Abstract
We have used paired-end sequencing of yeast nucleosomal DNA to obtain accurate genomic maps of nucleosome positions and occupancies in control cells and cells treated with 3-aminotriazole (3AT), an inducer of the transcriptional activator Gcn4. In control cells, 3AT-inducible genes exhibit a series of distinct nucleosome occupancy peaks. However, the underlying position data reveal that each nucleosome peak actually consists of a cluster of mutually exclusive overlapping positions, usually including a dominant position. Thus, each nucleosome occupies one of several possible positions and consequently, different cells have distinct local chromatin structures. Induction results in a major disruption of nucleosome positioning, sometimes with altered spacing and a dramatic loss of occupancy over the entire gene, often extending into a neighbouring gene. Nucleosome-depleted regions are generally unaffected. Genes repressed by 3AT show the same changes, but in reverse. We propose that yeast genes exist in one of several alternative nucleosomal arrays, which are disrupted by activation. We conclude that activation results in gene-wide chromatin remodelling and that this remodelling can even extend into the chromatin of flanking genes.
INTRODUCTION
The DNA of eukaryotic cells is organized into chromatin to facilitate packaging into the nucleus and to regulate access to genetic information. The basic structural unit of chromatin is the nucleosome, which includes the nucleosome core, the linker DNA between nucleosomes and histone H1 (1). The nucleosome core is composed of an octamer of the four core histones (H2A, H2B, H3 and H4), around which is wrapped ∼147 bp of DNA in 1.75 negative superhelical turns. The nucleosome core can be isolated as a metastable intermediate, the ‘core particle’, by digesting chromatin with micrococcal nuclease (MNase). Indeed, a low-resolution crystal structure of native core particles has been described (2). High-resolution structures were obtained later using core particles reconstituted with defined DNA (3,4).
Digestion of chromatin by MNase proceeds through several stages. Initially, MNase cuts the relatively unprotected linker DNA, resulting in a series of discrete DNA fragments corresponding to integral numbers of nucleosomes (appearing as a nucleosome ‘ladder’ in an agarose gel). Thus, nucleosomes are regularly spaced along the DNA in vivo. In most cell types, the average length of DNA associated with a nucleosome is ∼190 bp (5), but in budding yeast it is only ∼165 bp (6,7). Later in digestion, MNase begins to trim the ends of the linker DNA, eventually reaching a transient block in the form of the chromatosome, a particle containing ∼165 bp of DNA and H1 (8). Finally, MNase removes the remaining ∼20 bp of linker DNA to yield the core particle. The core particle is relatively stable, but MNase destroys it eventually.
In vitro, the histone octamer binds more strongly to some DNA sequences than to others; the strongest of these are referred to as positioning sequences [reviewed in (9)]. In vivo, the distribution of nucleosomes on DNA is also strongly dependent on the underlying sequence (10–14), suggesting that eukaryotic DNA possesses a nucleosome positioning code (12,13). However, the requirement for nucleosome spacing, the presence or otherwise of sequence-specific transcription factors (14,15) and the activities of ATP-dependent nucleosome mobilizing complexes (11,16) are expected to modify how the information specified by the nucleosome code is used (17).
The biological role of nucleosome positioning is a subject of intense interest and some controversy [reviewed in (18)]. The position of a nucleosome is defined by the DNA sequence it occupies i.e. the DNA within the core particle. Therefore, all nucleosomes have a position with respect to the genomic sequence. A useful concept is to imagine the genome as a series of overlapping ∼147-bp windows, each of which has the potential to be occupied by a nucleosome (19). The occupancy of each potential position might be very low (very few cells in a population have a nucleosome at this position, e.g. a nucleosome-depleted region), or very high (maximum occupancy is when all cells in a population have a nucleosome at this position). Thus, a strong position is one with a high occupancy (i.e. the same nucleosome is present in most cells).
The advent of DNA microarrays and massively parallel sequencing has revolutionized the study of positioning. In the first of these pioneering studies, hybridization of nucleosomal DNA to microarrays was used to measure average occupancies over an entire yeast chromosome (20) and later for the entire genome (21). However, this method cannot determine nucleosome positions very accurately because the precise borders of the nucleosomes cannot be ascertained by hybridization. Sequencing of nucleosomal DNA can determine positions very accurately (22,23) and is now possible on a genome-wide scale. Recent genome-wide single-end nucleosome sequencing studies have resulted in important insights into nucleosome positioning (13,14,24–27). There is, however, a significant drawback to this approach: only one end of each nucleosome is determined. This end sequence might be derived from a fully trimmed nucleosome (a core particle), thereby providing an accurate position, or it might be derived from an incompletely trimmed nucleosome (containing residual linker DNA), or from an over-digested nucleosome (cut internally), resulting in an inaccurate position. This problem is resolved by paired-end sequencing, a refinement of next-generation sequencing, which provides sequence reads from both ends of the same DNA molecule. Accordingly, after alignment with the genome sequence, the exact length of the DNA fragment can be deduced. Paired-end sequencing has been used recently to investigate the genomic distributions of several classes of MNase-resistant particle derived from chromatin (28).
Accurate positions are crucial for an understanding of the sequence determinants of nucleosome positioning. A fully trimmed nucleosome core particle should contain ∼147 bp, as in the crystal structures (3,4,29). Here, we describe the results of a paired-end nucleosome sequencing study aimed at defining accurate positions for nucleosomes in the budding yeast, Saccharomyces cerevisiae. We compare nucleosome positions in control cells with those in 3-aminotriazole (3AT)-treated cells. 3AT inhibits the enzyme encoded by HIS3, which is required for histidine biosynthesis, resulting in induction of the amino acid starvation pathway through translational control of the transcriptional activator Gcn4 (30).
We show that mono-nucleosome preparations are composed of a mixture of particles containing DNA of different lengths, as expected. We determine accurate nucleosome positions by considering the subset of nucleosome sequences derived from core particles (145–150 bp in length). Our results are best described using the concept of ‘nucleosome position clusters’, which specify sets of mutually exclusive overlapping positions and usually include a dominant position (11,16,17,31). Thus, each nucleosome can adopt one of the several alternative positions. To account for position clusters, we propose that yeast genes exist in one of several alternative, overlapping, nucleosomal arrays.
Activation by 3AT results in a dramatic loss of canonical nucleosomes from some genes and the position cluster organization of the remaining nucleosomes is disrupted. Furthermore, chromatin disruption often extends into neighbouring genes. Thus, activation-induced chromatin remodelling events are gene wide and can even spread farther, disturbing the chromatin of flanking genes.
MATERIALS AND METHODS
Preparation of core particle DNA
YDC111 (MATa ade2-1 can1-100 leu2-3,112 trp1-1 ura3-1 RAD5+) (16) was grown to late log phase in synthetic complete (SC) medium (control) or SC medium lacking histidine to which 3AT was added to 10 mM for 20 min just before harvesting (3AT treated). Core particle DNA was prepared by MNase digestion of nuclei, gel purified, repaired and checked for DNA size and quality as described (Figure 1) (32). Paired-end sequencing was performed as described (33). Control cells yielded 16.6 and 12.3 million aligned paired reads of 40 nt each for the first and second experiments, respectively; 3AT-treated cells gave 13.1 and 13.5 million aligned paired reads, respectively. Paired reads were aligned to the S. cerevisiae genome using ELAND. Reads with mis-matches were excluded from the analysis. The GEO accession number for the data presented here is GSE26493.
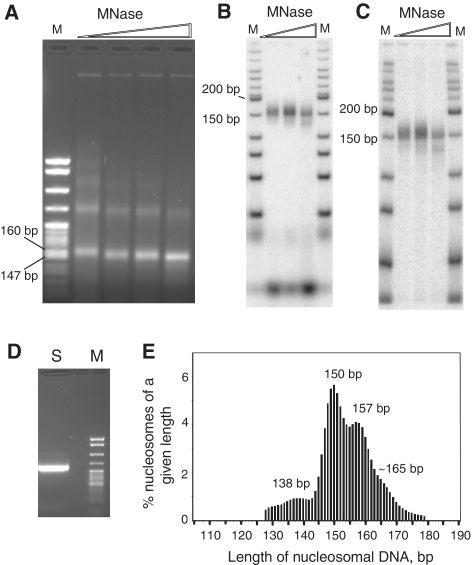
Figure 1.
Length distribution of nucleosomal DNA. Shown are data for 3AT-treated cells. (A) Digestion of yeast nuclei with MNase (30, 60, 120 and 240 Worthington units). DNA was analysed in an agarose gel stained with ethidium bromide. M, pBR322 digested with MspI. Mono-nucleosomal DNA in the samples obtained using 60, 120 and 240 U of MNase was gel purified and repaired. An aliquot was end-labelled using T4 polynucleotide kinase and analysed in (B) a native polyacrylamide gel and (C) a denaturing polyacrylamide gel; M: 50-bp ladder (NEB). The sample obtained using 120 U MNase was ligated to paired-end adaptors and amplified by PCR (D). The purified product was used for sequencing (M: pBR322 MspI). (E) Nucleosome sequence length distribution. All sequences are included, except those derived from the yeast 2-µm plasmid. The fraction of sequences of a given length is expressed as a percentage of the total. The numbers indicate peak values. The scale is single nucleotide resolution.
Nucleosome positioning
An algorithm was written to extract nucleosome positioning information from the sequence data. First, it was assumed that all nucleosome sequences between 145 and 150 bp represent accurate positions. These sequences were used to define a set of accurate positions (SAC) adopted by nucleosomes, represented by their midpoint coordinates (i.e. nucleosome dyad axis). Secondly, to include as much of the data as possible, the midpoints of all remaining reads (those <145 bp and >150 bp) were calculated and then these reads were allocated to the nucleosome in SAC with the closest midpoint, provided that the two midpoints were <10 bp apart. Finally, the data were smoothed using a 6-bp window. Nucleosome position maps were obtained in which the number of sequences corresponding to a specific nucleosome position is plotted against the chromosomal coordinate of the dyad axis. The scripts written to analyse the data are given in Supplementary Data.
RESULTS
Paired-end sequencing of yeast nucleosomes reveals complex length distributions
Nucleosome core particles were prepared by MNase digestion of nuclei prepared from control and 3AT-treated cells (Figure 1). We obtained between 12 and 17 million aligned paired reads per sample. The yeast genome can accommodate approximately 75 000 nucleosomes, given that the haploid genome is ∼12.1 Mb and the nucleosome spacing is ∼165 bp. Consequently, approximately 200 sequences per nucleosome should be expected. This read depth should provide data with low statistical sampling error.
To maximize the fraction of fully trimmed core particles, a balance must be struck between the full trimming required to obtain accurate position data and the tendency for MNase to begin cutting within the core particle. A typical nucleosome length distribution (Figure 1E) suggested the presence of three different populations: (i) core particles (peaking at 149 bp); these are mono-nucleosomes with little or no linker DNA remaining. Consequently, their DNA content defines accurate positions; (ii) mono-nucleosomes with residual linker DNA (peaking at ∼157 bp and at ∼165 bp). Incomplete trimming might reflect the binding of H1, which is present at relatively low levels in yeast and so only some nucleosomes would be expected to contain it (34), although poorly trimmed nucleosomes of about chromatosome size have been observed even in the absence of H1 (5); and (iii) subnucleosomal particles containing less than ∼140 bp. These probably derive primarily from internal cleavage of core particles by MNase, perhaps following spontaneous uncoiling of DNA from the ends of the nucleosome (35). Alternatively, some might represent remodelled nucleosomes or transcribed nucleosomes lacking an H2A-H2B dimer (36,37). The data presented below belong to the first of two independent experiments, which gave essentially the same results.
Position clusters on the PHO5 promoter
The PHO5 promoter was chosen as a control region because it is one of the best studied loci in the chromatin literature. Mapping of the PHO5 promoter by indirect end-labelling has established that the repressed promoter is organized into an array of positioned nucleosomes numbered −1 to −5 (38). There is a gap between nucleosomes −2 and −3, where binding sites for the transcription factors Pho4 and Pho2 are located. Induction disrupts this ordered chromatin structure and increases accessibility of the promoter DNA (38). In our experiments, cells were grown under conditions such that PHO5 should be repressed.
The nucleosome occupancy profile is a plot of the chromosome base coordinate versus the number of nucleosome sequences that contain that particular base. It is therefore a measure of the probability of a base being contained within a nucleosome. Occupancy profiles for the PHO5 promoter in control and 3AT-treated cells are shown (Figure 2A); all aligned nucleosome sequences were included. The data were not subjected to mathematical manipulation, except that the 3AT data were multiplied by 1.27 to compensate for the fact that fewer total sequences were obtained relative to the control. The agreement between the profiles for control and 3AT-treated cells was excellent; the traces superimposed in places and showed limited quantitative variation.
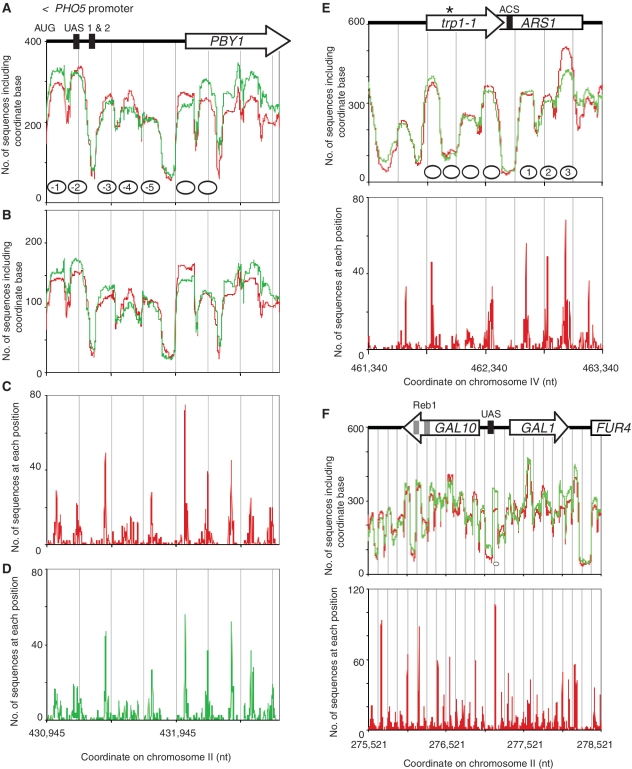
Figure 2.
Nucleosome position clusters on the PHO5 promoter, TRP1 ARS1 and GAL1-GAL10. Chromatin structures of well-studied genes. Control cells: red trace; 3AT-treated cells: green trace. Grey lines indicate 250-bp intervals. (A) Nucleosome occupancy of the PHO5 promoter and upstream PBY1 gene. All sequences are included. Nucleosomes are indicated by ovals drawn to scale and numbered (38). (B) Nucleosome occupancy as in A, except that only reads of 145–155 bp were included. Nucleosome positioning analysis for (C) control and (D) 3AT-treated cells. The PHO5 coding region was omitted because it is very homologous to other yeast genes (PHO3, PHO12 and DIA3) resulting in removal of some sequences because their origin is uncertain. (E) TRP1 ARS1. Occupancy profiles for control and 3AT-treated cells (all sequences included) and position analysis for control cells. Nucleosome ovals are drawn to scale and numbered (39). Our strain is trp1-1, which corresponds to a nonsense mutation (Asterisk) covered by the second nucleosome peak on TRP1, which is much reduced relative to the others, because sequences containing this mutation are rejected as they do not match the wild-type sequence. (F) GAL1-GAL10. Occupancy profiles for control and 3AT-treated cells (all sequences included) and position analysis for control cells. Black boxes: Gal4-binding sites. Grey boxes: Reb1 sites involved in expression of a ncRNA beginning within GAL10 (40). The nucleosome oval is drawn to scale.
The occupancy profiles for the PHO5 promoter exhibited peaks corresponding to the five reported nucleosomes (Figure 2A). Importantly, although the peaks were quite obvious, the troughs between them did not dip close to the baseline, indicating that many nucleosome sequences included what should be linker DNA between the reported nucleosomes. To assess whether this was due to poorly trimmed nucleosomes (i.e. from nucleosomes significantly > 150 bp and therefore including some linker DNA), the plot was restricted to data for nucleosome sequences 145–155 bp in length (Figure 2B), corresponding to 50% of all nucleosomes. The occupancy profiles were marginally sharper (Figure 2B), but not essentially different from the profiles corresponding to all nucleosomes (Figure 2A). This suggests that the excluded nucleosomes (those >155 bp and <145 bp) are derived from the same nucleosomes in the restricted data set (145–155 bp).
The accurate nucleosome positions are those between 145 and 150 bp, corresponding to core particles (comprising ∼30% of nucleosome sequences from control and 3AT-treated cells). These positions were defined by their midpoints (dyad axes). To include as much data as possible, the midpoints of all remaining sequences (those <145 bp and >150 bp) were calculated and then assigned to the accurate position with the closest midpoint. These simple rules yield a position map in which the number of sequences corresponding to the midpoint of each specific nucleosome position is plotted against the chromosomal coordinate of the dyad axis.
The position map for the PHO5 promoter showed that some positions were strongly favoured, particularly those corresponding to nucleosome −3 and the PBY1 coding region (Figure 2C, D). However, in all cases, there were some less prominent midpoints near each major midpoint, corresponding to positions which overlap the major position to different extents. Thus, each dominant midpoint was associated with a cluster of midpoints, which we term a ‘position cluster’. Each position cluster specifies a set of overlapping positions, which are mutually exclusive because canonical nucleosomes cannot physically overlap on the same DNA molecule. Therefore, in some cells the nucleosome is present at the dominant position in each cluster while in other cells, the nucleosome is at one of the alternative positions. All of the PHO5 promoter nucleosomes corresponded to clusters including a dominant position, except nucleosome −4, which was a cluster of alternative positions with similar probabilities. PBY1 also exhibited position clusters with a particularly dominant position for the first nucleosome. In conclusion, the chromatin structure of the PHO5 promoter is best described in terms of nucleosome position clusters, rather than uniquely positioned nucleosomes.
Position clusters on TRP1 ARS1 and at the GAL1–GAL10 locus
Immediately downstream of TRP1 is ARS1, a well-studied replication origin. Examination of the chromatin structure of a TRP1 ARS1 plasmid by indirect end-labelling has revealed a hypersensitive site at the ARS consensus sequence (ACS), where the origin recognition complex binds, and three well-positioned nucleosomes (39). Our occupancy profile for TRP1 ARS1 showed the expected nucleosome-depleted region at the ACS and three clear nucleosome peaks downstream (Figure 2E). Once again, the agreement between the profiles for control and 3AT-treated cells was excellent. As for the PHO5 promoter, analysis of nucleosome positioning on TRP1 and ARS1 indicated the presence of position clusters, rather than unique positions (Figure 2E).
The GAL1 and GAL10 genes are transcribed from a divergent promoter and should be repressed under our growth conditions. A quite regular series of nucleosome occupancy peaks was observed across both genes, which corresponded to a series of position clusters (Figure 2F). There were nucleosome-depleted regions in the divergent promoter [corresponding to Gal4-binding sites), within the GAL10-coding region (corresponding to Reb1 sites required for activation of a ncRNA gene (40)] and at the FUR4 promoter.
In conclusion, nucleosome position clusters were detected at all three regions examined (Figure 2), indicating that these genes can exist in any of several alternative chromatin structures.
Altered position clusters on HIS3 in 3AT-treated cells
Previously, we mapped nucleosome positions on HIS3 at high resolution using the monomer extension technique (16). In the absence of the Gcn4 activator, HIS3 is organized into a dominant array of five nucleosomes, D1–D5, with a background of alternative, overlapping positions. Activation by Gcn4 results in increased occupancy of the alternative positions, and the D-positions are no longer dominant. This study (16) provides positioning data of sufficiently high resolution for direct comparison with the current study.
Both HIS3 and the neighbouring PET56 gene are induced by 3AT (30). The occupancy profiles indicated that HIS3 is flanked by nucleosome-depleted regions, corresponding to the HIS3-PET56 and DED1 promoters (Figure 3A). The profile for control cells indicated five nucleosome peaks, although the separation between the third and fourth peaks was relatively indistinct and their occupancies were lower. The profile for 3AT-treated cells was somewhat different: the distinction between the third and fourth nucleosome peaks was even less clear, the fifth nucleosome peak was shifted a little upstream, and the overall occupancy was lower. The effect of 3AT on PET56 was more subtle, with slightly reduced occupancy at both ends of the coding region, but no obvious change in the fairly regular set of nucleosome peaks.
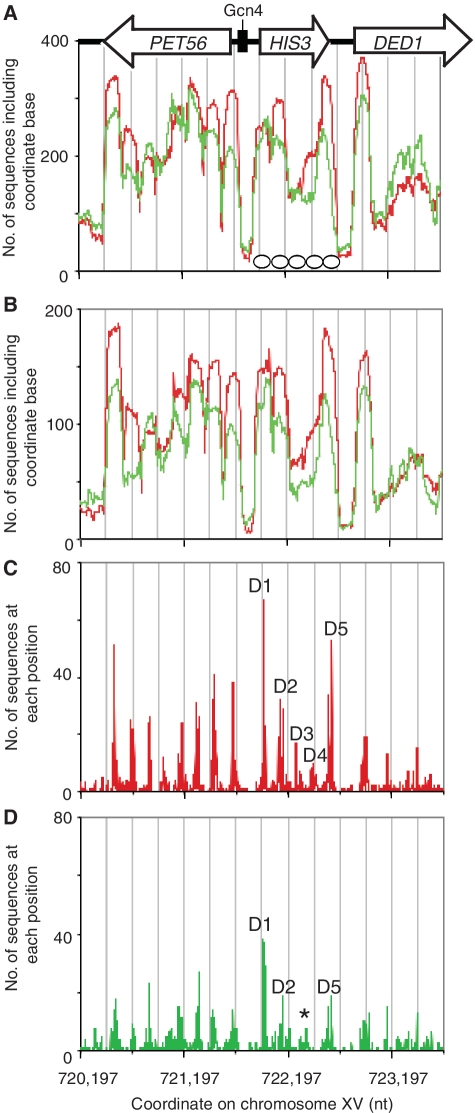
Figure 3.
Altered nucleosome position clusters on HIS3 in response to 3AT. (A) Nucleosome occupancy on HIS3 and flanking sequences. Control cells: red trace; 3AT-treated cells: green trace. All sequences are included. Nucleosomes are indicated by ovals drawn to scale. Grey lines indicate 250-bp intervals. (B) Nucleosome occupancy using only reads of 145–155 bp. Positioning analysis for (C) control cells and (D) 3AT-treated cells. D1–D5: dominant positions adopted by nucleosomes in cells lacking the Gcn4 activator (16). The asterisk indicates the new position cluster formed in place of D3 and D4 in 3AT-treated cells.
Five position clusters were present on HIS3 in control cells (Figure 3C). The midpoints of the most prominent peaks were +15, +179 (with slightly weaker peaks at +156 and +203), +327, +491 and +642/+672. These midpoints predict an array with a range of linker lengths and an average spacing of 164 bp (typical of yeast). They correspond reasonably well to the five dominant positions mapped previously (16): +8, +163, +327, +527 and +683, except for D4 that was mapped at +527 rather than at +491. In the paired-end data, the strongest peak in the cluster was at +491, but there was a smaller peak at +516.
There were significant changes in the position clusters on HIS3 in 3AT-treated cells (Figure 3D). In the first cluster, the D1 position remained the most probable but its dominance was reduced relative to an overlapping position ∼20-bp downstream. The same was true of the fifth cluster in which the dominance of the +672 position was reduced relative to that at +642. The major changes occurred in the D2, D3 and D4 clusters: the dominant position in the D2 cluster was shifted downstream to the +203 position; the D3 and D4 clusters were replaced by a very weak cluster centred on +425. Thus, the 3AT-induced HIS3 gene had only four nucleosomes on average, rather than the array of five in control cells. Since the positions adopted by the nucleosome at each end of the array (D1 and D5) did not change significantly, the average spacing of the nucleosomes was much greater on the 3AT-induced gene (218 bp) than in control cells (164 bp).
In conclusion, the paired-end data are in good agreement with our previous monomer extension studies, providing some validation. Induction with 3AT reduced the occupancies of the dominant positions relative to the alternative positions and resulted in removal of one nucleosome and some re-positioning of the remaining nucleosomes.
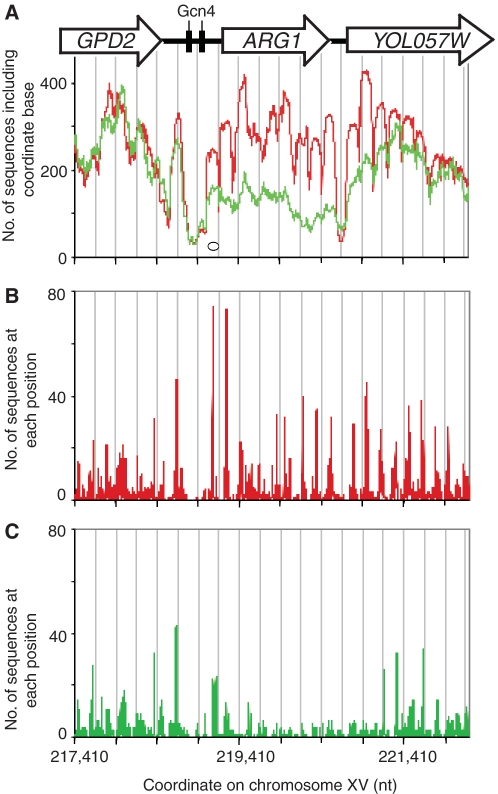
Activation disrupts the chromatin structures of ARG1 and the neighbouring YOL057W gene
ARG1, another Gcn4-dependent gene, is strongly induced by 3AT (30). In control cells, ARG1 was organized into nine nucleosome peaks, flanked by nucleosome-depleted regions corresponding to the ARG1 and YOL057W promoters (Figure 4A). The 5′ half of YOL057W was also well organized, displaying six nucleosome peaks before becoming more irregular. In 3AT-treated cells, there was a massive loss of occupancy across the entire ARG1 gene, extending into the 3′-flanking gene (YOL057W) (Figure 4A), even though its expression is not affected by 3AT (30). Furthermore, the regular nucleosome peaks observed on ARG1 in control cells merged into one another in 3AT-treated cells. The position clusters present on ARG1 and YOL057W in control cells were heavily disrupted in 3AT-treated cells (Figure 4B, C). Thus, ARG1 induction was associated with loss of more than half of its canonical nucleosomes; those remaining were no longer organized into clusters with dominant positions. Furthermore, these effects were propagated downstream into YOL057W.
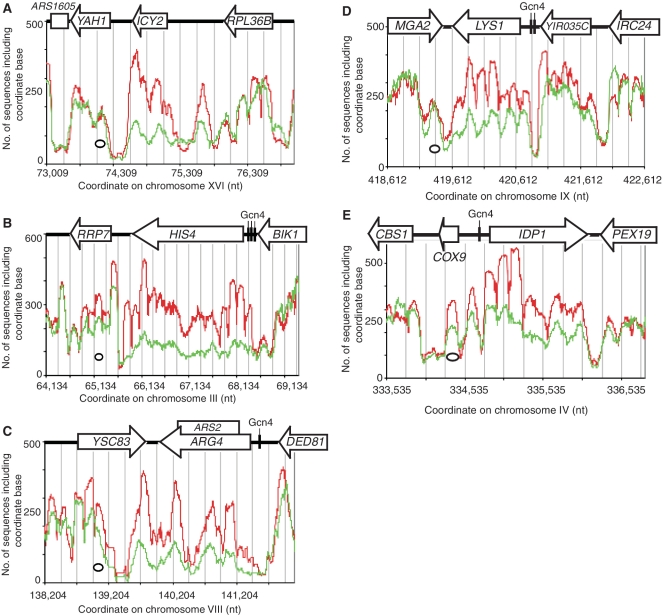
Figure 4.
Induction of ARG1 results in reduced occupancy on the coding region, extending into YOL057W downstream. ARG1 encodes an enzyme involved in arginine biosynthesis. (A) Nucleosome occupancy of the ARG1 gene and flanking sequences. Control cells: red trace; 3AT-treated cells: green trace. All nucleosome sequences are included. The oval indicates the size of a nucleosome. Grey lines indicate 250-bp intervals. Nucleosome positioning analysis for control cells (B) and 3AT-treated cells (C).
More genes exhibiting 3AT-induced disruption of position clusters
To find other genes displaying similarly dramatic, 3AT-induced effects on chromatin structure, the numbers of nucleosome sequences per coding region in control and 3AT-treated cells were compared using a whole-genome survey. This analysis ranked all genes using a ‘disruption score’, corresponding to the ratio of nucleosome sequences in 3AT-treated cells to sequences in control cells (after adjustment for the difference in the total number of nucleosome sequences obtained for control and 3AT-treated cells). A disruption score of <1 indicates that a gene has fewer nucleosome sequences in 3AT-treated cells, like ARG1. A cut-off score of 0.75 was set, requiring that a gene has >25% fewer sequences in 3AT-treated cells. Forty-nine genes, including ARG1, had an average disruption score of <0.75 (Table 1). In addition, 13 genes showed the reverse effect, with an equivalent cut-off score of >1.32 (25% fewer nucleosome sequences in control cells than in 3AT-treated cells).
Table 1.
Genes with disrupted chromatin structure in 3AT-treated or control cells
| Gene Ranka | Gene | Disruption score, average b | Disruption score, SD b | Ln(Average Disruption score)c | 3AT inductiond | Gcn4 targete | Notes |
|---|---|---|---|---|---|---|---|
| 1 | YPR036W-A | 0.26 | 0.05 | −1.35 | ND | ND | 3′ of ERV2 |
| 2 | ICY2 | 0.4 | 0.03 | −0.92 | 5.3 | Y | |
| 3 | ARG1 | 0.43 | 0.03 | −0.84 | 11.6 | Y | |
| 4 | HIS4 | 0.44 | 0.05 | −0.82 | 8.0 | Y | |
| 5 | ARG4 | 0.48 | 0.07 | −0.73 | 9.5 | Y | |
| 6 | LYS1 | 0.55 | 0.01 | −0.60 | 20.6 | Y | |
| 7 | PCL5 | 0.57 | 0.03 | −0.56 | 6.6 | Y | |
| 8 | LYS20 | 0.58 | 0.04 | −0.54 | 9.8 | Y | |
| 9 | YSC83 | 0.6 | 0.08 | −0.51 | 1.8 | N | 3′ of ARG4 |
| 10 | IDP1 | 0.6 | 0.03 | −0.51 | 3.2 | Y | |
| 11 | ASN1 | 0.6 | 0.08 | −0.51 | 3.0 | Y | |
| 12 | SNZ1 | 0.61 | 0.01 | −0.49 | 47.0 | Y | |
| 13 | MET6 | 0.63 | 0.03 | −0.46 | 1.0 | N | |
| 14 | CIT2 | 0.64 | 0.01 | −0.45 | 0.3 | Y | |
| 15 | YGL117W | 0.64 | 0.01 | −0.45 | 5.6 | Y | |
| 16 | MET17 | 0.66 | 0.01 | −0.42 | 1.8 | Y | |
| 17 | LEU4 | 0.66 | 0.07 | −0.42 | 1.4 | Y | |
| 18 | CPA2 | 0.66 | 0.03 | −0.42 | 4.5 | Y | |
| 19 | ADE3 | 0.67 | 0.02 | −0.40 | 2.8 | Y | |
| 20 | TRP3 | 0.67 | 0.08 | −0.40 | 3.2 | Y | |
| 21 | ARG7/ECM40 | 0.68 | 0.05 | −0.39 | 1.1 | Y | |
| 22 | SWF1 | 0.69 | 0.02 | −0.37 | 3.3 | Y | |
| 23 | CPA1 | 0.69 | 0.01 | −0.37 | 3.4 | Y | |
| 24 | DDR48 | 0.69 | 0.14 | −0.37 | 2.1 | Y | 5′ of PAI3 |
| 25 | YHR162W | 0.7 | 0.00 | −0.36 | 1.8 | Y | |
| 26 | TMT1 | 0.71 | 0.04 | −0.34 | 8.1 | Y | |
| 27 | YAT2 | 0.71 | 0.02 | −0.34 | 6.5 | Y | |
| 28 | YKL106C-A | 0.71 | 0.01 | −0.34 | ND | ND | |
| 29 | COX9 | 0.71 | 0.04 | −0.34 | 1.3 | Y | 5′ of IDP1 |
| 30 | YBR147W | 0.71 | 0.06 | −0.34 | 13.3 | Y | |
| 31 | ISU1 | 0.72 | 0.02 | −0.33 | 4.5 | Y | |
| 32 | YDR524C-B | 0.72 | 0.21 | −0.33 | ND | ND | |
| 33 | DIP5 | 0.72 | 0.01 | −0.33 | 0.4 | N | |
| 34 | RIB5 | 0.72 | 0.10 | −0.33 | 7.4 | Y | |
| 35 | TPO2 | 0.72 | 0.06 | −0.33 | 2.5 | N | |
| 36 | COX17 | 0.73 | 0.02 | −0.31 | 1.0 | N | |
| 37 | LYS2 | 0.73 | 0.02 | −0.31 | 7.0 | Y | |
| 38 | MET13 | 0.73 | 0.00 | −0.31 | 1.9 | Y | |
| 39 | BIO4 | 0.74 | 0.08 | −0.30 | 1.9 | Y | |
| 40 | ERV2 | 0.74 | 0.04 | −0.30 | 1.0 | N | 5′ YPR036W-A |
| 41 | MCH4 | 0.74 | 0.04 | −0.30 | 13.4 | Y | |
| 42 | VHT1 | 0.74 | 0.02 | −0.30 | 2.1 | Y | |
| 43 | PAI3 | 0.74 | 0.08 | −0.30 | 0.7 | Y | 3′ of DDR48 |
| 44 | GGC1 | 0.75 | 0.04 | −0.29 | 6.2 | Y | |
| 45 | SER33 | 0.75 | 0.05 | −0.29 | 1.1 | Y | |
| 46 | HOM2 | 0.75 | 0.06 | −0.29 | 4.5 | Y | |
| 47 | SHM2 | 0.75 | 0.13 | −0.29 | 3.5 | N | |
| 48 | STP22 | 0.75 | 0.03 | −0.29 | 1.8 | Y | |
| 49 | YIR035C | 0.75 | 0.10 | −0.29 | 1.1 | N | 5′ of LYS1 |
| 93 | HIS3 | 0.8 | 0.08 | −0.22 | 10.3 | Y | |
| 107 | RRP7 | 0.81 | 0.09 | −0.21 | 0.7 | N | |
| 358 | TRP1 | 0.88 | 0.03 | −0.13 | 0.9 | N | |
| 1571 | PHO5 | 0.96 | 0.04 | −0.04 | 0.8 | N | |
| 1822 | GAL10 | 0.97 | 0.01 | −0.03 | 1.1 | N | |
| 2271 | GAL1 | 0.99 | 0.06 | −0.01 | 0.9 | N | |
| 5274 | OPI3 | 1.14 | 0.12 | 0.13 | 0.3 | N | |
| 5529 | HXK2 | 1.33 | 0.01 | 0.29 | 0.6 | N | |
| 5530 | COX6 | 1.34 | 0.16 | 0.29 | 0.7 | N | |
| 5531 | STE18 | 1.35 | 0.09 | 0.30 | 1.0 | N | |
| 5532 | YDR366C | 1.35 | 0.15 | 0.30 | 1.7 | N | |
| 5533 | TMA7 | 1.35 | 0.04 | 0.30 | ND | N | |
| 5534 | YGR169C-A | 1.35 | 0.08 | 0.30 | ND | N | |
| 5535 | YOR072W-B | 1.36 | 0.48 | 0.31 | ND | N | |
| 5536 | MOG1 | 1.36 | 0.16 | 0.31 | 0.4 | N | |
| 5537 | OLE1 | 1.37 | 0.04 | 0.31 | 0.3 | N | |
| 5538 | YBR230W-A | 1.37 | 0.37 | 0.31 | ND | N | |
| 5539 | QCR8 | 1.39 | 0.06 | 0.33 | 0.8 | N | |
| 5540 | MRPS8 | 1.41 | 0.21 | 0.34 | 0.6 | N | |
| 5541 | URA1 | 1.53 | 0.13 | 0.43 | 0.2 | N |
aA total of 5541 genes were ranked according to their average disruption scores (tRNA genes and some very short ORFs <170 bp were eliminated from the ranking, because they are too short).
bDisruption score = no. of sequence reads in 3AT-treated cells/ no. of reads in control cells (normalized for the difference in the total number of reads). Average disruption scores for two independent experiments are given with standard deviation (SD) in the next column. Genes with average scores ≤0.75 (25% fewer reads in 3AT-treated cells; 49 genes), or ≥1.33 (25% more reads in 3AT-treated cells; 13 genes) are shown. Also included are the values for other genes described in the text.
cThe average disruption score is expressed as a natural log to facilitate comparison of the extent of disruption in 3AT treated and control genes. The cut-off scores are −0.28 for genes with disrupted chromatin in 3AT-treated cells and +0.28 for genes with disrupted chromatin in control cells.
dInterpolated from expression microarray data (30). ND: no data reported for this gene. Although most genes with disruption scores equal to or <0.75 are induced by 3AT, two genes (CIT2 and DIP5) are repressed by 3AT; the explanation for this is unknown.
eGcn4 target gene in a set of 539 genes (30).
ND, no data reported.
The expression microarray study (30) found 305 genes that are induced >2-fold by 10 mM 3AT and 104 genes that are repressed >2-fold. Of the 49 genes with disruption scores equal to or <0.75, 29 were induced >2-fold [(30), Table 1]. If the three genes for which there are no data are excluded, 63% of genes with heavily disrupted chromatin are induced by 3AT. In addition, four of the genes unaffected by 3AT are located next to genes that are induced by 3AT (ERV2/YPR036W-A, YSC83/ARG4, YIR035C/LYS1 and COX9/IDP1). Of the 13 genes with heavily disrupted chromatin in control cells, three are repressed by 3AT (33% of genes for which data are available; Table 1). Genome-wide, there was a good correlation between the disruption score and the fold induction by 3AT (Supplementary Figure S1). A small fraction of genes which were strongly induced or repressed by 3AT showed only weak chromatin disruption. This could be because the fold-change in expression of these genes is high but the absolute level of transcription is not very high (see ‘Discussion’ section). It should also be noted that the 3AT expression data are for a different yeast strain grown in a different medium (30) and so some genes might be affected differently by 3AT.
By comparing changes in expression in wild-type and gcn4Δ cells, Natarajan et al. (30) reported a list of 539 Gcn4 target genes. Of the 46 genes with disrupted chromatin structure in 3AT-treated cells and for which there are expression data, 38 are Gcn4 targets (83%). Only eight genes are not Gcn4 targets and three of these are neighbours of affected genes (Table 1). As expected, none of the genes with disrupted chromatin in control cells are Gcn4 targets. Thus, most of the genes identified by the disruption survey are known Gcn4 targets and are induced by 3AT.
Induction with 3AT results in loss of canonical nucleosomes from coding regions and rearrangement of the remaining clusters, with little effect on nucleosome-depleted regions
Occupancy profiles for some genes with heavily disrupted chromatin structures identified by the whole-genome survey (Table 1) are shown (Figure 5). In all cases, there was a dramatic loss of occupancy over the coding region in 3AT-treated cells. In control cells, the chromatin structures of LYS1 (Figure 5D), the 5′- and 3′-ends of HIS4 (Figure 5B) and the 5′-half of IDP1 (Figure 5E) were quite regular, displaying well-defined nucleosome peaks, corresponding to position clusters with dominant positions. The chromatin structures of ARG4 (Figure 5C), ICY2 (Figure 5A), the central region of HIS4 and the 3′ half of IDP1 were less regular, indicating a more complex position cluster organization. The occupancy profiles of these genes in 3AT-treated cells were very different from those of control cells, indicating that the remaining nucleosomes had been rearranged. In the case of HIS4, all regularity was lost, indicating the absence of dominant positions (Figure 5B). The occupancy profile of LYS1 remained quite regular in 3AT-treated cells, but there were only six clear peaks, which were out of phase with the peaks in control cells, with the exception of the sixth peak (Figure 5D). This indicated a change in the average positions and spacing of the nucleosomes, as observed for HIS3 (Figure 3).
Figure 5.
Nucleosome occupancy profiles of 3AT-induced genes with severely disrupted chromatin structure. Occupancy profiles of genes with the most disrupted chromatin structure in 3AT-treated cells (Table 1). Control cells: red trace; 3AT-treated cells: green trace. All nucleosome sequences are included. Nucleosomes are indicated by ovals drawn to scale. Grey lines indicate 250-bp intervals. (A) ICY2 (ranked second). (B) HIS4 (ranked fourth). (C) ARG4 (fifth) and YSC83 (ninth). The disorganized central region of ARG4 most likely reflects the presence of an origin of replication (ARS2) within the coding region. (D) LYS1 (sixth) and YIR035C (49th). (E) IDP1 (10th). All of these genes are induced by 3AT and are Gcn4 targets (30) (Table 1). All have canonical Gcn4-binding sites in their promoters except ICY2, which has two non-canonical sites.
In striking contrast to the effects of 3AT on nucleosome occupancy of the coding regions, there was little effect on occupancy at the promoters and 3′-ends of these genes, which were all significantly depleted of nucleosomes in both control and 3AT-treated cells.
Extension of chromatin disruption into flanking genes
The disruption of ARG1 chromatin in 3AT-treated cells extended far into the gene downstream (Figure 4A). This was also true for HIS4 and ARG4 (Figure 5). None of these downstream genes are Gcn4 targets and all are unaffected by 3AT (30). Indeed, nucleosome occupancy on YSC83, downstream of ARG4, was reduced so strongly that it was scored as a gene with heavily disrupted chromatin structure (Table 1). In all three cases, occupancy of the downstream gene at the end farthest from the target gene was similar to that in control cells, revealing that the disruptive effect diminished with distance from the target gene.
The chromatin structures of the genes downstream of ICY2, LYS1 and IDP1 were unaffected (Figure 5). However, in these cases, the chromatin structure of the upstream gene was disrupted. Most strikingly, YIR035C, upstream of LYS1 (Figure 5D), was heavily disrupted, even though it is not a Gcn4 target gene and is unaffected by 3AT (30).
In summary, the chromatin structures of 49 genes were heavily disrupted in 3AT-treated cells: the entire coding region was heavily depleted of canonical nucleosomes. In some cases, this disruption extended to flanking genes, either upstream or downstream. Nucleosome positioning was heavily disrupted with major reductions in occupancies of dominant positions observed in control cells. At HIS3 and LYS1, the average number of nucleosomes on the gene was reduced, implying changes in nucleosome spacing.
The chromatin of genes repressed by 3AT is more ordered in 3AT-treated cells
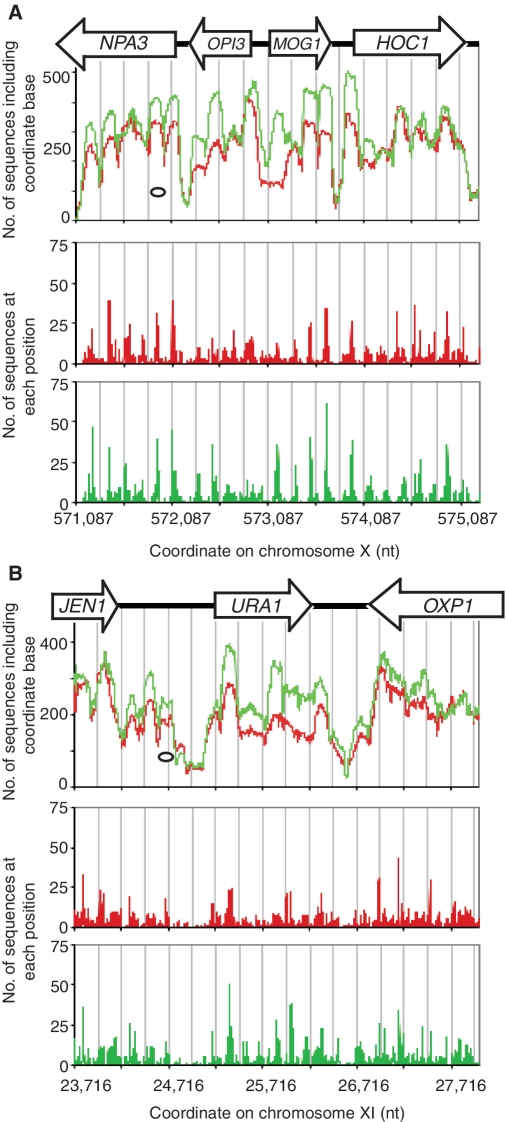
There were 13 genes with extreme disruption scores in the opposite sense: more nucleosome sequences were obtained from 3AT-treated cells than from control cells (Table 1). Three of these genes (URA1, OLE1 and MOG1) are repressed by 3AT (30). MOG1 had a more ordered chromatin structure in 3AT-treated cells than in control cells (Figure 6A), with four nucleosome peaks corresponding to position clusters (Figure 6A). In control cells, the first nucleosome peak on MOG1 and the corresponding position cluster were greatly diminished. MOG1 shares a divergent promoter with OPI3, the coding region of which was somewhat depleted of nucleosomes (Table 1), but it is unclear whether this effect was communicated from MOG1, because OPI3 is also repressed by 3AT (30). The gene with the most disrupted chromatin in control cells was URA1. There was a major loss of occupancy over the coding region in control cells relative to 3AT-treated cells (Figure 6B). All nine position clusters located between the nucleosome-depleted regions flanking URA1 in 3AT-treated cells were disrupted in control cells (Figure 6B). In conclusion, MOG1 and URA1 are repressed by 3AT (30) and their chromatin structures showed the opposite transition from 3AT-induced genes.
Figure 6.
Repression of MOG1 and URA1 by 3AT results in re-ordering of disrupted chromatin structure. Occupancy profiles and position cluster analysis for two genes with disrupted chromatin structure in control cells (Table 1) that are repressed by 3AT (30) (Table 1). Control cells: red trace; 3AT-treated cells: green trace. All sequences are included. Nucleosomes are indicated by ovals drawn to scale. Grey lines indicate 250-bp intervals. (A) Nucleosome occupancy and positioning on MOG1. (B) Nucleosome occupancy and positioning on URA1.
DISCUSSION
Advantage of paired-end sequencing for mapping nucleosome positions
Nucleosome length distributions indicate that each sample contains fully trimmed nucleosome core particles, together with some incompletely trimmed nucleosomes and damaged core particles. This was expected because of MNase digestion kinetics, the possible influence of H1, and slower trimming of the final ∼20 bp of linker DNA. DNA length is essential information for determining accurate nucleosome positions. If the DNA is significantly >150 bp, there is uncertainty in the position of the nucleosome, because it occupies only 145–150 bp. If the DNA is significantly <145 bp, the position is also unclear, because the nucleosome from which it is derived must have been cleaved internally or trimmed excessively from one or both ends. Sequences which are too long or too short can be selectively excluded from paired-end data, but not from single-end data.
In our analysis, we consider only DNA fragments of approximately mono-nucleosome size (any protected DNA fragments much larger or much smaller than the nucleosome are not present in our data sets, because the DNA was gel purified). Thus, we are considering only canonical nucleosomes, which are really defined by their ability to protect ∼147 bp. We expect that there might be a small fraction of ∼147-bp sequences scored as canonical nucleosomes that are not canonical nucleosomes. Some of these sequences might correspond to internal cleavage sites in neighbouring nucleosomes. If so, such sequences would have to contain an intact linker, which is unlikely and cannot be quantitatively very significant because such cleavages would smear the nucleosomal repeat pattern. Another possible problem is that a transcription factor bound adjacent to a nucleosome might protect some linker DNA after the very extensive digestion used to make core particles, but we are not aware of any studies indicating that transcription factors offer strong protection against MNase digestion. It is worth noting that transcription factors bind reversibly to DNA (unlike histones in the nucleosome) and so would be expected to offer less protection. In addition, even histone H1, which binds tightly to the nucleosome, offers only transient protection from MNase under these conditions. Overall, we believe that the vast majority of ∼147-bp sequences are indeed canonical nucleosomes.
There is potential for bias in genome-wide sequencing studies, particularly because two different DNA amplification steps are involved. We do not believe that bias is a major problem in our study because: (i) we have validated our current mapping data by comparison with some famous examples in the classical literature (Figures 2 and 3); (ii) the average nucleosome occupancy is very consistent across the genome; and (iii) our data are very reproducible.
Biological implications of position clusters
The chromatin structures reported here for the PHO5 promoter and TRP1 ARS1 (Figure 2) are consistent with previous studies using low-resolution indirect end-labelling (38,39). However, the higher resolution provided by paired-end sequencing reveals that each positioned nucleosome reported by indirect end-labelling is in fact an average of several overlapping positions (a position cluster). We and others have described similarly complex chromatin structures previously (11,16,23,31,39), but it has been generally assumed that these are atypical. More recently, complex chromatin structures have been noted genome wide in Caenorhabditis elegans (27). The present study demonstrates that complex chromatin structures are the rule in yeast chromatin, not the exception.
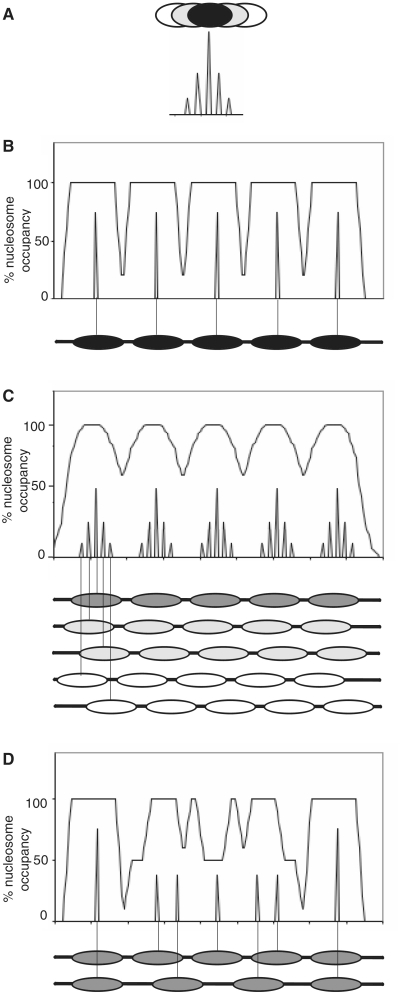
We define a position cluster as a set of overlapping positions, usually including a dominant position (Figure 7A). These must be alternative positions, because canonical nucleosomes cannot physically occupy the same DNA. In this context, it is worth noting that in vitro, a nucleosome can invade the territory of a neighbouring nucleosome, resulting in the loss of one H2A–H2B dimer and forming a particle that protects ∼250 bp from MNase digestion (42). If such coalesced nucleosomes are present in yeast, they would not appear in our maps because they protect much >147 bp.
Figure 7.
Alternative arrays can account for position clusters. (A) A position cluster with a central dominant position and four alternative positions. Peaks indicate nucleosome dyad positions. (B) Occupancy profile and position midpoints for an array of five perfectly positioned nucleosomes. Nucleosome: 145 bp; linker: 20 bp. Smoothed with a 25-bp moving average. (C) Occupancy profile and position midpoints for five alternative arrays of five nucleosomes: a dominant array (dark grey ovals; relative occupancy = 1); two arrays shifted by 20-bp upstream and downstream of the dominant array (light grey ovals; relative occupancy = 0.5); two arrays shifted by 40-bp upstream and downstream of the dominant array (white ovals; relative occupancy = 0.2). (D) Occupancy profile and position midpoints for two arrays of equal occupancy but different spacing, beginning and ending with the same nucleosome: upper array: five nucleosomes with 20-bp linker (165-bp repeat); lower array: four nucleosomes with 75-bp linker (220-bp repeat).
In a particular cell at a given moment, the nucleosome represented by a position cluster occupies one of the positions within the cluster. Thus, in some cells, the nucleosome will occupy the dominant position; in other cells, it will be at one of the alternative positions. This observation has important biological implications. For example, many models proposed for the regulation of specific genes depend on precise positions adopted by nucleosomes at the promoter, with critical transcription factor-binding sites located in the linker DNA, or just inside the nucleosome core, rather than in the inaccessible centre. Our data imply that factor-binding sites at nucleosomal promoters (e.g. PHO5), might be accessible in some cells, but not in others. It seems likely that remodelling machines will play critical roles here, because they are able to move nucleosomes along the DNA, perhaps from one position in a cluster to another, perhaps exposing or obscuring specific factor-binding sites. Furthermore, there is potential for stochastic effects, given that apparently identical cells can have different chromatin structures.
Position clusters and nucleosome spacing
Although the existence of position clusters indicates that chromatin structure is more complex than has been generally acknowledged, a significant simplifying factor is that nucleosomes in yeast are regularly spaced with an average linker length of 15–20 bp (a 160- to 165-bp repeat). Consequently, we propose that each position cluster corresponds to positions belonging to alternative arrays with the same spacing (17). An array of perfectly positioned nucleosomes predicts a ‘square-wave’ occupancy profile (Figure 7B), which is not generally observed. The only obvious example of a square nucleosome occupancy peak that we have found in our data is the single nucleosome located over each centromere, which is therefore perfectly positioned, but this nucleosome is unusual in that it contains CenH3 (Cse4), a variant of H3 (33).
A set of five overlapping arrays with the same spacing (165 bp) predicts a profile similar to the more regular profiles and position clusters observed experimentally (Figure 7C); this example is just one of many possibilities. Most arrays must have the same spacing to yield the observed bulk chromatin repeat of 165 bp, but quantitatively rare arrays could have quite different spacing. An interesting example is the square wave interference pattern generated in the case where half the cells have an array of five nucleosomes on gene X (165-bp spacing) and the other half have an array of four nucleosomes (220-bp spacing), beginning and ending with the same nucleosome (Figure 7D): both outermost nucleosomes give rise to a clear nucleosome peak in the occupancy profile, but the inner nucleosomes contribute an irregular pattern, including sharp spikes. Counter-intuitively, a well-positioned nucleosome is located below the central trough in the occupancy profile (Figure 7D). Thus, both regular and irregular occupancy profiles could be accounted for by overlapping regular arrays.
If a spacing factor begins at one nucleosome-depleted region and terminates at the next, then nucleosomes on a gene might be subjected to spacing from both ends, resulting in at least two alternative arrays. Little is known about how nucleosomes are spaced in yeast in vivo. In vitro, the yeast ISW1 and INO80 complexes can create arrays with ∼175-bp spacing, and ISW2 can assemble arrays with ∼200-bp spacing (43,44). In Drosophila, there are two well-characterized nucleosome spacing factors, ACF and CHD1 (45,46). How the activities of spacing factors interact in terms of array formation in vivo is an important question.
Low occupancy and disrupted position clusters on transcriptionally active genes
The effect of 3AT on the chromatin structures of some induced genes is dramatic: nucleosome occupancy is heavily reduced over the entire coding region. Moreover, some 3AT-repressed genes show the opposite trend: occupancy increases to normal levels in 3AT-treated cells. Thus, reduced occupancy correlates with transcriptional activation. In addition, nucleosome spacing is altered after induction on at least two genes (HIS3 and LYS1). Altered nucleosome spacing might reflect an intermediate chromatin state corresponding to a level of disruption in between the resting state and a major loss of canonical nucleosomes. Only a subset of 3AT-induced genes show extreme loss of canonical nucleosomes. These are probably the most transcriptionally active genes, since single gene and microarray studies indicate that the extent of histone loss from coding regions correlates with heavy transcription (47–51).
The mono-nucleosome sequencing approach identifies only canonical nucleosomes (i.e. those which protect ∼147 bp of DNA). Consequently, reduced occupancy on coding regions and at nucleosome-depleted regions could reflect actual loss of nucleosomes (resulting in free DNA), or the presence of ‘non-canonical’ nucleosomes which have been remodelled such that they no longer adequately protect their DNA from MNase. Thus, reduced occupancy on coding regions might reflect loss of the entire histone octamer, or of just one or both H2A–H2B dimers (48). Histone hexamers and H3-H4 tetramers protect less DNA than the octamer and DNA in this size range (∼80 to ∼120 bp) is not present in our preparations of core particle DNA. Alternatively, the histones might still be bound to the DNA, but present in remodelled nucleosomes, as we have suggested previously (16,52).
Loss of canonical nucleosomes can extend into flanking genes
In the cases of the genes most strongly affected by 3AT, loss of canonical nucleosomes occurs not just over the coding region, but extends into neighbouring genes (Figure 5). It seems unlikely that this is a direct effect of transcription, involving RNA polymerase II ploughing on into the chromatin of the downstream gene after release of the mRNA, because in some cases upstream genes are affected. Previously, we have observed disruption of nucleosome positioning on the flanking TRP1 gene in CUP1 or HIS3 plasmid chromatin after induction (11,16). However, since CUP1 and HIS3 were not in their native chromosomal contexts and the TRP1 gene was also active, the biological significance of the disruption of flanking chromatin structure is unclear. More recently, in Drosophila, a single-gene nucleosome scanning study has shown that heat shock induces nucleosome loss over a pair of divergently transcribed Hsp70 genes, extending in both directions into the flanking sequences as far as the scs and scs′ insulating elements (50). This effect does not depend on transcription, but on poly(ADP-ribose) polymerase (50). Since there is no evidence for this enzyme in yeast, the mechanism in yeast must be different, perhaps involving remodelling by SWI/SNF, as we have observed previously for HIS3 (16,52). We are currently investigating this possibility.
In summary, our genome-wide nucleosome sequence data show not only that there is a major loss of canonical nucleosomes from the coding regions of some 3AT-induced genes, but also that the positioning of the remaining nucleosomes is heavily disrupted. Thus, the chromatin structure of the coding region undergoes major remodelling on activation, with disruption of the dominant nucleosomal array and loss of canonical nucleosomes. This disruptive effect can be communicated to flanking genes through nucleosome-depleted promoters and 3′-regions that are seemingly unaffected, indicating that they do not act as strict boundaries. The factors that direct the formation of these domains of altered chromatin structure and determine their boundaries are currently under investigation.
ACCESSION NUMBER
We have submitted four sets of paired-end sequencing data to the GEO database; these correspond to two independent experiments: control cells and cells treated with 3AT. The accession number is GSE26493. These data are available to the public as of 20 July 2011.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health (National Institute for Child Health and Human Development).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Gary Felsenfeld, Alan Hinnebusch, Rohinton Kamakaka and Victor Zhurkin for helpful comments on the manuscript. The authors thank Kip Bodi and Michael Berne at the Tufts University Core Facility for paired-end sequencing.
REFERENCES
- 1.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 Å resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 5.van Holde KE. Chromatin. New York Inc, NY: Springer; 1988. [Google Scholar]
- 6.Thomas JO, Furber V. Yeast chromatin structure. FEBS Lett. 1976;66:274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- 7.Lohr D, Kovacic RT, van Holde KE. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochem. 1977;16:463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- 8.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochem. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 9.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen CH, Clark DJ. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 2001;276:35209–35216. doi: 10.1074/jbc.M104733200. [DOI] [PubMed] [Google Scholar]
- 11.Shen CH, Leblanc BP, Alfieri JA, Clark DJ. Remodelling of yeast CUP1 chromatin involves activator-dependent re-positioning of nucleosomes over the entire gene and flanking sequences. Mol. Cell. Biol. 2001;21:534–547. doi: 10.1128/MCB.21.2.534-547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JZ, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organisation of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struc. Mol. Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedor MJ, Lue NF, Kornberg RD. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J. Mol. Biol. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell. Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark DJ. Nucleosome positioning, nucleosome spacing and the nucleosome code. J. Biomol. Struc. Dyn. 2010;27:781–793. doi: 10.1080/073911010010524945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travers A, Hiriart E, Churcher M, Caserta M, di Mauro E. The DNA sequence-dependence of nucleosome positioning in vivo and in vitro. J. Biol. Struc. Dyn. 2010;27:713–724. doi: 10.1080/073911010010524942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowary PT, Widom J. Nucleosome packaging and nucleosome positioning of genomic DNA. Proc. Natl Acad. Sci. USA. 1997;94:1183–1188. doi: 10.1073/pnas.94.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan G, Liu Y, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 21.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 22.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 23.Ambrose C, Lowman H, Rajadhyaksha A, Blasquez V, Bina M. Location of nucleosomes in Simian Virus 40 chromatin. J. Mol. Biol. 1990;214:875–884. doi: 10.1016/0022-2836(90)90342-J. [DOI] [PubMed] [Google Scholar]
- 24.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A-Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 25.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comp. Biol. 2008;4:1–25. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Gen. Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Gen. Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent NA, Adams S, Moorhouse A, Paszkiewicz K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 2011;39:e26. doi: 10.1093/nar/gkq1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the “601” strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdone L, Camilloni G, di Mauro E, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell. Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaykalova DA, Nagarajavel V, Bondarenko VA, Bartholomew B, Clark DJ, Studitsky VM. A polar barrier to transcription can be circumvented by remodeller-induced nucleosome translocation. Nucleic Acids Res. 2011;39:3520–3528. doi: 10.1093/nar/gkq1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl Acad Sci. USA. 2011;108:12687–12692. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freidkin I, Katcoff DJ. Specific distribution of the Saccharomyces cerevisiae linker histone homologue Hho1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 36.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodelling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 37.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svaren J, Hörz W. Transcription factors vs. nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 39.Thoma F, Bergman LW, Simpson RT. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol. 1984;177:715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- 40.Houseley J, Rubbi L, Grunstein M, Tollervy D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fragoso G, John S, Roberts MS, Hager GL. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- 42.Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen Hughes T. Nucleosomes can invade DNA territories occupied by their neighbours. Nat. Struc. Mol. Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Udugama M, Sabri A, Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struc. Mol. Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 46.Maier VK, Chioda M, Rhodes D, Becker PB. ACF catalyses chromatosome movements in chromatin fibres. EMBO J. 2008;27:817–826. doi: 10.1038/sj.emboj.7601902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee C, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 49.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y, Clark DJ. SWI/SNF-dependent long-range remodelling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. USA. 2002;99:15381–15386. doi: 10.1073/pnas.242536699. [DOI] [PMC free article] [PubMed] [Google Scholar]