Abstract
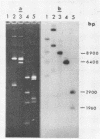
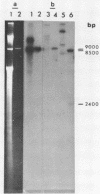
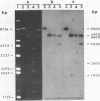
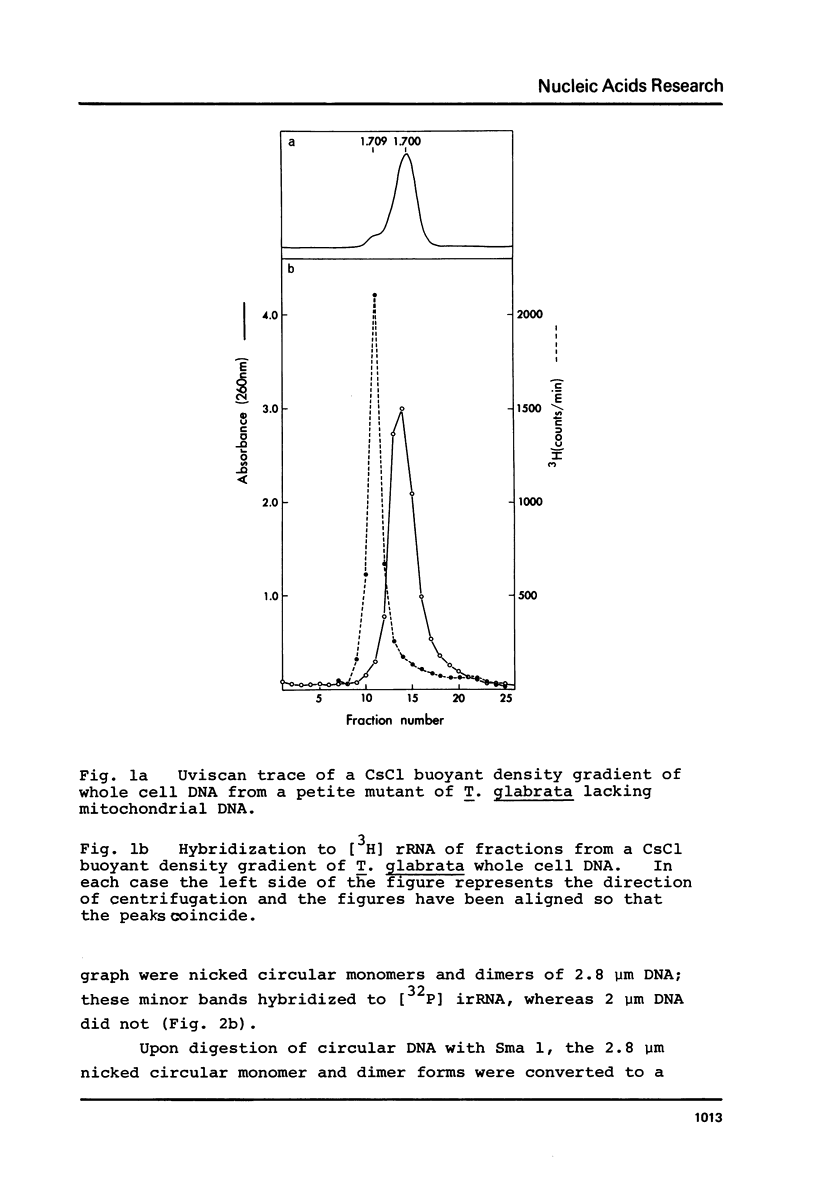
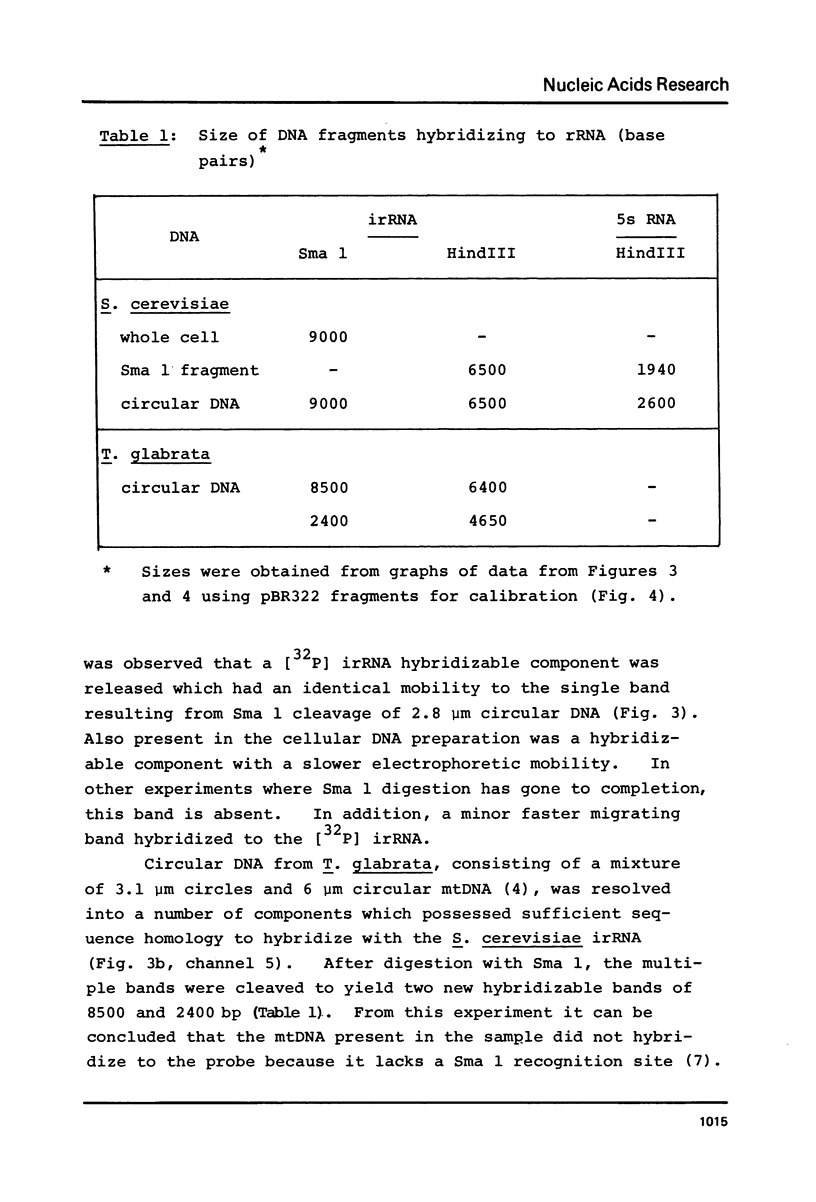
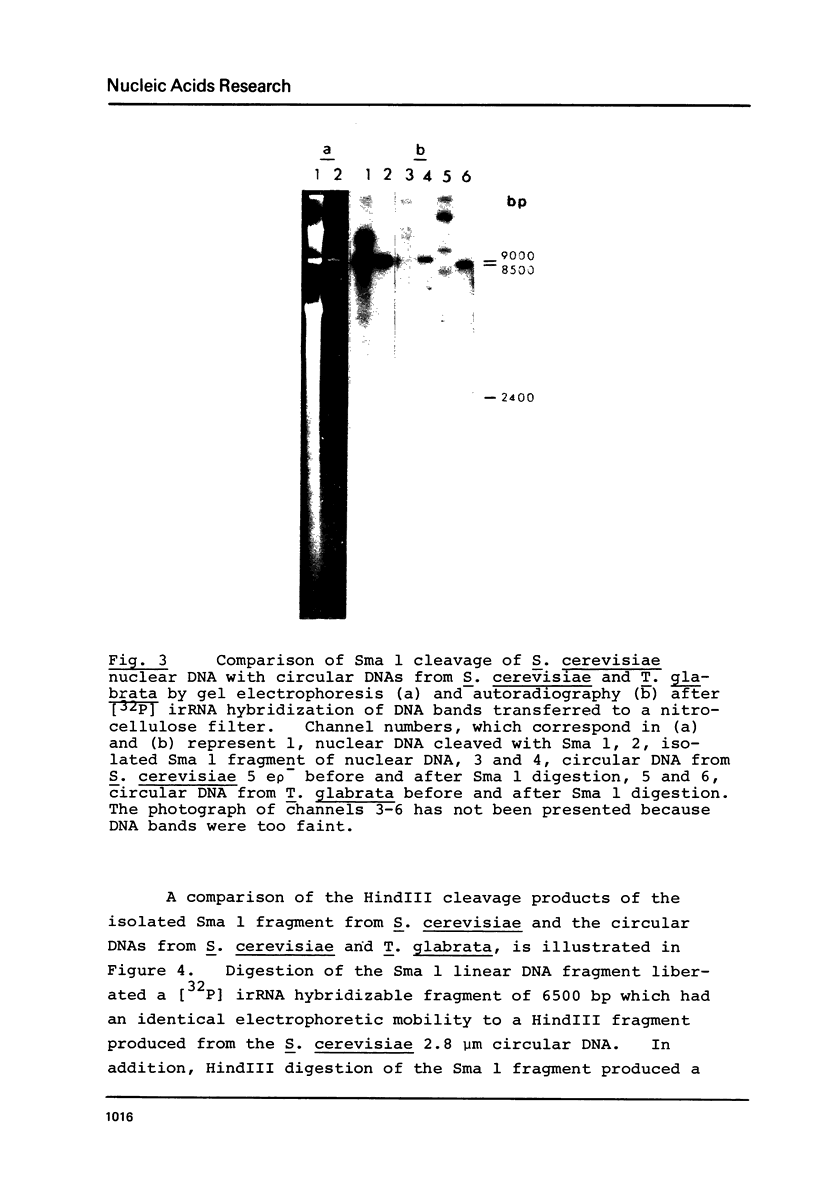
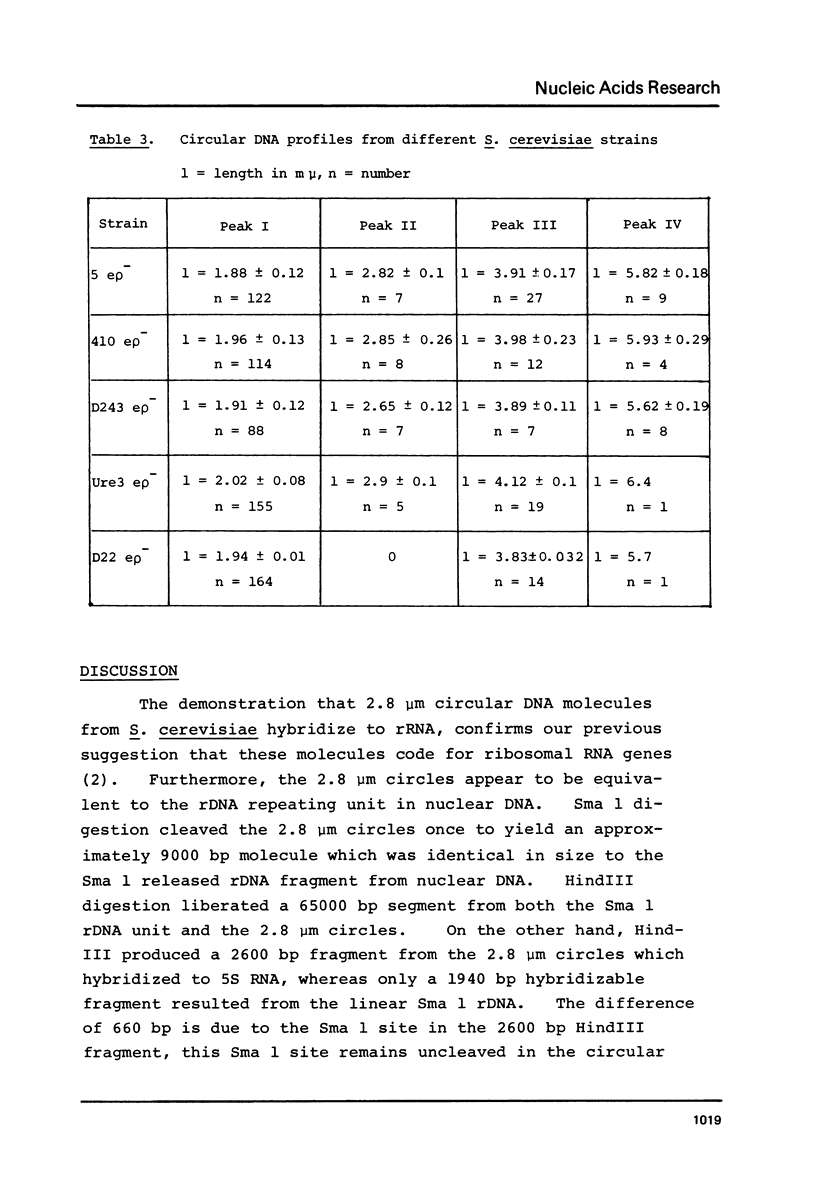
We have shown that 2.8 and 3.1 micron circular DNA molecules, previously reported to be present in Saccharomyces cerevisiae and Torulopsis glabrata respectively, contain sequences hybridizing to cytoplasmic ribosomal RNAs. In S. cerevisiae the 2.8 micron circular DNA appears to be identical to the rDNA repeating unit from nuclear DNA, both in length (approximately 9000 base pairs) and in the location of the 25, 18 and 5.8S rRNA sequences on the large HindIII fragment (6500 bp) and the presence of the 5S rRNA sequence on the small HindIII fragment. The 3.1 micron molecule from T. glabrata is approximately 2000 base pairs longer than the S. cerevisiae molecule and in addition, one of the HindIII sites lies within the region hybridizing to 25, 18 and 5.8S rRNAs. In S. cerevisiae the 4-5 copies of the 2.8 micron circular DNA molecules per cell, which have an extra-nuclear location, do not appear to be essential for cell viability as in one strain they were undetectable.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azad A. A. Cytoplasmic RNA from hen reticulocytes, mouse sarcoma 180 ascites cells, rat liver and barley embryos. Their preparation and purification by a standard procedure and characterization by polyacrylamide gel electrophoresis. Comp Biochem Physiol B. 1978;61(2):213–218. doi: 10.1016/0305-0491(78)90163-3. [DOI] [PubMed] [Google Scholar]
- Azad A. A., Lane B. G. Wheat-embryo ribonucleates. IV. Factors that influence the formation and stability of a complex between 5S rRNA and 18S rRNA. Can J Biochem. 1975 Mar;53(3):320–327. doi: 10.1139/o75-045. [DOI] [PubMed] [Google Scholar]
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J. R., Philippsen P., Davis R. W. Analysis of chromosomal integration and deletions of yeast plasmids. Nucleic Acids Res. 1977;4(5):1429–1448. doi: 10.1093/nar/4.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D. Isolation of circular DNA from a mitochondrial fraction from yeast. Proc Natl Acad Sci U S A. 1972 Feb;69(2):388–392. doi: 10.1073/pnas.69.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., Miklos G. L. Localization and quantification of circular DNA in yeast. Eur J Biochem. 1974 Jan 16;41(2):359–365. doi: 10.1111/j.1432-1033.1974.tb03278.x. [DOI] [PubMed] [Google Scholar]
- Cramer J. H., Bhargava M. M., Halvorson H. O. Isolation and characterization of DNA of Saccharomyces cerevisiae. J Mol Biol. 1972 Oct 28;71(1):11–20. doi: 10.1016/0022-2836(72)90396-8. [DOI] [PubMed] [Google Scholar]
- Cramer J. H., Farrelly F. W., Rownd R. H. Restriction endonuclease analysis of ribosomal DNA from Saccharomyces cerevisiae. Mol Gen Genet. 1976 Nov 17;148(3):233–241. doi: 10.1007/BF00332897. [DOI] [PubMed] [Google Scholar]
- Culbertson M. R., Charnas L., Johnson M. T., Fink G. R. Frameshifts and frameshift suppressors in Saccharomyces cerevisiae. Genetics. 1977 Aug;86(4):745–764. doi: 10.1093/genetics/86.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G. Free ribosomal RNA genes in the macronucleus of Tetrahymena. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3078–3081. doi: 10.1073/pnas.71.8.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Rochaix J. D. The amplified ribosomal DNA of dytiscid beetles. Proc Natl Acad Sci U S A. 1974 May;71(5):1819–1823. doi: 10.1073/pnas.71.5.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Graziani F., Caizzi R., Gargano S. Circular ribosomal DNA during ribosomal magnification in Drosophila melanogaster. J Mol Biol. 1977 May 5;112(1):49–63. doi: 10.1016/s0022-2836(77)80155-1. [DOI] [PubMed] [Google Scholar]
- Gubbins E. J., Newlon C. S., Kann M. D., Donelson J. E. Sequence organization and expression of a yeast plasmid DNA. Gene. 1977 May;1(3-4):185–207. doi: 10.1016/0378-1119(77)90045-2. [DOI] [PubMed] [Google Scholar]
- Guerineau M., Grandchamp C., Slonimski P. P. Circular DNA of a yeast episome with two inverted repeats: structural analysis by a restriction enzyme and electron microscopy. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3030–3034. doi: 10.1073/pnas.73.9.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg C. P., Degelmann A., Kustermann-Kuhn B., Royer H. D. Characterization of 2-mum DNA of Saccharomyces cerevisiae by restriction fragment analysis and integration in an Escherichia coli plasmid. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2072–2076. doi: 10.1073/pnas.73.6.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourcade D., Dressler D., Wolfson J. The amplification of ribosomal RNA genes involves a rolling circle intermediate. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2926–2930. doi: 10.1073/pnas.70.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston D. M., Klein H. L. Deoxyribonucleic acid sequence organization of a yeast plasmid. J Bacteriol. 1977 Jan;129(1):472–481. doi: 10.1128/jb.129.1.472-481.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready S. J., Cox B. S., McLaughlin C. S. The extrachromosomal control of nonsense suppression in yeast: an analysis of the elimination of [psi+] in the presence of a nuclear gene PNM. Mol Gen Genet. 1977 Feb 15;150(3):265–270. doi: 10.1007/BF00268125. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr, Beatty B. R. Extrachromosomal nucleolar genes in amphibian oocytes. Genetics. 1969;61(1 Suppl):133–143. [PubMed] [Google Scholar]
- O'Connor R. M., McArthur C. R., Clark-Walker G. D. Respiratory-deficient mutants of Torulopsis glabrata, a yeast with circular mitochondrial deoxyribonucleic acid of 6 mu m. J Bacteriol. 1976 May;126(2):959–968. doi: 10.1128/jb.126.2.959-968.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley K. M., Clark-Walker G. D. Abnormal mitochondrial genomes in yeast restored to respiratory competence. Genetics. 1978 Nov;90(3):517–530. doi: 10.1093/genetics/90.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer E., Wilhelm J. M., Sherman F. Variation of phenotypic suppression due to the psi+ and psi- extrachromosomal determinants in yeast. J Mol Biol. 1979 Feb 15;128(1):107–110. doi: 10.1016/0022-2836(79)90311-5. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Retèl J., Planta R. J. The investigation of the ribosomal RNA sites in yeast DNA by the hybridization technique. Biochim Biophys Acta. 1968 Dec 17;169(2):416–429. doi: 10.1016/0005-2787(68)90050-6. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D., Bird A., Barkken A. Ribosomal RNA gene amplification by rolling circles. J Mol Biol. 1974 Aug 15;87(3):473–487. doi: 10.1016/0022-2836(74)90098-9. [DOI] [PubMed] [Google Scholar]
- Schweizer E., MacKechnie C., Halvorson H. O. The redundancy of ribosomal and transfer RNA genes in Saccharomyces cerevisiae. J Mol Biol. 1969 Mar 14;40(2):261–277. doi: 10.1016/0022-2836(69)90474-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trendelenburg M. F., Scheer U., Zentgraf H., Franke W. W. Heterogeneity of spacer lengths in circles of amplified ribosomal DNA of two insect species, Dytiscus marginalis and Acheta domesticus. J Mol Biol. 1976 Dec;108(2):453–470. doi: 10.1016/s0022-2836(76)80130-1. [DOI] [PubMed] [Google Scholar]