
There are essentially two types of approach to overcome these problems. First, one could try to decrease error rates, either experimentally or through the application of in silico filters. For example, polymerase errors that arise during template preparation (during the pre-amplification step required for library preparation) or the solid-phase amplification on the instrument can be reduced by using high-fidelity enzymes, such as the phusion DNA polymerase. Also machine errors can be expected to decrease further in the future by advances in chemistry and fluidics. As mentioned above, to reduce false positive genome rearrangement calls in the form of chimeras between two unrelated fragments it is possible to apply a more stringent size selection. Ultimately, the solution to sequencing-related errors is single molecule sequencing; multiple passes of the same molecule will quickly eliminate random errors. Such a system is now available in the form of PacBio’s single molecule, real-time sequencing technology, but is as yet immature for this purpose (see below). To eliminate or greatly reduce errors it is also possible to apply sophisticated algorithms that filter out errors. This can only be done with errors that exhibit some consistent patterns related to the library preparation and sequencing steps[116] and not feasible for errors that appear at random. Interestingly, a recent approach to the identification of rare variants utilized the PCR amplification step in the (Illumina) library preparation to identify families of templates carrying the same mutation, which then could be inferred to have been present in the original template molecule as a real mutation[115]
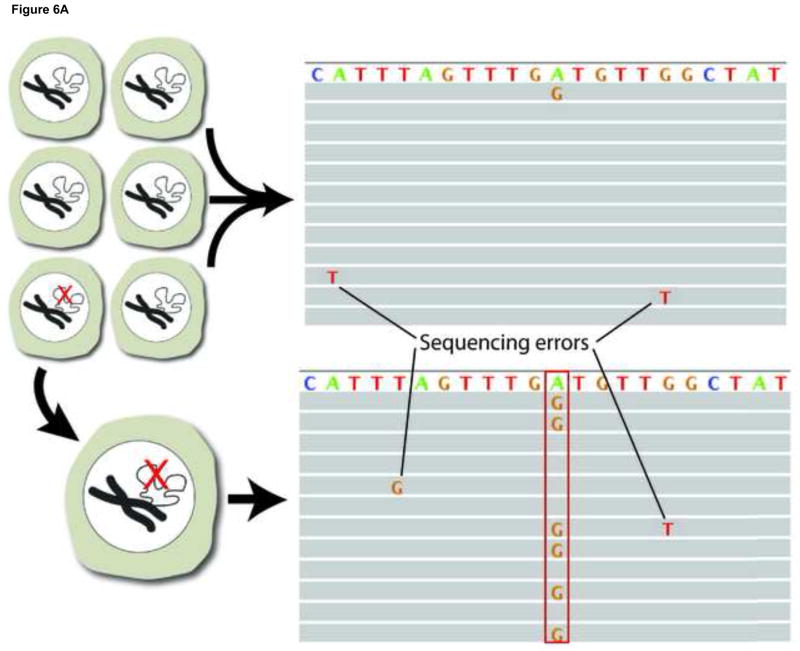
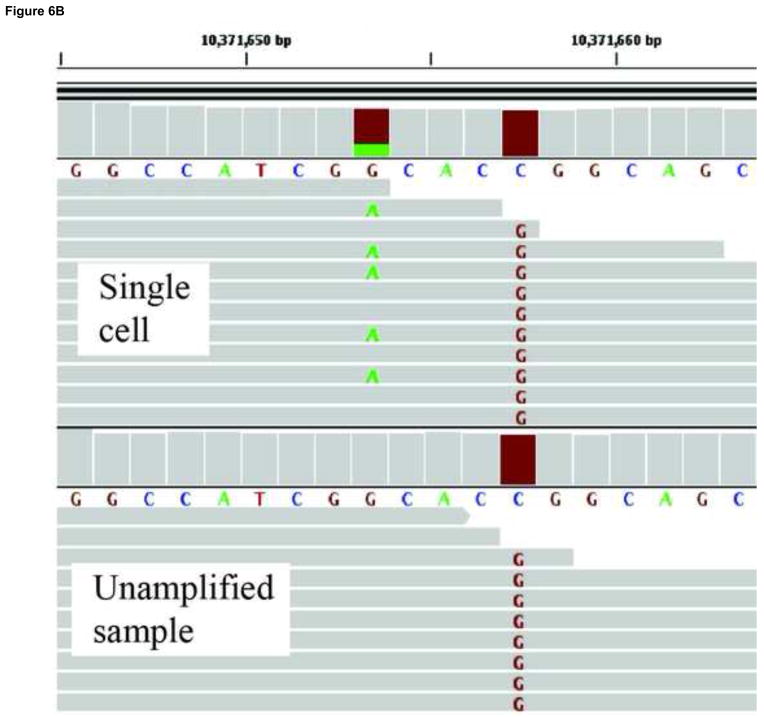
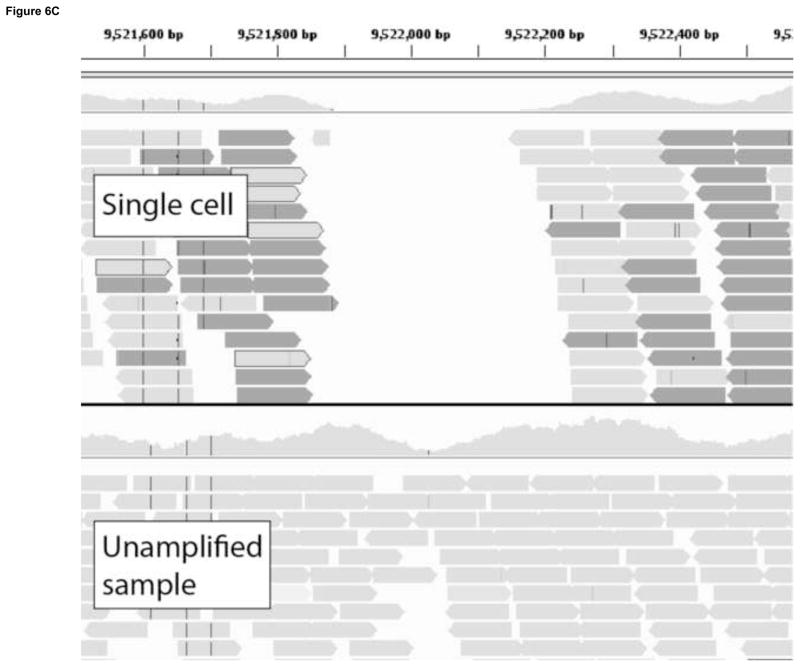
A second approach for avoiding sequencing errors as confounders in the identification of low-abundant mutations is to use single cells. To sequence the genomes of single cells rather than mixtures of genomes from whole cell populations or tissues is critically important in its own right[117]. However, it also allows one to circumvent the problem of sequencing errors by adopting the consensus model after single-cell, whole-genome amplification (WGA). Multiple protocols exist for whole genome amplification[118, 119], the most common of which is Multiple Displacement Amplification (MDA), which uses a high-fidelity isothermal polymerase (phi29) to generate up to 20μg of DNA product after starting from a single cell. Working with single cells, it is possible to follow the consensus model in next-generation sequencing in which mutations can be called in the single cell on the basis of their occurrence in multiple reads (Fig. 6A), not unlike current procedures for identifying mutations in tumors. In our lab, we use an MDA-based protocol where single cells are subjected to whole genome amplification followed by paired-end sequencing. An unamplified sample, representing the population as a whole, is also sequenced. After the alignment to a reference sequence, a three-way comparison is made between the reference sequence, the unamplified sample and the amplified single cells. Both germline and somatic mutations can be recorded(Fig. 6B,C).
Figure 6.
Somatic mutation detection using single cell sequencing. a. Somatic mutations in tissues are rare and therefore found only in single sequencing reads from which they are routinely filtered out as sequencing errors during post-alignment processing. Adopting a single cell approach overcomes this limitation by transforming each somatic event into a consensus variant call. b. An example of a somatically acquired G->A point mutation identified using single cell sequencing. The top panel shows sequencing reads obtained from a single cell, a fraction of which contain the mutant allele. The bottom panel shows sequencing reads obtained from the unamplified population, which do not show evidence of the mutant base. A homozygous SNP specific to the cell-line (C->G) is also shown, and as expected, is found in all reads in both the single cell and the cell population samples. c. An example of a somatically acquired deletion identified using single cell sequencing. The top panel shows sequencing reads from the single cell, with shaded box-arrows representing reads that map across the deleted segment. The bottom panel shows sequencing reads obtained from the unamplified population, which do not show evidence of any deletion.
The success of any single cell genomics approach is heavily dependent on the development of accurate variant calling algorithms that can correct for the vast differences in coverage, allele dropout, and locus dropout that are produced by whole genome amplification procedures. Multiple aCGH experiments[36, 37], and recently a MPS experiment[104], have used single-cell approaches to profile CNVs in individual human normal and tumor cells, but base-pair resolution genotyping of single eukaryotic cells has not yet been shown. Massively parallel sequencing of single-cells has the potential to revolutionize reproductive medicine, cancer research, developmental biology and aging research.
4. Future prospects
Almost immediately following the release of the Illumina and SOLiD platforms, reports appeared regarding third-generation sequencing. Pacific Biosciences’ PacBio RS detects fluorescently labeled nucleotides in real time as they are incorporated during a second-strand synthesis (Fig. 3)[63, 120]. Oxford Nanopore’s sequencing platform uses an exonuclease cleavage reaction and a protein nanopore to sequence individual cleaved bases by a unique electrical signature as they are transported through the pore[121].
The Pacific Biosciences instrument was made available to early access customers in September 2010. The instrument has a quicker turn-around time (approximately 100MB of sequence is produced during a 90 minute run), and longer read lengths (average of 2000bp, with 5% exceeding 5000bp) than the Illumina and SOLiD platforms, but the throughput is only 1 Gb/day and the error rate is approximately 20%. To compensate for the high per base error rate, the sample preparation, which is similar to the Illumina protocol but uses hairpin adapters, was designed to allow for multiple sequencing rounds across the same site in the template molecule, lowering the error rate to 4% with two passes, 0.8% with three passes and 0.16% with 4 passes. When designing experiments with this platform, a trade-off must be made between sequencing 2000bp fragments at 80% accuracy or 500bp fragments at 99.8% accuracy[122]
The limiting factor in the length of PacBio RS sequencing reads is the half-life of the polymerase, which is damaged by the laser-induced photochemistry used in base calling. An alternative protocol, termed “strobe sequencing” has been revealed by the company that would allow for gapped sequences covering up to 10kb. Results from a simulation[123] showed that using strobe reads instead of classical paired-end reads for calling inversions and deletions larger than 120bp improved the sensitivity while reducing the false positive rate by more than 50%. Because the experiment was run under the assumption that the base-pair error rate was 5%, instead of 20%, it will be necessary to validate the results by repeating the experiment.
The SOLiD and Illumina platforms, which can produce up to 75GB a day, will remain the first choice for the majority of applications. For genome-wide mutation detection in mammalian species, high throughput and high accuracy is a prerequisite. Although the PacBio RS instrument could help identify germline structural variants as a supplement to a whole genome run with the Illumina or SOLiD, it does not have a high enough throughput to be used for the detection of random somatic variants.
Oxford Nanopore has two sequencing strategies in development, both of which will likely enable almost unlimited read lengths. The technology closest to the market is exonuclease-based[121] and will be marketed, sold, distributed, and serviced by Illumina. The alternative technology, termed ‘strand sequencing’ [124] may be capable of rereading the same strand multiple times. It remains to be seen what the throughput and error rate of the instruments will be, but due to a lower reagent cost and the absence of a library preparation, the cost of the assay will most likely be significantly lower than current technologies.
The study of genetic variation has progressed rapidly over the last twenty years and because of recent advances in sequencing technology, the rate of discovery is faster than ever. Classical tools for the detection and characterization of germline and clonally amplified somatic variants are being replaced by a new array of massively parallel sequencing based methods. The characterization of genome-wide genetic variants in individual humans has become an important tool in the diagnosis of congenital malformations[125, 126], especially with recent advances in non-invasive prenatal genetic testing[127], and has led to the discovery of novel genes and pathways associated with human disease[128, 129]. Applications in cancer research and clinical oncology are even more pertinent, where MPS has already impacted oncogene discovery, clinical diagnosis, pharmacogenomics, and the monitoring of disease progression. As we enter the age of personalized genomics there remain gaps in our knowledge, none greater than a lack of understanding of the rate and spectra of random somatic variation in our tissues. The accumulation of random alterations in the genome sequence of our cells might have profound functional consequences that have been ignored because of a focus on the average (or consensus) level of gene and protein expression in our tissues. Although there are limitations with the technology as it stands, advances in single-cell genomics techniques and the development of methods to lower the intrinsic error rates of MPS technology and filter out false positives will lead to increasingly sensitive and specific assays for measuring low-abundant somatic mutations. Additionally, single-molecule sequencing technologies have begun to emerge that may have lower intrinsic error rates than the current platforms, which would not interfere with the detection of low-abundant somatic variants.
Acknowledgments
This work was supported by AG034421, ES019520 and AG17242.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch M. The cellular, developmental and population-genetic determinants of mutation-rate evolution. Genetics. 2008;180:933–943. doi: 10.1534/genetics.108.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic acids research. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castle WE. The Mutation Theory of Organic Evolution, from the Standpoint of Animal Breeding. Science. 1905;21:521–525. doi: 10.1126/science.21.536.521. [DOI] [PubMed] [Google Scholar]
- 5.Muller HJ. Further changes in the white-eye series of Drosophila and their bearing on the manner of occurrence of mutation. Journal of Experimental Zoology. 1920;31:443–474. [Google Scholar]
- 6.Muller HJ. The Production of Mutations by X-Rays. Proc Natl Acad Sci U S A. 1928;14:714–726. doi: 10.1073/pnas.14.9.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haldane JBS. A Mathematical Theory of Natural and Artificial Selection, Part V: Selection and Mutation. Mathematical Proceedings of the Cambridge Philosophical Society. 1927;23:838–844. [Google Scholar]
- 8.Haldane JB. The rate of spontaneous mutation of a human gene. J Genet. 1935;83:235–244. doi: 10.1007/BF02717892. [DOI] [PubMed] [Google Scholar]
- 9.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 10.Yang HP, Tanikawa AY, Kondrashov AS. Molecular nature of 11 spontaneous de novo mutations in Drosophila melanogaster. Genetics. 2001;157:1285–1292. doi: 10.1093/genetics/157.3.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Human mutation. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 12.Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 13.Neel JV, Satoh C, Goriki K, Asakawa J, Fujita M, Takahashi N, Kageoka T, Hazama R. Search for mutations altering protein charge and/or function in children of atomic bomb survivors: final report. American journal of human genetics. 1988;42:663–676. [PMC free article] [PubMed] [Google Scholar]
- 14.Russell LB, Russell WL. Spontaneous mutations recovered as mosaics in the mouse specific-locus test. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13072–13077. doi: 10.1073/pnas.93.23.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller HJ. The Measurement of Gene Mutation Rate in Drosophila, Its High Variability, and Its Dependence upon Temperature. Genetics. 1928;13:279–357. doi: 10.1093/genetics/13.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukai T. The Genetic Structure of Natural Populations of Drosophila Melanogaster. I. Spontaneous Mutation Rate of Polygenes Controlling Viability. Genetics. 1964;50:1–19. doi: 10.1093/genetics/50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keightley PD, Eyre-Walker A. Terumi Mukai and the riddle of deleterious mutation rates. Genetics. 1999;153:515–523. doi: 10.1093/genetics/153.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vrana M, Rudikoff S, Potter M. Sequence variation among heavy chains from inulin-binding myeloma proteins. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:1957–1961. doi: 10.1073/pnas.75.4.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:5631–5635. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson DA, Symons RH, Berg P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:2904–2909. doi: 10.1073/pnas.69.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullrich A, Dull TJ, Gray A, Brosius J, Sures I. Genetic variation in the human insulin gene. Science. 1980;209:612–615. doi: 10.1126/science.6248962. [DOI] [PubMed] [Google Scholar]
- 22.Eng C, Vijg J. Genetic testing: the problems and the promise. Nat Biotechnol. 1997;15:422–426. doi: 10.1038/nbt0597-422. [DOI] [PubMed] [Google Scholar]
- 23.Vijg JSY. Screening for mutations in cancer predisposition genes. In: Eeles RA, editor. Genetic predisposition to cancer, Arnold. Oxford University Press; London New York, NY: 2004. p. xxii.p. 441. Distributed in the U.S.A. [Google Scholar]
- 24.Fischer SG, Lerman LS. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983;80:1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riesner D, Steger G, Zimmat R, Owens RA, Wagenhofer M, Hillen W, Vollbach S, Henco K. Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis. 1989;10:377–389. doi: 10.1002/elps.1150100516. [DOI] [PubMed] [Google Scholar]
- 26.Hestekin CN, Barron AE. The potential of electrophoretic mobility shift assays for clinical mutation detection. Electrophoresis. 2006;27:3805–3815. doi: 10.1002/elps.200600421. [DOI] [PubMed] [Google Scholar]
- 27.Bounpheng M, McGrath S, Macias D, van Orsouw N, Suh Y, Rines D, Vijg J. Rapid, inexpensive scanning for all possible BRCA1 and BRCA2 gene sequence variants in a single assay: implications for genetic testing. J Med Genet. 2003;40:e33. doi: 10.1136/jmg.40.4.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD. Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic acids research. 1998;26:1396–1400. doi: 10.1093/nar/26.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Human mutation. 2009;30:857–859. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- 30.Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- 31.Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- 32.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, Hansmann ML, Haralambieva E, Harder L, Hasenclever D, Kuhn M, Lenze D, Lichter P, Martin-Subero JI, Moller P, Muller-Hermelink HK, Ott G, Parwaresch RM, Pott C, Rosenwald A, Rosolowski M, Schwaenen C, Sturzenhofecker B, Szczepanowski M, Trautmann H, Wacker HH, Spang R, Loeffler M, Trumper L, Stein H, Siebert R. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 33.Przybytkowski E, Ferrario C, Basik M. The use of ultra-dense array CGH analysis for the discovery of micro-copy number alterations and gene fusions in the cancer genome. BMC Med Genomics. 2011;4:16. doi: 10.1186/1755-8794-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 35.Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, Gordon A, Wells D. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod. 2011;26:480–490. doi: 10.1093/humrep/deq344. [DOI] [PubMed] [Google Scholar]
- 36.Le Caignec C, Spits C, Sermon K, De Rycke M, Thienpont B, Debrock S, Staessen C, Moreau Y, Fryns JP, Van Steirteghem A, Liebaers I, Vermeesch JR. Single-cell chromosomal imbalances detection by array CGH. Nucleic acids research. 2006;34:e68. doi: 10.1093/nar/gkl336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiegler H, Geigl JB, Langer S, Rigler D, Porter K, Unger K, Carter NP, Speicher MR. High resolution array-CGH analysis of single cells. Nucleic acids research. 2007;35:e15. doi: 10.1093/nar/gkl1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordling CO. A new theory on cancer-inducing mechanism. Br J Cancer. 1953;7:68–72. doi: 10.1038/bjc.1953.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szilard L. On the Nature of the Aging Process. Proceedings of the National Academy of Sciences of the United States of America. 1959;45:30–45. doi: 10.1073/pnas.45.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijg J. Aging of the genome: the dual role of the DNA in life and death. Oxford University Press; Oxford; New York: 2007. [Google Scholar]
- 42.Erickson R. Somatic gene mutation and human disease other than cancer. Mutation Research/Reviews in Mutation Research. 2003;543:125–136. doi: 10.1016/s1383-5742(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 43.McClintock B. A Correlation of Ring-Shaped Chromosomes with Variegation in Zea Mays. Proceedings of the National Academy of Sciences of the United States of America. 1932;18:677–681. doi: 10.1073/pnas.18.12.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vijg J, van Steeg H. Transgenic assays for mutations and cancer: current status and future perspectives. Mutation research. 1998;400:337–354. doi: 10.1016/s0027-5107(98)00030-x. [DOI] [PubMed] [Google Scholar]
- 45.O’Neill JP, Sullivan LM, Albertini RJ. In vitro induction, expression and selection of thioguanine-resistant mutants with human T-lymphocytes. Mutation research. 1990;240:135–142. doi: 10.1016/0165-1218(90)90017-v. [DOI] [PubMed] [Google Scholar]
- 46.Albertini RJ. HPRT mutations in humans: biomarkers for mechanistic studies. Mutation research. 2001;489:1–16. doi: 10.1016/s1383-5742(01)00064-3. [DOI] [PubMed] [Google Scholar]
- 47.Cervantes RB, Stringer JR, Shao C, Tischfield JA, Stambrook PJ. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gossen JA, de Leeuw WJ, Tan CH, Zwarthoff EC, Berends F, Lohman PH, Knook DL, Vijg J. Efficient rescue of integrated shuttle vectors from transgenic mice: a model for studying mutations in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boerrigter ME, Dolle ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377:657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- 50.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, Putman DL, Short JM. Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahabir AG, Zwart E, Schaap M, van Benthem J, de Vries A, Hernandez LG, Hendriksen CF, van Steeg H. lacZ mouse embryonic fibroblasts detect both clastogens and mutagens. Mutation research. 2009;666:50–56. doi: 10.1016/j.mrfmmm.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Vijg J, Dolle ME. Large genome rearrangements as a primary cause of aging. Mech Ageing Dev. 2002;123:907–915. doi: 10.1016/s0047-6374(02)00028-3. [DOI] [PubMed] [Google Scholar]
- 53.Dolle ME, Vijg J. Genome dynamics in aging mice. Genome Res. 2002;12:1732–1738. doi: 10.1101/gr.125502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Busuttil RA, Garcia AM, Reddick RL, Dolle ME, Calder RB, Nelson JF, Vijg J. Intra-organ variation in age-related mutation accumulation in the mouse. PLoS One. 2007;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan EY. Advances in sequencing technology. Mutation research. 2005;573:13–40. doi: 10.1016/j.mrfmmm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic acids research. 2011;39:D32–37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J. Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2011;39:D38–51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227–240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 62.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 63.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 64.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen L, Chernikoff D, Cheung A, Chirita R, Curson B, Ebert JC, Hacker CR, Hartlage R, Hauser B, Huang S, Jiang Y, Karpinchyk V, Koenig M, Kong C, Landers T, Le C, Liu J, McBride CE, Morenzoni M, Morey RE, Mutch K, Perazich H, Perry K, Peters BA, Peterson J, Pethiyagoda CL, Pothuraju K, Richter C, Rosenbaum AM, Roy S, Shafto J, Sharanhovich U, Shannon KW, Sheppy CG, Sun M, Thakuria JV, Tran A, Vu D, Zaranek AW, Wu X, Drmanac S, Oliphant AR, Banyai WC, Martin B, Ballinger DG, Church GM, Reid CA. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 65.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 66.Quail MA, Swerdlow H, Turner DJ. Curr Protoc Hum Genet. Unit 18. Chapter 18. 2009. Improved protocols for the illumina genome analyzer sequencing system; p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6:S6–S12. doi: 10.1038/nmeth.1376. [DOI] [PubMed] [Google Scholar]
- 68.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr, Velculescu VE. Development of personalized tumor biomarkers using massively parallel sequencing. Science Translational Medicine. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, Hamalainen E, Stebbings LA, Andersson LC, Flanagan AM, Durbecq V, Ignatiadis M, Kallioniemi O, Heckman CA, Alitalo K, Edgren H, Futreal PA, Stratton MR, Campbell PJ. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer. 2010;49:1062–1069. doi: 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clark MJ, Homer N, O’Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeitouni B, Boeva V, Janoueix-Lerosey I, Loeillet S, Legoix-ne P, Nicolas A, Delattre O, Barillot E. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics. 2010;26:1895–1896. doi: 10.1093/bioinformatics/btq293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lam HY, Mu XJ, Stutz AM, Tanzer A, Cayting PD, Snyder M, Kim PM, Korbel JO, Gerstein MB. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sindi S, Helman E, Bashir A, Raphael BJ. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25:i222–230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Medvedev P, Fiume M, Dzamba M, Smith T, Brudno M. Detecting copy number variation with mated short reads. Genome Res. 2010;20:1613–1622. doi: 10.1101/gr.106344.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hajirasouliha I, Hormozdiari F, Alkan C, Kidd JM, Birol I, Eichler EE, Sahinalp SC. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics. 2010;26:1277–1283. doi: 10.1093/bioinformatics/btq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong K, Keane TM, Stalker J, Adams DJ. Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome Biol. 2010;11:R128. doi: 10.1186/gb-2010-11-12-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, Thomas WK. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledo JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M, Baer CF. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16310–16314. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keightley PD, Trivedi U, Thomson M, Oliver F, Kumar S, Blaxter ML. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 90.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, deBakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu Y, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang S, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HY, Leng J, Li R, Li Y, Lin CY, Luo R, Mu XJ, Nemesh J, Peckham HE, Rausch T, Scally A, Shi X, Stromberg MP, Stutz AM, Urban AE, Walker JA, Wu J, Zhang Y, Zhang ZD, Batzer MA, Ding L, Marth GT, McVean G, Sebat J, Snyder M, Wang J, Eichler EE, Gerstein MB, Hurles ME, Lee C, McCarroll SA, Korbel JO. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 95.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, Guttmacher A, Guyer M, Hemsley FM, Jennings JL, Kerr D, Klatt P, Kolar P, Kusada J, Lane DP, Laplace F, Youyong L, Nettekoven G, Ozenberger B, Peterson J, Rao TS, Remacle J, Schafer AJ, Shibata T, Stratton MR, Vockley JG, Watanabe K, Yang H, Yuen MM, Knoppers BM, Bobrow M, Cambon-Thomsen A, Dressler LG, Dyke SO, Joly Y, Kato K, Kennedy KL, Nicolas P, Parker MJ, Rial-Sebbag E, Romeo-Casabona CM, Shaw KM, Wallace S, Wiesner GL, Zeps N, Lichter P, Biankin AV, Chabannon C, Chin L, Clement B, de Alava E, Degos F, Ferguson ML, Geary P, Hayes DN, Johns AL, Kasprzyk A, Nakagawa H, Penny R, Piris MA, Sarin R, Scarpa A, van de Vijver M, Futreal PA, Aburatani H, Bayes M, Botwell DD, Campbell PJ, Estivill X, Grimmond SM, Gut I, Hirst M, Lopez-Otin C, Majumder P, Marra M, McPherson JD, Ning Z, Puente XS, Ruan Y, Stunnenberg HG, Swerdlow H, Velculescu VE, Wilson RK, Xue HH, Yang L, Spellman PT, Bader GD, Boutros PC, Flicek P, Getz G, Guigo R, Guo G, Haussler D, Heath S, Hubbard TJ, Jiang T, Jones SM, Li Q, Lopez-Bigas N, Luo R, Muthuswamy L, Ouellette BF, Pearson JV, Quesada V, Raphael BJ, Sander C, Speed TP, Stein LD, Stuart JM, Teague JW, Totoki Y, Tsunoda T, Valencia A, Wheeler DA, Wu H, Zhao S, Zhou G, Lathrop M, Thomas G, Yoshida T, Axton M, Gunter C, Miller LJ, Zhang J, Haider SA, Wang J, Yung CK, Cross A, Liang Y, Gnaneshan S, Guberman J, Hsu J, Chalmers DR, Hasel KW, Kaan TS, Lowrance WW, Masui T, Rodriguez LL, Vergely C, Bowtell DD, Cloonan N, deFazio A, Eshleman JR, Etemadmoghadam D, Gardiner BA, Kench JG, Sutherland RL, Tempero MA, Waddell NJ, Wilson PJ, Gallinger S, Tsao MS, Shaw PA, Petersen GM, Mukhopadhyay D, DePinho RA, Thayer S, Shazand K, Beck T, Sam M, Timms L, Ballin V, Lu Y, Ji J, Zhang X, Chen F, Hu X, Yang Q, Tian G, Zhang L, Xing X, Li X, Zhu Z, Yu Y, Yu J, Tost J, Brennan P, Holcatova I, Zaridze D, Brazma A, Egevard L, Prokhortchouk E, Banks RE, Uhlen M, Viksna J, Ponten F, Skryabin K, Birney E, Borg A, Borresen-Dale AL, Caldas C, Foekens JA, Martin S, Reis-Filho JS, Richardson AL, Sotiriou C, Thoms G, van’t Veer L, Birnbaum D, Blanche H, Boucher P, Boyault S, Masson-Jacquemier JD, Pauporte I, Pivot X, Vincent-Salomon A, Tabone E, Theillet C, Treilleux I, Bioulac-Sage P, Decaens T, Franco D, Gut M, Samuel D, Zucman-Rossi J, Eils R, Brors B, Korbel JO, Korshunov A, Landgraf P, Lehrach H, Pfister S, Radlwimmer B, Reifenberger G, Taylor MD, von Kalle C, Majumder PP, Pederzoli P, Lawlor RA, Delledonne M, Bardelli A, Gress T, Klimstra D, Zamboni G, Nakamura Y, Miyano S, Fujimoto A, Campo E, de Sanjose S, Montserrat E, Gonzalez-Diaz M, Jares P, Himmelbaue H, Bea S, Aparicio S, Easton DF, Collins FS, Compton CC, Lander ES, Burke W, Green AR, Hamilton SR, Kallioniemi OP, Ley TJ, Liu ET, Wainwright BJ. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 99.Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maher B. Exome sequencing takes centre stage in cancer profiling. Nature. 2009;459:146–147. doi: 10.1038/459146b. [DOI] [PubMed] [Google Scholar]
- 101.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A, Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC, Graubert TA, DiPersio JF, Wilson RK. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Salk JJ, Salipante SJ, Risques RA, Crispin DA, Li L, Bronner MP, Brentnall TA, Rabinovitch PS, Horwitz MS, Loeb LA. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20871–20876. doi: 10.1073/pnas.0909428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: origin and consequences. Annu Rev Pathol. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet. 2010;19:R188–196. doi: 10.1093/hmg/ddq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, O’Neill K, Sasaki H, Lindeman N, Wong KK, Borras AM, Gutmann EJ, Dragnev KH, DeBiasi R, Chen TH, Glatt KA, Greulich H, Desany B, Lubeski CK, Brockman W, Alvarez P, Hutchison SK, Leamon JH, Ronan MT, Turenchalk GS, Egholm M, Sellers WR, Rothberg JM, Meyerson M. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 110.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 115.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y, Ishikawa S, Linak MC, Hirai A, Takahashi H, Altaf-Ul-Amin M, Ogasawara N, Kanaya S. Sequence-specific error profile of Illumina sequencers. Nucleic acids research. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kalisky T, Quake SR. Single-cell genomics. Nat Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 118.Panelli S, Damiani G, Espen L, Micheli G, Sgaramella V. Towards the analysis of the genomes of single cells: further characterisation of the multiple displacement amplification. Gene. 2006;372:1–7. doi: 10.1016/j.gene.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 119.Geigl JB, Speicher MR. Single-cell isolation from cell suspensions and whole genome amplification from single cells to provide templates for CGH analysis. Nat Protoc. 2007;2:3173–3184. doi: 10.1038/nprot.2007.476. [DOI] [PubMed] [Google Scholar]
- 120.Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic acids research. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 122.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. Origins of the E. coli Strain Causing an Outbreak of Hemolytic-Uremic Syndrome in Germany. N Engl J Med. 2011 doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ritz A, Bashir A, Raphael BJ. Structural variation analysis with strobe reads. Bioinformatics. 2010;26:1291–1298. doi: 10.1093/bioinformatics/btq153. [DOI] [PubMed] [Google Scholar]
- 124.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maxmen A. Exome sequencing deciphers rare diseases. Cell. 2011;144:635–637. doi: 10.1016/j.cell.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 127.Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RW. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Science Translational Medicine. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 128.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]