Abstract
Background
Excessive alcohol consumption contributes to significant morbidity and mortality. Heritable influences contribute to 50% of the variation in alcohol consumption, suggesting the important role of genes. We used data on a previously defined alcohol consumption factor score in a sample of 827 young women to investigate association with 1014 single nucleotide polymorphisms in genes related to addiction.
Methods
Data were drawn from the Missouri Adolescent Female Twin Study (MOAFTS) with replication in the College Drinking Sample (CDS). Genotypic and phenotypic data were available on 827 MOAFTS and 100 CDS women of European- American ancestry. Data on 1014 SNPs across 130 genes related to addiction were utilized. Association was conducted in QTDT, which allows for identity-by-descent information to account accurately for twin status in the analysis. The total association variance components model was used, with specification of variance components for relatedness in MOAFTS.
Results
The top signals included clusters of SNPs in TPH2 (e.g. rs1386496, p=0.0003) and DDC (e.g. rs3779084, p=0.0008), genes that encode proteins responsible for serotonin synthesis. Additional polymorphisms in ADH1B, ADH1C, ADH7 and ADH1A1 were also associated at p < 0.05. The FDR for the top signal (p=0.0003) was 0.15 suggesting nominal significance only. Replication was limited and noted for 2 SNPs in ADH1C.
Conclusions
While no results survive the burden of multiple testing, nominal findings in TPH2 and DDC suggest the potential role of the serotonin synthesis pathway in alcohol consumption.
Introduction
Excessive alcohol consumption is among the leading contributors to preventable deaths worldwide (Centers for Disease Control (CDC), 2004). Excessive alcohol consumption is associated with traffic-related fatalities and other injuries to self and others as well as adverse health consequences (Rehm et al., 2009). Every year, in the U.S. alone, over 20 billion dollars are invested to ameliorate the burden imposed by alcohol-related negative health outcomes (Rice, 1999).
The etiology of excessive alcohol consumption is complex. Family and twin studies show that most quantitative indices of alcohol consumption (e.g. quantity × frequency, maximum drinks) are highly heritable (Heath and Martin, 1994). In a prior study (Agrawal et al., 2009), we demonstrated that genetic factors account for 34–47% of the variance in alcohol consumption indices with the remainder arising from individual-specific environmental factors. We also provided the rationale and measurement characteristics of a composite measure – an alcohol consumption factor score, with a heritability of 50%. This factor score accounts for 50–100% of the genetic influences on individual indices of alcohol consumption. The overlap between this factor and alcohol dependence was confirmed in an independent sample of adult Australian twins (Grant et al., 2009) and more recently, in a sample of Virginia twins (Kendler et al., 2010).
In contrast to commonly used symptom counts of alcohol dependence symptomatology, the alcohol consumption factor score has the favorable characteristic of being more representative of a greater majority of exposed drinkers. Its distribution and measurement invariance, tested by comparing factor loadings across samples, (Agrawal et al., 2009) make it a highly suitable phenotype for genomic studies. Further, using this quantitative measure allows us to use data on all subjects, not just those with diagnostic abuse or dependence (e.g. classical transmission disequilibrium test (TDT) where only transmission to an affected offspring is studied), which increases sample size.
There are currently several strategies for association studies of alcohol consumption – the two most frequently used techniques are (i) genomewide association studies (GWAS) where markers are placed across the length of the entire genome, ranging in density from 370,000 to over a million (Bierut et al., 2010; Edenberg et al., 2010; Treutlein et al., 2009) and (ii) candidate gene studies (Dick and Bierut, 2006; Edenberg and Foroud, 2006), where polymorphisms are typed in genes of putative biological relevance. Both techniques have strengths and weakness – for instance, GWAS allows for gene discovery by taking a somewhat agnostic approach to SNP selection – it is limited, however, by difficulty in explaining association results when identified SNPs are intergenic. Candidate gene studies, on the other hand, rely on fewer SNPs in genes of interest – while this limits the ability to discover novel polymorphisms, it provides interpretability and potential for replication – we selected the latter approach.
For this study, we selected 1014 SNPs genotyped in addiction-related genes. The selection of genes that may influence addiction to a variety of psychoactive substances and also be relevant for alcohol consumption is well supported by twin studies demonstrating a high degree of genetic overlap across use and misuse of alcohol, tobacco and illicit drugs (Dick et al., 2009b; Kendler et al., 2007; Krueger et al., 2002). The selection of these genes is based on a prior report by Hodgkinson and colleagues (Hodgkinson et al., 2008)– based on a review of the literature, the authors designed a 1536 SNP array mapping to 1350 tagging SNPs (and 186 ancestry informative SNPs) in 130 candidate genes that were selected across the following domains: GABA (e.g. GABRA2), Serotonin (e.g. TPH2), Dopamine (e.g. DRD2), NMDA (e.g. GRIN2B), Opioid (e.g. OPRM1), Glycine (e.g. GLRA1), Cannabinoid (e.g. CNR1), Cholinergic (e.g. CHRNA4), Adrenergic (e.g. ADRA1A), Drug Metabolism (e.g. ADH1A), Signal Transduction (e.g. FOS), Stress (e.g. CRHR1) and Other (e.g. BDNF). SNPs were selected from within the gene as well as upstream and downstream regions and care was taken to ensure the prioritization of exonic polymorphisms while keeping inter-marker linkage disequilibrium low. The panel was validated across 7 independent samples.
The aim of this study is to investigate the association between the alcohol consumption factor score and 1014 SNPs from an array designed (as described above) for coverage of addiction-related genes, in a sample of 827 young adult twin women from the Missouri Adolescent Female Twin Registry, with an effort to replicate findings in an independent sample of 100 European American women from the College Drinking Study. The study of alcohol consumption in women is of particular interest as excessive alcohol consumption is relatively less common in women than in men and genetic pathways mediating risk for excessive alcohol consumption may vary across the sexes.
Materials and Methods
Sample
The primary sample for the candidate gene study was drawn from a large prospective cohort study of women born in a Midwestern state of the United States. The Missouri Adolescent Female Twin Study (MOAFTS) consists of a cohort of female same-sex twin pairs born between July 1st 1975 – June 30th 1985 who were identified from birth records (Heath et al., 2002). Twins were eligible to participate if both members of the twin pair had survived past infancy, were not adopted and if their biological parents were Missouri residents at the time of the twins’ birth. Using a cohort-sequential sampling design, twins and at least one biological parent (typically the biological mother), were invited to participate in the baseline interviews during 1994–1999, when the twins were 13, 15, 17 or 19 years old. Recruitment of additional 13 year olds continued over a two-year period as twins became age-eligible. A telephone diagnostic interview was administered, first to the parents, and subsequently, after obtaining parental permission, to the twins (minors). Further details regarding sample recruitment and characteristics of this first wave of interview data, which are not utilized in the current study, are given elsewhere (Heath et al., 1999; Knopik et al., 2005). Subsequently, participants were invited to participate in several full-length and mini follow-up interviews and respond to mailed questionnaires. During 2002–2005, the first full-length follow-up interview was completed, which included a detailed assessment of substance involvement. All eligible twins, including those who may not have completed a baseline assessment, were invited to participate in the follow-up, provided that they, or their parents, had not previously indicated an unwillingness to participate in future studies. A total of 3,787 twins, aged 18–29 years, completed follow-up interviews. Twins were also invited, during this period, to provide permission for future blood sample collection for genomic studies. Of those contacted, 1191 subjects, including one or both members of monozygotic and dizygotic twin pairs provided a sample – collection of samples from remaining subjects is ongoing and this study reports on genotyping efforts completed on 1,188 subjects (with 3 dropped due to genotyping problems) of whom 827 also had phenotypic data.
Replication sample
For this study, we used data on 100 women from the College Drinking study who were also genotyped with the MOAFTS subjects on the Addiction array. The CDS (Sher et al., 1991; Sher et al., 1996) comprised of 489 freshmen (46% male, mean age = 18.2) from a large Midwestern University, who were assessed prospectively, using interviews and questionnaires, seven times (Years 1, 2, 3, 4, 7, 11, and 16) over a 16 year period. Of the 489 subjects in the baseline sample, 51% could be classified as family history positive for alcoholism. In all, over 84% of participants were retained over the first 11 years of the study and over 78% were retained through Year 16 (mean age = 34.5 years). 203 (108 women) subjects, of which 175 reported European-American ancestry, had relevant phenotypic data, provided a viable DNA sample and were successfully genotyped.
Genotyping
As part of a broader study of addiction genetics, 1536 single nucleotide polymorphisms (SNPs) that comprise the custom ‘Addiction’ array were genotyped on all subjects. Genotyping was completed by Illumina and of the 1536 SNPs, 1441 (success rate of 93.8%) were successfully typed on 1,188 MOAFTS and 203 CDS subjects. These included 23 of the 186 SNPs included as ancestry informative markers (AIMs)(Enoch et al., 2006). A list of genes that encompass the Addiction array may be found in another publication (Hodgkinson et al., 2008). The genotyping success rate was greater than 99.5% with 100% reproducibility in 220 plated duplicate samples.
Data cleaning and quality control
Upon receipt of genotypic data, internal checks to verify reproducibility and call frequencies were completed. Additional protocols for management of genetic data were also completed:
Checks were completed, in PLINK, (Purcell et al., 2007) for low call rates (less than 95%, 24 SNPs) and deviations from Hardy-Weinberg equilibrium (p < .00001, 123 SNPs). For the present analyses, SNPs with minor allele frequency of less than 5% were also excluded (286 SNPs). This led to 1032 SNPs (including 18 AIMs) (Enoch et al., 2006) to be included in data analysis.
Verification of relatedness in GRR (Abecasis et al., 2001b) led to exclusion of a twin pair that appeared genetically unrelated and one sample that appeared to be a duplicate. GRR also detected 250 MZ twins pairs – of these, 35 pairs had been previously classified as DZ twins based on self-reported parental and respondent phenotypic data. There was also very modest discordance in MZ pair genotypes – of the 250 × 1441 genotypes, only 54 were set to missing due to differing within-pair genotypes.
Based on self-reported ethnicity, African-American twins were excluded from analysis.
A detailed characterization of the 1014 SNPs and 18 AIMs used for these analyses, in the European-American participants from MOAFTS, is available in Supplemental Table 1.
Measures
An alcohol consumption factor score was created by factor analyzing (a) maximum typical consumption (quantity × frequency), (b) maximum drinks in 24 hours, (c) frequency of heavy (5 or more) drinking and (d) frequency of drinking to intoxication during heaviest use. We have previously demonstrated the psychometric properties and heritability of this measure – furthermore, we have shown that the alcohol consumption factor largely passes tests of measurement invariance, with evidence for differential factor loadings for typical consumption and 5 or more, across MOAFTS and CDS (Agrawal et al., 2009). Factor loadings for the four indices ranged from 0.61 (maximum drinks) to 0.90 (5 or more drinks) with individual heritabilities ranging from 0.34 to 0.47. The overall heritability of the factor score was 50%, and a majority of the genetic influences on the individual indices (76–100%) overlapped with the genetic influences on the overall factor score, except those on maximum drinks (37%). These indices were only available in individuals who reported a lifetime history of consuming 6 or more alcohol beverages and thus, the final number of subjects analyzed was 827.
Data Analysis
Association analyses were conducted in QTDT (Abecasis et al., 2000; Abecasis et al., 2001a), which allows for variance-components modeling. The pedigree structure required for analysis is family-based and required parents to be included in each pedigree. However, parental phenotypic and genotypic data is not required – as this was not available for our study, they were set to missing. For the MOAFTS data, MZ twins were retained in the data (although re-doing analyses by randomly excluding one MZ twin did not change results) and a variance components model, including components for additive genetic (a), polygenic effects (g), twin environment (t) and unique environment (e) were modeled. Total association, which is distinct from classical transmission disequilibrium test (TDT), was modeled while accounting for sibling resemblance via identity-by-descent (calculated in MERLIN (Abecasis et al., 2002)) and the variance specification (polygenic, additive, environmental and twin environmental components). For the replication, total association was modeled but as all subjects were unrelated, variance-components were not specified.
Results
Representativeness of genotyped sample
The subset of genotyped twins did not differ from their counterparts who did not provide a DNA sample (Table 1). When compared with those who had phenotypic but no genotypic data, genotyped subjects were similarly aged and had comparable rates of alcohol abuse/dependence, nicotine dependence, psychopathology, lifetime cannabis use and self-reported exposure to childhood sexual and physical abuse. Also as shown in Table 1, the MOAFTS sample did not ascertain for any psychiatric or environmental measures, being largely representative of the Midwestern female population (Heath et al., 2002). Further, in the genotyped sample, the alcohol consumption factor score and its components were strongly correlated (r=0.33–0.44) to DSM-IV alcohol abuse/dependence underscoring their relevance as indices of problematic drinking.
TABLE 1.
Psychiatric characteristics of MOAFTS twins who were genotyped versus those not currently genotyped.
| Genotyped (N=1178) | Not Genotyped (2609) | |
|---|---|---|
| Mean age | 21.7 [18–28 years] | 21.7 [18–28 years] |
| Cannabis use | 43.8% | 45.5% |
| Childhood sexual abuse | 6.4% | 8.0% |
| Childhood physical abuse | 6.8% | 7.1% |
| Panic attack | 22.2% | 25.0% |
| Depressed mood | 57.8% | 55.9% |
| Suicidal ideation | 26.3% | 24.6% |
| Nicotine dependence | 18.8% | 20.7% |
| Alcohol abuse/dependence | 18.0% | 17.5% |
| Family history of drinking problems | 32.5% | 30.3% |
Note: None the differences across the genotyped and non-genotyped groups are statistically significant at p < 0.05.
Association analysis in MOAFTS
Of the 1014 SNPs genotyped, 67 SNPs had p-values less than 0.05 (Table 2). The top two SNPs (p=0.0003), in high linkage disequilibrium (LD) with each other, were in the tryptophan hydroxylase 2 (TPH2) gene followed by 4 SNPs in the dopa decarboxylase (DDC) gene – all these polymorphisms were intronic. In total, across the significant findings, 6/12 TPH2 and 5/12 DDC SNPs were associated with the alcohol consumption factor score at p < 0.05. Other prominent associations included a cluster of SNPs within the cluster of alcohol dehydrogenase (ADH) genes of chromosome 4 (2/4 in ADH1B, 2/6 in ADH1C and 2/9 in ADH7) as well as 3 (of 8) SNPs in ALDH1A1 on chromosome 9. Other suggestive association in putative signal transduction genes neurotensin 1 (NTSR1) and protein kinase C, epsilon (PRKCE) as well as in the GABA-ergic (GABRB2 and GABRB3) and glutamatergic-NMDA (GRIN2B) was also noted.
TABLE 2.
Association results (p<0.05) for 1014 SNPs associated with addiction and alcohol consumption in young women.
| Chromosome | SNP | Gene | Position (bp) |
MOAFTS p-value |
CDS replication p-value |
FDR p-value |
|---|---|---|---|---|---|---|
| 4 | rs1229967 | ADH1A | 100426601 | 0.0077 | 0.30 | 0.6006 |
| 4 | rs13134764 | ADH1A | 100431196 | 0.039 | 0.48 | 0.7323 |
| 4 | rs1159918 | ADH1B | 100462032 | 0.0236 | 0.23 | 0.7323 |
| 4 | rs1353621 | ADH1B | 100460598 | 0.0339 | 0.58 | 0.7323 |
| 4 | rs904096 | ADH1C | 100482607 | 0.0376 | 0.005 | 0.7323 |
| 4 | rs698 | ADH1C | 100479812 | 0.0408 | 0.005 | 0.7356 |
| 4 | rs729147 | ADH7 | 100552290 | 0.0267 | 0.96 | 0.7323 |
| 4 | rs894369 | ADH7 | 100552869 | 0.0267 | 0.96 | 0.7323 |
| 9 | rs1888202 | ALDH1A1 | 74709071 | 0.0307 | 0.38 | 0.7323 |
| 9 | rs2303317 | ALDH1A1 | 74731762 | 0.037 | 0.51 | 0.7323 |
| 9 | rs3764435 | ALDH1A1 | 74706696 | 0.0376 | 0.38 | 0.7323 |
| 4 | rs2000978 | CCKAR | 26097087 | 0.0387 | 0.99 | 0.7323 |
| 20 | rs6011776 | CHRNA4 | 61454200 | 0.0139 | 0.31 | 0.7323 |
| 20 | rs3787138 | CHRNA4 | 61449668 | 0.022 | 0.15 | 0.7323 |
| 6 | rs1049353 | CNR1 | 88910354 | 0.0313 | 0.78 | 0.7323 |
| 22 | rs174696 | COMT | 18333176 | 0.0479 | 0.84 | 0.7472 |
| 9 | rs6271 | DBH | 135512095 | 0.0189 | 0.99 | 0.7323 |
| 7 | rs3779084 | DDC | 50536229 | 0.0008 | 0.72 | 0.2231 |
| 7 | rs880028 | DDC | 50537630 | 0.001 | 0.45 | 0.2231 |
| 7 | rs732215 | DDC | 50511557 | 0.0011 | 0.91 | 0.2231 |
| 7 | rs2122822 | DDC | 50519646 | 0.0023 | 0.45 | 0.3332 |
| 7 | rs4490786 | DDC | 50511808 | 0.0031 | 0.36 | 0.3493 |
| 7 | rs9639213 | DGKB | 14707372 | 0.0286 | 0.90 | 0.7323 |
| 11 | rs4648318 | DRD2 | 112818599 | 0.0043 | 0.69 | 0.436 |
| 3 | rs2134655 | DRD3 | 115340891 | 0.0306 | 0.04 | 0.7323 |
| 22 | rs5753625 | EIF4ENIF1 | 30184846 | 0.0456 | 0.98 | 0.7356 |
| 2 | rs12624279 | FOSL2 | 28488294 | 0.0167 | 0.24 | 0.7323 |
| 5 | rs11949658 | GABRB2 | 160734301 | 0.0061 | 0.87 | 0.5623 |
| 15 | rs7179279 | GABRB3 | 24532939 | 0.0475 | 0.60 | 0.7472 |
| 4 | rs10517662 | GLRB | 158312511 | 0.0133 | 0.22 | 0.7323 |
| 1 | rs6424922 | GLT25D2 | 182240761 | 0.0113 | 0.21 | 0.7161 |
| 12 | rs2300238 | GRIN2B | 13704597 | 0.0091 | 0.08 | 0.6152 |
| 12 | rs220573 | GRIN2B | 13848001 | 0.0256 | 0.98 | 0.7323 |
| 12 | rs2268122 | GRIN2B | 13788728 | 0.0292 | 0.73 | 0.7323 |
| 12 | rs2284416 | GRIN2B | 13810481 | 0.037 | 0.84 | 0.7323 |
| 15 | rs10152453 | HERC1 | 61885666 | 0.0497 | 0.47 | 0.7636 |
| 11 | rs2276302 | HTR3A | 113355350 | 0.0437 | 0.43 | 0.7356 |
| 12 | rs6582086 | INTERGENIC | 70784458 | 0.007 | 0.45 | 0.5915 |
| 8 | rs2609998 | INTERGENIC | 57522588 | 0.009 | 0.36 | 0.6152 |
| 10 | rs10883533 | INTERGENIC | 102462646 | 0.0185 | 0.71 | 0.7323 |
| 8 | rs3808627 | INTERGENIC | 54327355 | 0.0195 | 0.61 | 0.7323 |
| 6 | rs806377 | INTERGENIC | 88915442 | 0.0215 | 0.85 | 0.7323 |
| 20 | rs3830064 | INTERGENIC | 1923679 | 0.0242 | 0.62 | 0.7323 |
| 9 | rs4406477 | INTERGENIC | 74842278 | 0.0252 | 0.19 | 0.7323 |
| 7 | rs255102 | INTERGENIC | 30697689 | 0.0254 | 0.75 | 0.7323 |
| 5 | rs2416504 | INTERGENIC | 117436991 | 0.0337 | 0.33 | 0.7323 |
| 3 | rs6414248 | INTERGENIC | 112365016 | 0.0384 | 0.79 | 0.7323 |
| 20 | rs2427400 | INTERGENIC | 60806120 | 0.0429 | 0.49 | 0.7356 |
| 12 | rs2220159 | INTERGENIC | 70792554 | 0.0438 | 0.63 | 0.7356 |
| 6 | rs851011 | MAPK14 | 36145961 | 0.0129 | 0.35 | 0.7323 |
| 16 | rs7698 | MAPK3 | 30033301 | 0.0312 | 0.99 | 0.7323 |
| 9 | rs10733253 | MPDZ | 13095638 | 0.0419 | 0.29 | 0.7356 |
| 20 | rs3787535 | NTSR1 | 60823966 | 0.0263 | 0.92 | 0.7323 |
| 20 | rs6089930 | NTSR1 | 60829446 | 0.0344 | 0.13 | 0.7323 |
| 20 | rs3915568 | NTSR1 | 60839917 | 0.0423 | 0.82 | 0.7356 |
| 6 | rs2272381 | OPRM1 | 154584183 | 0.0181 | 0.28 | 0.7323 |
| 2 | rs1868389 | PRKCE | 45991373 | 0.0023 | 0.23 | 0.3332 |
| 2 | rs4953268 | PRKCE | 45922798 | 0.0346 | 0.95 | 0.7323 |
| 2 | rs921183 | PRKCE | 46143920 | 0.0373 | 0.88 | 0.7323 |
| 3 | rs2272399 | SLC6A11 | 10950745 | 0.0447 | 0.19 | 0.7356 |
| 12 | rs1386496 | TPH2 | 70637057 | 0.0003 | 0.55 | 0.1521 |
| 12 | rs1843809 | TPH2 | 70634965 | 0.0003 | 0.55 | 0.1521 |
| 12 | rs1386488 | TPH2 | 70630885 | 0.0031 | 0.73 | 0.3493 |
| 12 | rs6582078 | TPH2 | 70661158 | 0.0245 | 0.55 | 0.7323 |
| 12 | rs2171363 | TPH2 | 70646531 | 0.0321 | 0.98 | 0.7323 |
| 12 | rs1352250 | TPH2 | 70684051 | 0.0457 | 0.98 | 0.7356 |
Despite these intriguing findings, none of our signals would be considered statistically significant after adjustment for multiple testing, using Bonferroni corrections (.05/1041) or by the False Discovery Rate (FDR) (Benjamini and Hochberg, 1995), which was 0.15 for the top two SNPs with raw p-values of 0.0003. Thus, results should be interpreted with caution – keeping this in mind, we only present details on TPH2 and DDC which have the lowest FDR values.
Mean levels of alcohol consumption by genotype
As shown in Table 3, carriers of the minor allele in the TPH2 SNP, rs1386496, had lower alcohol consumption factor scores while carriers of the minor allele of the DDC SNP, rs3779084, had higher scores. However, as these SNPs are intronic, it is not possible to speculate whether the mechanism of gene action for TPH2 and DDC is similar or different.
TABLE 3.
Distribution of alcohol consumption factor scores in young women, by rs1386496 in TPH2 and by rs3779084 in DDC.
| Copies of minor allele |
MOAFTS | CDS | |||||
|---|---|---|---|---|---|---|---|
| N | Mean factor score [SD] |
p- value |
N | Mean factor score [SD] |
p- value |
||
| r1386496 | 0 | 572 | 0.0541 [0.93] | <0.0001 | 71 | −0.089 [0.79] | ns |
| 1 or 2 | 255 | −0.215 [0.78] | 29 | −0.040 [0.91] | |||
| rs3779084 | 0 | 502 | −0.106 [0.85] | <0.0001 | 53 | −0.1462 [0.76] | ns |
| 1 or 2 | 324 | 0.089 [0.94] | 47 | 0.006 [0.87] | |||
Linkage disequilibrium
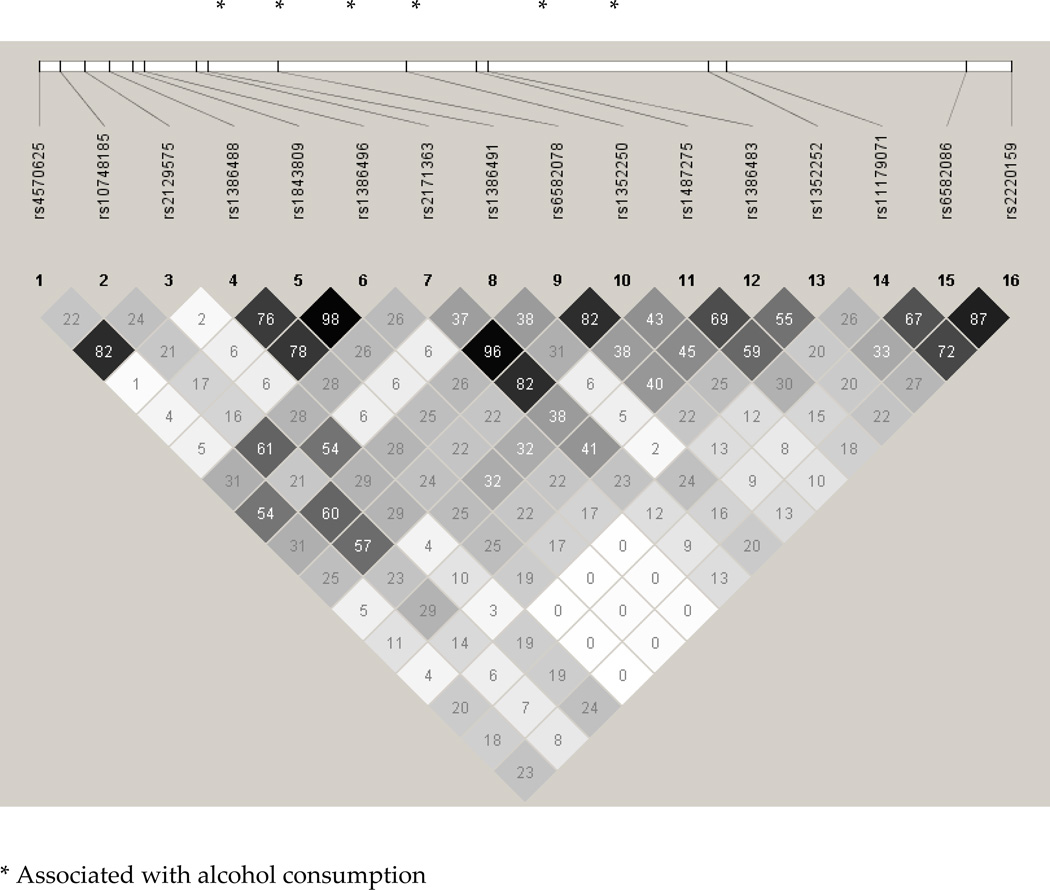
As shown in Figure 1, LD across genotyped markers in TPH2 varied substantially – while the three signals with the lowest p-values were in high LD, there was considerably less inter-marker LD across the other SNPs, suggesting a more diffuse association spreading across the gene. The top association signals in DDC, rs3779084 and rs880028, were correlated with each other and with rs4490786, but not with a majority of the remaining SNPs – these are shown in Figure 2.
FIGURE 1.
LD structure, as measured using r2, of Tryptophan Hydroxylase 2 (TPH2) SNPs genotyped in MOAFTS.
FIGURE 2.
LD structure, as measured using r2, of Dopa Decarbozylase (DDC) SNPs genotyped in MOAFTS.
Replication analysis in CDS
Of the signals associated at p < 0.05 in MOAFTS, only 2 SNPs in ADH1C were replicated in the CDS subset of 100 European-American women.
Discussion
This study sought to examine the association between 1014 SNPs from the Addiction array and an alcohol consumption factor score in 827 young adult female twin participants from Missouri. Several SNPs in TPH2 and DDC were associated with alcohol consumption at p < 0.05. TPH2 has previously been implicated as a gene potential associated with alcohol-related anxiety and suicidality. In an alcohol detoxification study, a functional polymorphism G-703T (rs4570625) as well as an intronic proxy (rs2129575) in TPH2 were found to be marginally associated with baseline Hamilton Depression and Anxiety ratings (Serretti et al., 2009). However, in two studies of alcohol dependence, no significant main or epistatic effects of TPH2 were noted (Drago et al., 2009; Zill et al., 2007). Furthermore, a study of alcohol consumption in college students found no association with G-703T (Gacek et al., 2008). The SNPs identified by us are not in LD with these markers (r2=0.01–0.40).
Multiple markers in DDC were also associated with alcohol consumption. The top association signals, rs3779084 and rs880028 were correlated with each other but not with rs732215 which was also in low LD (r2=0.37, see Figure 2), with rs2122822, which is in high LD (r2=0.97) with rs12718541 which was previously reported to be associated with the Fagerstrom Test for Nicotine Dependence in European- and African-American subjects (Ma et al., 2005; Yu et al., 2006; Zhang et al., 2006). Our study demonstrates that this association extends to alcohol consumption as well. However, another SNP (rs921451) which has also been found to be associated with smoking-related outcomes was not associated with alcohol consumption in our study.
What is particularly noteworthy about our findings is that TPH2 and DDC encode two critical steps in the production of serotonin (Haavik et al., 2008), a key neurotransmitter in humans. TPH2 is responsible for the rate-limiting step in the biosynthesis of serotonin – the conversion of dietary L-tryptophan to 5-hydroxytryptophan. DDC encodes a protein that is responsible for the next step in the biosynthesis pathway – the conversion of 5-hydroxytryptophan to serotonin. DDC is also involved in synthesis of dopamine from L-dopa. This provides some early, albeit nominal support, for a possible role of the serotonergic and dopaminergic system in alcohol consumption in women.
Our study also revealed nominal associations with several SNPs in alcohol dehydrogenase genes, ADH1B, ADH1C, ADH7 and ADH1A1 with SNPs in ADH1C showing replication in the CDS sample. The two SNPs that show replication are vital in the enzymatic activity (Edenberg, 2007; Hoog et al., 1986) of ADH1C. rs698 is a missense mutation (Ile349Val) and the other SNP, rs904096, while intronic, is in perfect linkage disequilibrium with rs1693482, which is another missense mutation (Arg271Gln). This underscores the robustness of the findings in the metabolism genes. SNPs in these genes have previously been implicated to be associated with rate of elimination of alcohol, alcohol reactions (flushing), consumption and dependence (Edenberg et al., 2006; Macgregor et al., 2009). The most promising finding with alcohol consumption (maxdrinks) was shown for rs1229984, a missense mutation (Arg48His) in ADH1B, where the G (48Arg) allele was associated with increased alcohol consumption – while this SNP was typed in our sample, its minor allele frequency was 0.036 and hence it was excluded from analyses. In light of the putative findings in ADH1B, we analyzed this SNP and found it to not be associated with alcohol consumption in our sample. One prior study has also noted associated between rs8187974 in ALDH1A1 and alcohol dependence, however our signals in the gene do not appear to be in strong LD with that SNP. We also did not note association signals for SNPs previously reported in ADH4 (Luo et al., 2006; Preuss et al., 2010).
Prior studies have found evidence for association between alcohol dependence and GABRA2 (Edenberg et al., 2004; Fehr et al., 2006; Lappalainen et al., 2005; Lind et al., 2008; Soyka et al., 2008). In the present study, no such evidence was found. A possible explanation for this lack of association may be that the links between GABRA2 and alcohol misuse have previously been attributed to a general predisposition to externalizing behaviors (Dick et al., 2009a), which may be somewhat less prevalent in this sample of young women.
Some limitations of this study are noteworthy. First, primary (MOAFTS) data for this study come from a sample of young women, aged 18–28 years, born in Missouri – results may not generalize to other samples. To what extent this would contribute to non-replication in a male sample, remains unknown, and is in some regards, dependent on the mechanisms governing the association between the SNPs identified in this sample and alcohol consumption. The twin literature (Heath et al., 1997; Prescott et al., 1999) and emerging genomewide association studies (Bierut et al., 2010; Edenberg et al., 2010; Treutlein et al., 2009) find limited evidence for gender heterogeneity attributable to genetic factors, therefore, it is more likely than not that some of these signals would also be noted in a similarly ascertained male cohort. Relatedly, other studies may wish to examine the role of these SNPs in older and younger cohorts – there is some evidence for genetic variability in alcohol consumption, particularly at younger ages/adolescence (Rose, 1998) and the effect of this variation on genotype remains unexplored. Second, due to inadequate numbers of genotyped African-American participants, they could not be analyzed jointly with the European-American subjects and hence, were excluded to avoid spurious stratification-related signals. Third, our lack of replication suggests caution in the interpretation of these results – to what extent this absence of replication is attributable to the relatively small size of the CDS sample or whether the association signals are highly specific to our cohort of young women from a Midwestern population (versus, women attending a college with a prominent Greek system (Bartholow et al., 2003; Park et al., 2008; Sher et al., 2001) remains to be explored. Additionally, the CDS selected a subset of their participants to be family history positive for alcoholism while MOAFTS is a general population sample with only 30% of the sample reporting parental drinking problems (Table 1) and this too may have contributed to non-replication. An examination of the data reveal that while CDS women report higher maximum typical consumption, MOAFTS women tend to report significantly higher drinking to intoxication and frequency of heavy (5+ drinking).
Perhaps the most critical contributor to such association studies and to subsequent replication is power attributable to sample size. Using Quanto (Guaderman and Morrison, 2006), we determined that, assuming non-relatedness, association analyses with a population standardized quantitative trait would identify effect sizes of R2 = 0.008 and greater in a sample of 827 individuals. However, only those explaining more than 6% of the variance, a rather optimistic expectation for complex behavioral traits such as alcohol consumption, would replicate in the smaller CDS sample. Undoubtedly, additional ongoing genotyping of these samples is required.
While there is some, related, concern regarding the rather modest p-values emerging from this study, it is important to note that our Bonferroni and FDR corrections apply to all genotypes that encompass this research project. This needs to be contrasted with other prominent candidate gene projects where corrections for multiple-testing are regularly applied at the gene level (i.e. correcting within a gene but not across all genes). For instance, if we had simply opted to report association results for TPH2 and DDC alone (i.e. 24 SNPs), our p-values would well have exceeded the Bonferroni corrected threshold of 0.002. We do not advocate for the superiority of one strategy versus another – indeed, in many studies with an ongoing genotyping component, it is often impossible to know, a priori, how many SNPs will eventually be genotyped and hence, preclude such an omnibus correction. However, the statistical significance of our results should be viewed in the context of these candidate gene studies, with the addition of a more stringent correction for all SNPs within all genes, and should not be compared to thresholds of statistical significance imposed on genomewide association efforts.
Finally, underscoring the complex genetic underpinnings of alcohol consumption, our top SNP has an R2 of 0.02, explaining a very modest proportion of variance. Despite the fairly modest effect size, we note promising findings for alcohol consumption via a serotonergic pathway. Given the high comorbidity between alcohol use and depression, anxiety and, particularly suicide, which are also linked to serotonin modulation, future efforts will aim to disentangle the effects of these genes on sub-types of alcoholism in this cohort of women.
Supplementary Material
Acknowledgments
Funding: AA07728, AA09022 & K05 AA17688 (ACH); 2P60AA011998, DA18660 & DA18267 (MTL); AA12640 (KKB) ; AA017242; AA013526, AA013987, & AA007231 (KJS); DA18823 & MH083823 (AAT); DA23668 & ABRMF/Foundation for Alcohol Research (AA).
References
- 1.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abecasis GR, Cardon LR, Cookson WO, Sham PC, Cherny SS. Association analysis in a variance components framework. Genet Epidemiol. 2001a;21(Suppl 1):S341–S346. S341–S346. doi: 10.1002/gepi.2001.21.s1.s341. [DOI] [PubMed] [Google Scholar]
- 3.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001b;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 4.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal A, Grant JD, Littlefield A, Waldron M, Pergadia ML, Lynskey MT, Madden PA, Todorov A, Trull T, Bucholz KK, Todd RD, Sher K, Heath AC. Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. J Stud Alcohol Drugs. 2009;70:157–168. doi: 10.15288/jsad.2009.70.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartholow BD, Sher KJ, Krull JL. Changes in heavy drinking over the third decade of life as a function of collegiate fraternity and sorority involvement: a prospective, multilevel analysis. Health Psychol. 2003;22:616–626. doi: 10.1037/0278-6133.22.6.616. [DOI] [PubMed] [Google Scholar]
- 7.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 8.Bierut L, Agrawal A, Bucholz K, Doheny KF, Laurie CC, Pugh EW, Fisher S, Fox L, Howells B, Bertelsen S, Hinrichs A, Almasy L, Breslau N, Culverhouse R, Dick D, Edenberg H, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson E, Kramer J, Krueger R, Kuperman S, Lynskey MT, Mann K, Neuman R, Nothen M, Nurnberger J, Jr, Porjesz B, Ridinger M, Saccone NL, Schuckit M, Tischfield J, Wang JC, Rietschel M, Goate A, Rice JP. A Genome-wide Association Study of Alcohol Dependence. Proc Natl Acad Sci U S A. 2010;34:364–372. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control (CDC) Alcohol-Attributable Deaths and Years of Potential Life Lost --- United States, 2001. MMWR. 2004;53:870. [PubMed] [Google Scholar]
- 10.Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep. 2006;8:151–157. doi: 10.1007/s11920-006-0015-1. [DOI] [PubMed] [Google Scholar]
- 11.Dick DM, Latendresse SJ, Lansford JE, Budde JP, Goate A, Dodge KA, Pettit GS, Bates JE. Role of GABRA2 in trajectories of externalizing behavior across development and evidence of moderation by parental monitoring. Arch Gen Psychiatry. 2009a;66:649–657. doi: 10.1001/archgenpsychiatry.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dick DM, Prescott CA, McGue M. The Genetics of Substance Use and Substance Use Disorders. In: Yong-Kyu K, editor. Handbook of Behavior Genetics. New York: Springer; 2009b. pp. 433–453. [Google Scholar]
- 13.Drago A, Liappas I, Petio C, Albani D, Forloni G, Malitas P, Piperi C, Politis A, Tzavellas EO, Zisaki KK, Prato F, Batelli S, Polito L, De RD, Paparrigopoulos T, Kalofoutis A, Serretti A. Epistasis between IL1A, IL1B, TNF, HTR2A, 5-HTTLPR and TPH2 variations does not impact alcohol dependence disorder features. Int J Environ Res Public Health. 2009;6:1980–1990. doi: 10.3390/ijerph6071980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 15.Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- 17.Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr, Rice JP, Schuckit MA, Taylor R, Todd WB, Tischfield JA, Porjesz B, Foroud T. Genome-Wide Association Study of Alcohol Dependence Implicates a Region on Chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 19.Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- 20.Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- 21.Gacek P, Conner TS, Tennen H, Kranzler HR, Covault J. Tryptophan hydroxylase 2 gene and alcohol use among college students. Addict Biol. 2008;13:440–448. doi: 10.1111/j.1369-1600.2008.00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant JD, Agrawal A, Bucholz KK, Madden PA, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, Whitfield JB, Martin NG, Heath AC. Alcohol consumption indices of genetic risk for alcohol dependence. Biol Psychiatry. 2009;66:795–800. doi: 10.1016/j.biopsych.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guaderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 http://hydra usc edu/gxe. [Google Scholar]
- 24.Haavik J, Blau N, Thony B. Mutations in human monoamine-related neurotransmitter pathway genes. Hum Mutat. 2008;29:891–902. doi: 10.1002/humu.20700. [DOI] [PubMed] [Google Scholar]
- 25.Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 26.Heath AC, Howells W, Bucholz KK, Glowinski AL, Nelson EC, Madden PA. Ascertainment of a mid-western US female adolescent twin cohort for alcohol studies: assessment of sample representativeness using birth record data. Twin Res. 2002;5:107–112. doi: 10.1375/1369052022974. [DOI] [PubMed] [Google Scholar]
- 27.Heath AC, Madden PA, Grant JD, McLaughlin TL, Todorov AA, Bucholz KK. Resiliency factors protecting against teenage alcohol use and smoking: influences of religion, religious involvement and values, and ethnicity in the Missouri Adolescent Female Twin Study. Twin Res. 1999;2:145–155. doi: 10.1375/136905299320566013. [DOI] [PubMed] [Google Scholar]
- 28.Heath AC, Martin NG. Genetic influences on alcohol consumption patterns and problem drinking: results from the Australian NH&MRC twin panel follow-up survey. Ann N Y Acad Sci. 1994;708:72–85. 72–85. doi: 10.1111/j.1749-6632.1994.tb24699.x. [DOI] [PubMed] [Google Scholar]
- 29.Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, Mann J, Riley B, Roy A, Tabakoff B, Todd RD, Zhou Z, Goldman D. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoog JO, Heden LO, Larsson K, Jornvall H, von Bahr-Lindstrom H. The gamma 1 and gamma 2 subunits of human liver alcohol dehydrogenase. cDNA structures, two amino acid replacements, and compatibility with changes in the enzymatic properties. Eur J Biochem. 1986;159:215–218. doi: 10.1111/j.1432-1033.1986.tb09855.x. [DOI] [PubMed] [Google Scholar]
- 31.Kendler KS, Myers J, Dick D, Prescott CA. The Relationship Between Genetic Influences on Alcohol Dependence and on Patterns of Alcohol Consumption. Alcohol Clin Exp Res. 2010;34:1058–1065. doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kendler KS, Myers J, Prescott CA. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch Gen Psychiatry. 2007;64:1313–1320. doi: 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- 33.Knopik VS, Sparrow EP, Madden PA, Bucholz KK, Hudziak JJ, Reich W, Slutske WS, Grant JD, McLaughlin TL, Todorov A, Todd RD, Heath AC. Contributions of parental alcoholism, prenatal substance exposure, and genetic transmission to child ADHD risk: a female twin study. Psychol Med. 2005;35:625–635. doi: 10.1017/s0033291704004155. [DOI] [PubMed] [Google Scholar]
- 34.Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: modeling the externalizing spectrum. J Abnorm Psychol. 2002;111:411–424. [PubMed] [Google Scholar]
- 35.Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- 36.Lind P, MacGregor S, Agrawal A, Heath AC, Martin NG, Whitfield JB. The Role of GABRA2 in Alcohol Dependence, Smoking and Illicit Drug Use in an Australian Population Sample. Alcohol Clin Exp Res. 2008;32:1721–1731. doi: 10.1111/j.1530-0277.2008.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31:1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- 38.Ma JZ, Beuten J, Payne TJ, Dupont RT, Elston RC, Li MD. Haplotype analysis indicates an association between the DOPA decarboxylase (DDC) gene and nicotine dependence. Hum Mol Genet. 2005;14:1691–1698. doi: 10.1093/hmg/ddi177. [DOI] [PubMed] [Google Scholar]
- 39.Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park A, Sher KJ, Krull JL. Risky drinking in college changes as fraternity/sorority affiliation changes: a person-environment perspective. Psychol Addict Behav. 2008;22:219–229. doi: 10.1037/0893-164X.22.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prescott CA, Aggen SH, Kendler KS. Sex differences in the sources of genetic liability to alcohol abuse and dependence in a population-based sample of U.S. twins. Alcohol Clin Exp Res. 1999;23:1136–1144. doi: 10.1111/j.1530-0277.1999.tb04270.x. [DOI] [PubMed] [Google Scholar]
- 42.Preuss UW, Ridinger M, Rujescu D, Samochowiec J, Fehr C, Wurst FM, Koller G, Bondy B, Wodarz N, Debniak T, Grzywacz A, Soyka M, Zill P. Association of ADH4 genetic variants with alcohol dependence risk and related phenotypes: results from a larger multicenter association study. Addict Biol. 2010 doi: 10.1111/j.1369-1600.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373:2223–2233. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- 45.Rice DP. Economic costs of substance abuse (1995) Proceedings of the Association of American Physicians. 1999;111:119–125. doi: 10.1046/j.1525-1381.1999.09254.x. [DOI] [PubMed] [Google Scholar]
- 46.Rose RJ. A developmental behavior-genetic perspective on alcoholism risk. Alcohol Health Res World. 1998;22:131–143. [PMC free article] [PubMed] [Google Scholar]
- 47.Serretti A, Liappas I, Mandelli L, Albani D, Forloni G, Malitas P, Piperi C, Politis A, Tzavellas EO, Papadopoulou-Daifoti Z, Zisaki A, Prato F, Batelli S, Polito L, De RD, Kalofoutis A. TPH2 gene variants and anxiety during alcohol detoxification outcome. Psychiatry Res. 2009;167:106–114. doi: 10.1016/j.psychres.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Sher KJ, Bartholow BD, Nanda S. Short- and long-term effects of fraternity and sorority membership on heavy drinking: a social norms perspective. Psychol Addict Behav. 2001;15:42–51. doi: 10.1037/0893-164x.15.1.42. [DOI] [PubMed] [Google Scholar]
- 49.Sher KJ, Walitzer KS, Wood PK, Brent EE. Characteristics of children of alcoholics: putative risk factors, substance use and abuse, and psychopathology. J Abnorm Psychol. 1991;100:427–448. doi: 10.1037//0021-843x.100.4.427. [DOI] [PubMed] [Google Scholar]
- 50.Sher KJ, Wood MD, Wood PK, Raskin G. Alcohol outcome expectancies and alcohol use: a latent variable cross-lagged panel study. J Abnorm Psychol. 1996;105:561–574. doi: 10.1037/0021-843X.105.4.561. [DOI] [PubMed] [Google Scholar]
- 51.Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res. 2008;42:184–191. doi: 10.1016/j.jpsychires.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Y, Panhuysen C, Kranzler HR, Hesselbrock V, Rounsaville B, Weiss R, Brady K, Farrer LA, Gelernter J. Intronic variants in the dopa decarboxylase (DDC) gene are associated with smoking behavior in European-Americans and African-Americans. Hum Mol Genet. 2006;15:2192–2199. doi: 10.1093/hmg/ddl144. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Ye Y, Wang X, Gelernter J, Ma JZ, Li MD. DOPA decarboxylase gene is associated with nicotine dependence. Pharmacogenomics. 2006;7:1159–1166. doi: 10.2217/14622416.7.8.1159. [DOI] [PubMed] [Google Scholar]
- 55.Zill P, Preuss UW, Koller G, Bondy B, Soyka M. SNP- and haplotype analysis of the tryptophan hydroxylase 2 gene in alcohol-dependent patients and alcohol-related suicide. Neuropsychopharmacology. 2007;32:1687–1694. doi: 10.1038/sj.npp.1301318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.