Abstract
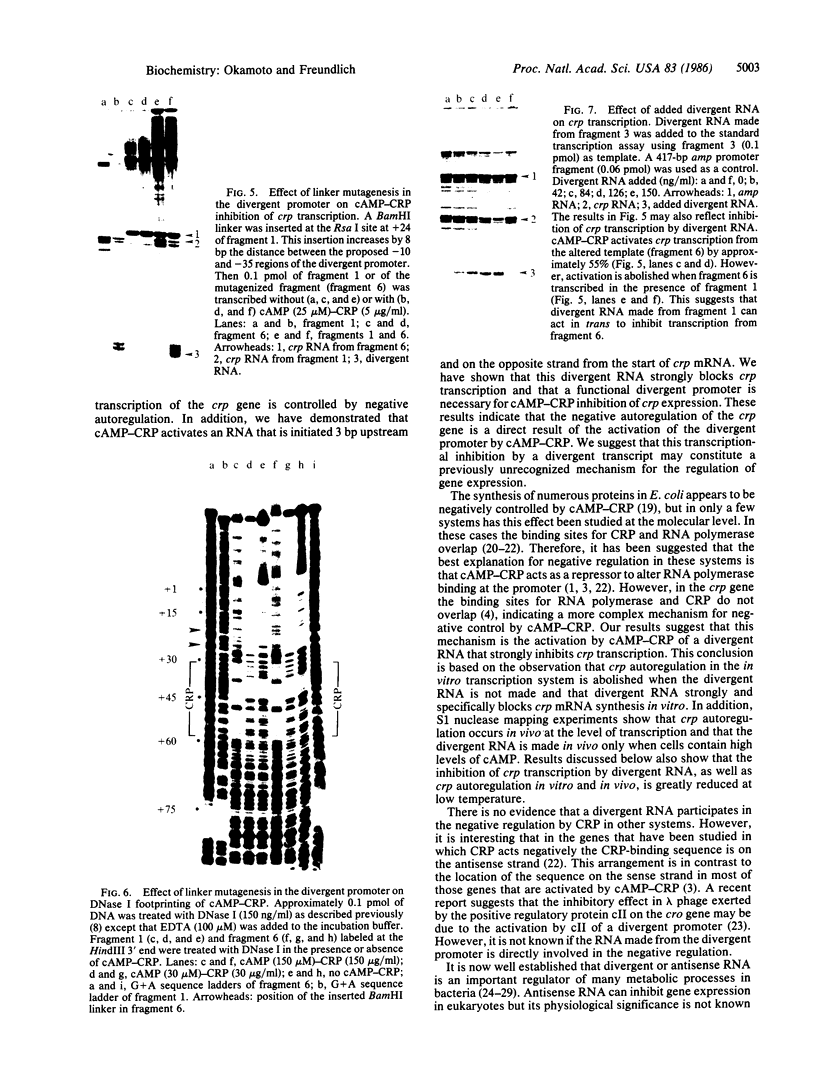
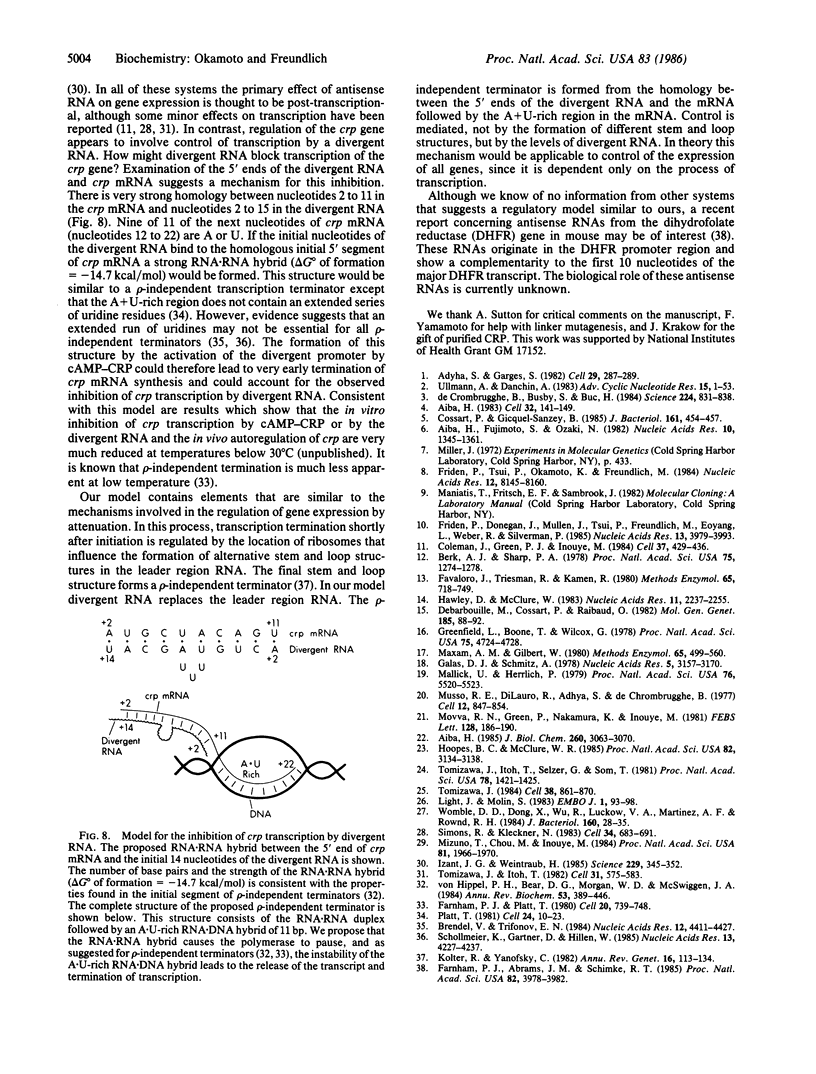
Expression of the crp gene is negatively autoregulated by the complex of cyclic AMP and its receptor protein (cAMP-CRP). We find a second promoter in this region that is strongly activated in vitro and in vivo by cAMP-CRP. Transcription from this promoter is initiated 3 nucleotides upstream and on the opposite strand from the start of crp mRNA. The addition of the purified 5' segment of the divergent RNA specifically inhibits crp transcription in vitro. cAMP-CRP does not block crp expression if the new promoter is altered so that divergent RNA cannot be made. The initial nucleotides of the divergent RNA are complementary to 10 of the first 11 nucleotides of the crp mRNA. Since the next 11 nucleotides of crp mRNA are A + U-rich, and RNA hybrid between the divergent RNA and the 5' end of crp mRNA could produce a structure similar to a rho-independent terminator, leading to inhibition of crp transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Garges S. How cyclic AMP and its receptor protein act in Escherichia coli. Cell. 1982 Jun;29(2):287–289. doi: 10.1016/0092-8674(82)90145-3. [DOI] [PubMed] [Google Scholar]
- Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983 Jan;32(1):141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- Aiba H., Fujimoto S., Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982 Feb 25;10(4):1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H. Transcription of the Escherichia coli adenylate cyclase gene is negatively regulated by cAMP-cAMP receptor protein. J Biol Chem. 1985 Mar 10;260(5):3063–3070. [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J., Green P. J., Inouye M. The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell. 1984 Jun;37(2):429–436. doi: 10.1016/0092-8674(84)90373-8. [DOI] [PubMed] [Google Scholar]
- Cossart P., Gicquel-Sanzey B. Regulation of expression of the crp gene of Escherichia coli K-12: in vivo study. J Bacteriol. 1985 Jan;161(1):454–457. doi: 10.1128/jb.161.1.454-457.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Cossart P., Raibaud O. A DNA sequence containing the control sites for gene malT and for the malPQ operon. Mol Gen Genet. 1982;185(1):88–92. doi: 10.1007/BF00333795. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Abrams J. M., Schimke R. T. Opposite-strand RNAs from the 5' flanking region of the mouse dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3978–3982. doi: 10.1073/pnas.82.12.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980 Jul;20(3):739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Friden P., Donegan J., Mullen J., Tsui P., Freundlich M., Eoyang L., Weber R., Silverman P. M. The ilvB locus of Escherichia coli K-12 is an operon encoding both subunits of acetohydroxyacid synthase I. Nucleic Acids Res. 1985 Jun 11;13(11):3979–3993. doi: 10.1093/nar/13.11.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden P., Tsui P., Okamoto K., Freundlich M. Interaction of cyclic AMP receptor protein with the ilvB biosynthetic operon in E. coli. Nucleic Acids Res. 1984 Nov 12;12(21):8145–8160. doi: 10.1093/nar/12.21.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield L., Boone T., Wilcox G. DNA sequence of the araBAD promoter in Escherichia coli B/r. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4724–4728. doi: 10.1073/pnas.75.10.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes B. C., McClure W. R. A cII-dependent promoter is located within the Q gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1985 May;82(10):3134–3138. doi: 10.1073/pnas.82.10.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izant J. G., Weintraub H. Constitutive and conditional suppression of exogenous and endogenous genes by anti-sense RNA. Science. 1985 Jul 26;229(4711):345–352. doi: 10.1126/science.2990048. [DOI] [PubMed] [Google Scholar]
- Kolter R., Yanofsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. EMBO J. 1983;2(1):93–98. doi: 10.1002/j.1460-2075.1983.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick U., Herrlich P. Regulation of synthesis of a major outer membrane protein: cyclic AMP represses Escherichia coli protein III synthesis. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5520–5523. doi: 10.1073/pnas.76.11.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Chou M. Y., Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A. 1984 Apr;81(7):1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movva R. N., Green P., Nakamura K., Inouye M. Interaction of cAMP receptor protein with the ompA gene, a gene for a major outer membrane protein of Escherichia coli. FEBS Lett. 1981 Jun 15;128(2):186–190. doi: 10.1016/0014-5793(81)80077-4. [DOI] [PubMed] [Google Scholar]
- Musso R. E., Di Lauro R., Adhya S., de Crombrugghe B. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell. 1977 Nov;12(3):847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Schollmeier K., Gärtner D., Hillen W. A bidirectionally active signal for termination of transcription is located between tetA and orfL on transposon Tn10. Nucleic Acids Res. 1985 Jun 25;13(12):4227–4237. doi: 10.1093/nar/13.12.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Itoh T. The importance of RNA secondary structure in CoIE1 primer formation. Cell. 1982 Dec;31(3 Pt 2):575–583. doi: 10.1016/0092-8674(82)90313-0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Dong X., Wu R. P., Luckow V. A., Martinez A. F., Rownd R. H. IncFII plasmid incompatibility product and its target are both RNA transcripts. J Bacteriol. 1984 Oct;160(1):28–35. doi: 10.1128/jb.160.1.28-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]










