Abstract
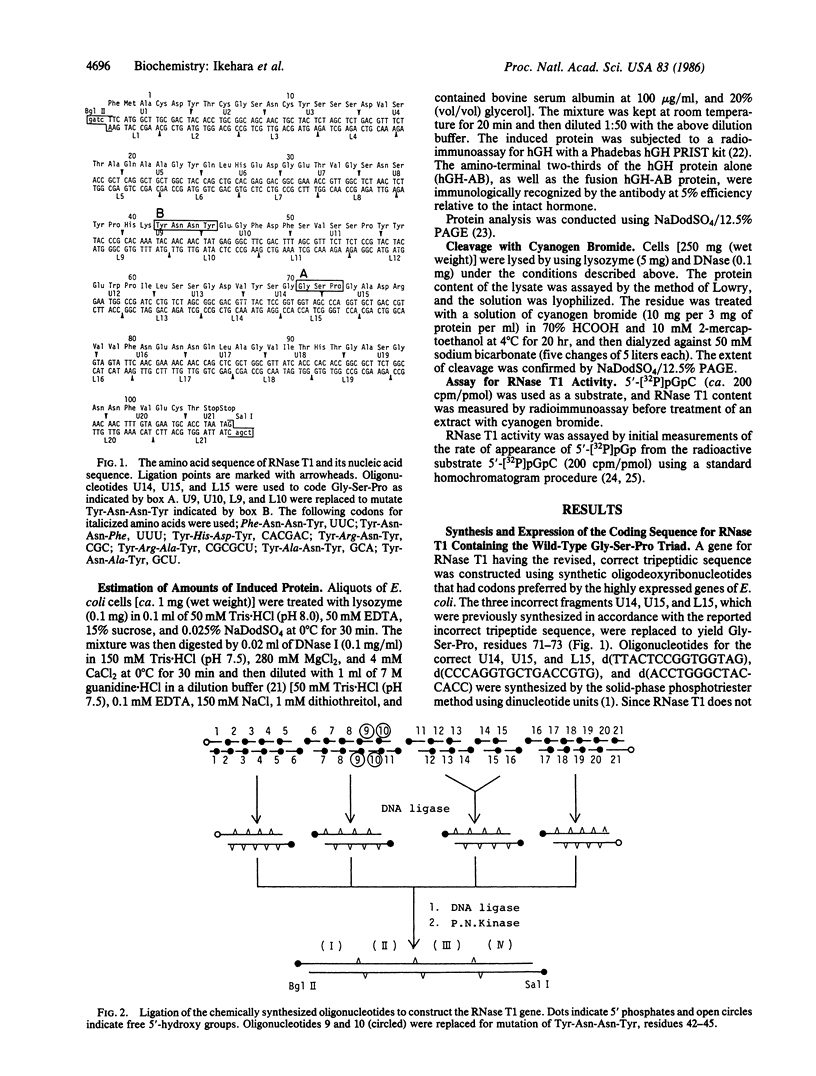
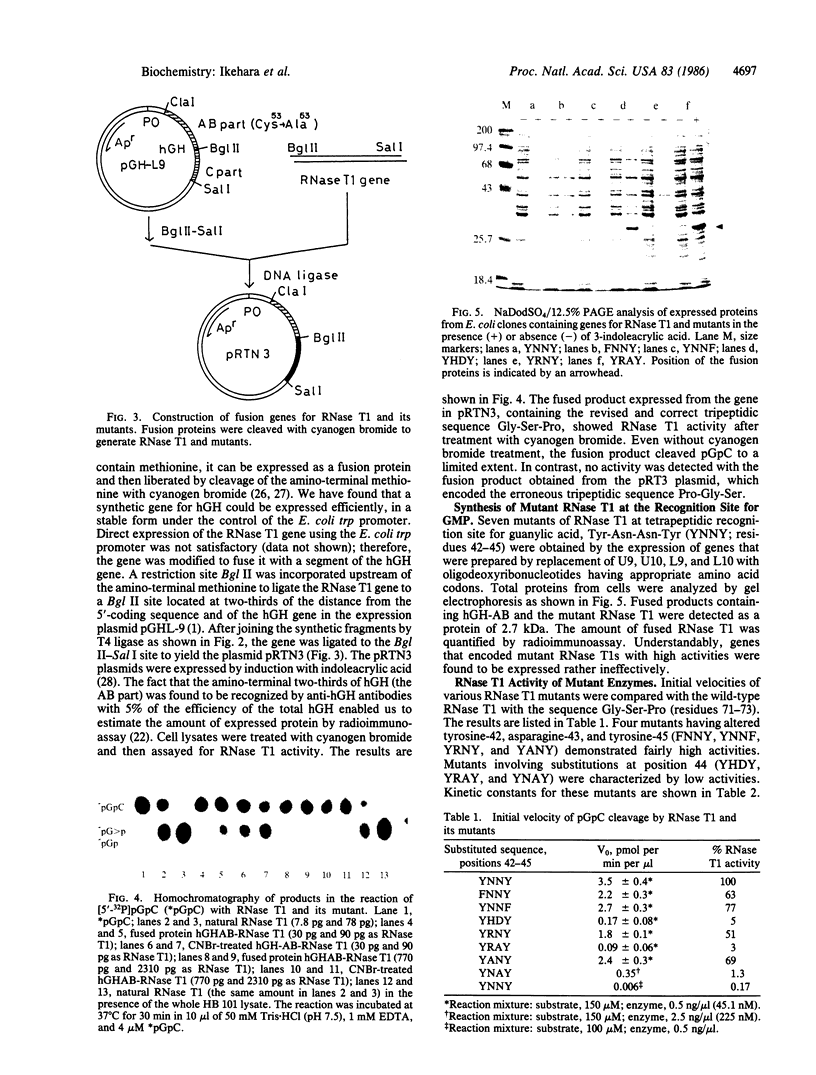
The genes for ribonuclease T1 and its site-specific mutants were chemically synthesized and introduced to Escherichia coli. All enzymes were fusion products produced by joining the synthetic gene at specific restriction sites to the synthetic gene for human growth hormone in a plasmid containing the E. coli trp promoter. The fusion protein from this plasmid contained 66% of the amino-terminal sequences of the human growth hormone, which were recognizable immunologically. RNase T1 or its mutants were cleaved from the fusion protein with cyanogen bromide. The synthetic RNase T1 endowed with the revised wild-type triad Gly-Ser-Pro, residues 71-73, was fully functional, readily hydrolyzing pGpC bonds, whereas a mutant enzyme having the originally reported, erroneous triad Pro-Gly-Ser was totally inactive. Various amino acid substitutions were also introduced to the guanosine recognition region comprised of residues 42-45, Tyr-Asn-Asn-Tyr. Substitution of either of the tyrosine residues noted above with phenylalanine had no dramatic effect on the enzyme's function. Replacement of asparagine-43 with arginine or alanine also caused only a small change in the hydrolyzing activity--a mutant enzyme maintained greater than 50% of the wild-type activity. In sharp contrast, when aspartic acid or alanine was substituted for asparagine-44, the activity was dramatically reduced to a few percent of the wild-type activity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arima T., Uchida T., Egami F. Studies on extracellular ribonucleases of Ustilago sphaerogena. Characterization of substrate specificity with special reference to purine-specific ribonucleases. Biochem J. 1968 Feb;106(3):609–613. doi: 10.1042/bj1060609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima T., Uchida T., Egami F. Studies on extracellular ribonucleases of Ustilago sphaerogena. Purification and properties. Biochem J. 1968 Feb;106(3):601–607. doi: 10.1042/bj1060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- GROSS E., WITKOP B. Nonenzymatic cleavage of peptide bonds: the methionine residues in bovine pancreatic ribonuclease. J Biol Chem. 1962 Jun;237:1856–1860. [PubMed] [Google Scholar]
- Goeddel D. V., Heyneker H. L., Hozumi T., Arentzen R., Itakura K., Yansura D. G., Ross M. J., Miozzari G., Crea R., Seeburg P. H. Direct expression in Escherichia coli of a DNA sequence coding for human growth hormone. Nature. 1979 Oct 18;281(5732):544–548. doi: 10.1038/281544a0. [DOI] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Saenger W. Specific protein-nucleic acid recognition in ribonuclease T1-2'-guanylic acid complex: an X-ray study. Nature. 1982 Sep 2;299(5878):27–31. doi: 10.1038/299027a0. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J., Yoshida H. The primary structure of ribonuclease F1 from Fusarium moniliforme. Biochem Int. 1983 Aug;7(2):255–262. [PubMed] [Google Scholar]
- Ikehara M., Ohtsuka E., Tokunaga T., Taniyama Y., Iwai S., Kitano K., Miyamoto S., Ohgi T., Sakuragawa Y., Fujiyama K. Synthesis of a gene for human growth hormone and its expression in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5956–5960. doi: 10.1073/pnas.81.19.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F., Shimada I., Miyazawa T. Binding modes of inhibitors to ribonuclease T1 as studied by nuclear magnetic resonance. Biochemistry. 1985 Feb 12;24(4):1013–1020. doi: 10.1021/bi00325a031. [DOI] [PubMed] [Google Scholar]
- Itakura K., Hirose T., Crea R., Riggs A. D., Heyneker H. L., Bolivar F., Boyer H. W. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977 Dec 9;198(4321):1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku Y., Watanabe M., Kainosho M., Oshima T. A 15N-NMR study on ribonuclease T1-guanylic acid complex. J Biochem. 1982 Feb;91(2):675–679. doi: 10.1093/oxfordjournals.jbchem.a133740. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Mosteller R. D., Yanofsky C. Dynamics of synthesis, translation, and degradation of trp operon messenger RNA in E. coli. Cold Spring Harb Symp Quant Biol. 1969;34:725–740. doi: 10.1101/sqb.1969.034.01.082. [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Morioka H., Tokunaga T., Aoyama Y., Kikyotani S., Fujimoto K., Yanase K., Tanaka T., Uesugi S., Ohtsuka E. Synthesis and expression of the native RNase T1 gene and several mutant genes. Nucleic Acids Symp Ser. 1985;(16):287–290. [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Uchida T. The amino acid sequence of ribonuclease U2 from Ustilago sphaerogena. Biochem J. 1975 Feb;145(2):353–360. doi: 10.1042/bj1450353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio S., Oka K., Ohishi H., Tomita K., Saenger W. Three-dimensional structure of the ribonuclease T1 X 3'-guanylic acid complex at 2.6 A resolution. FEBS Lett. 1985 Apr 8;183(1):115–118. doi: 10.1016/0014-5793(85)80966-2. [DOI] [PubMed] [Google Scholar]
- Takahashi K. A revision and confirmation of the amino acid sequence of ribonuclease T1. J Biochem. 1985 Sep;98(3):815–817. doi: 10.1093/oxfordjournals.jbchem.a135339. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Stein W. H., Moore S. The identification of a glutamic acid residue as part of the active site of ribonuclease T-1. J Biol Chem. 1967 Oct 25;242(20):4682–4690. [PubMed] [Google Scholar]
- Takahashi K. The amino acid sequence of ribonuclease T-1. J Biol Chem. 1965 Oct;240(10):4117–4119. [PubMed] [Google Scholar]
- Watanabe H., Ohgi K., Irie M. Primary structure of a minor ribonuclease from Aspergillus saitoi. J Biochem. 1982 May;91(5):1495–1509. doi: 10.1093/oxfordjournals.jbchem.a133841. [DOI] [PubMed] [Google Scholar]