Abstract
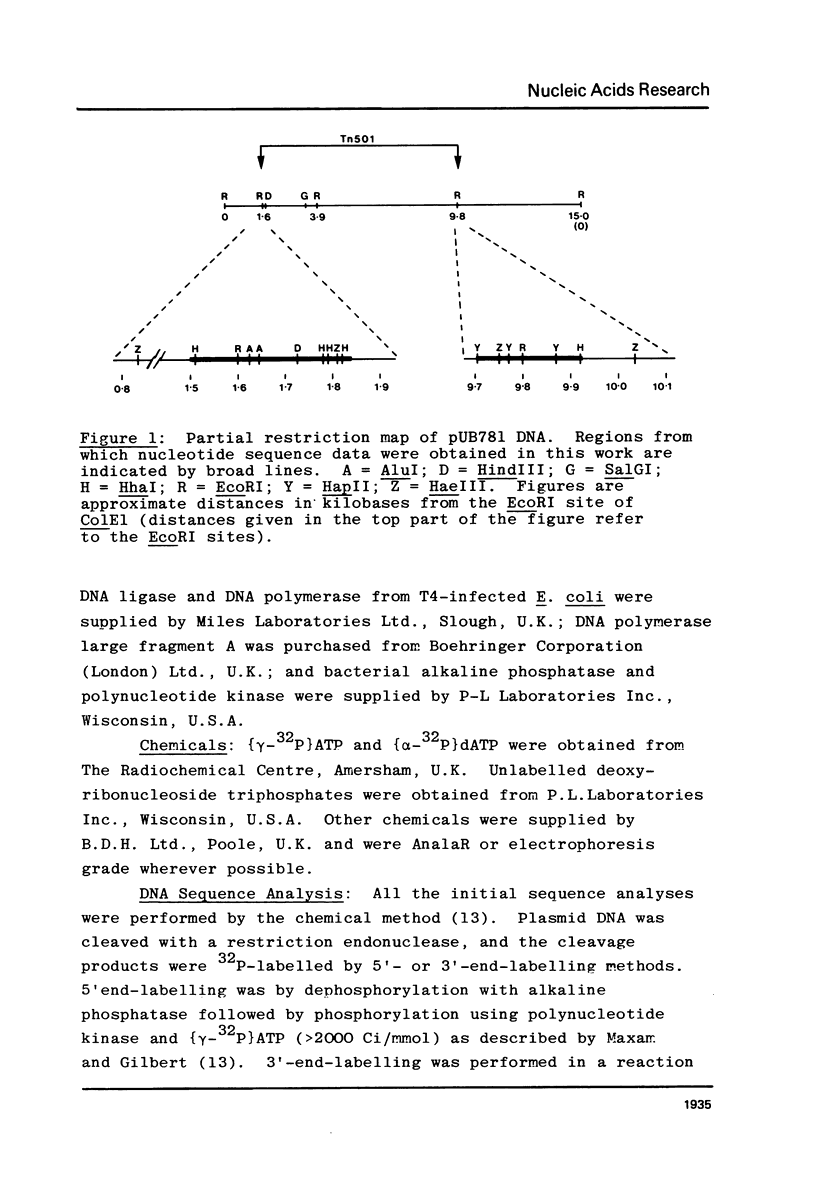
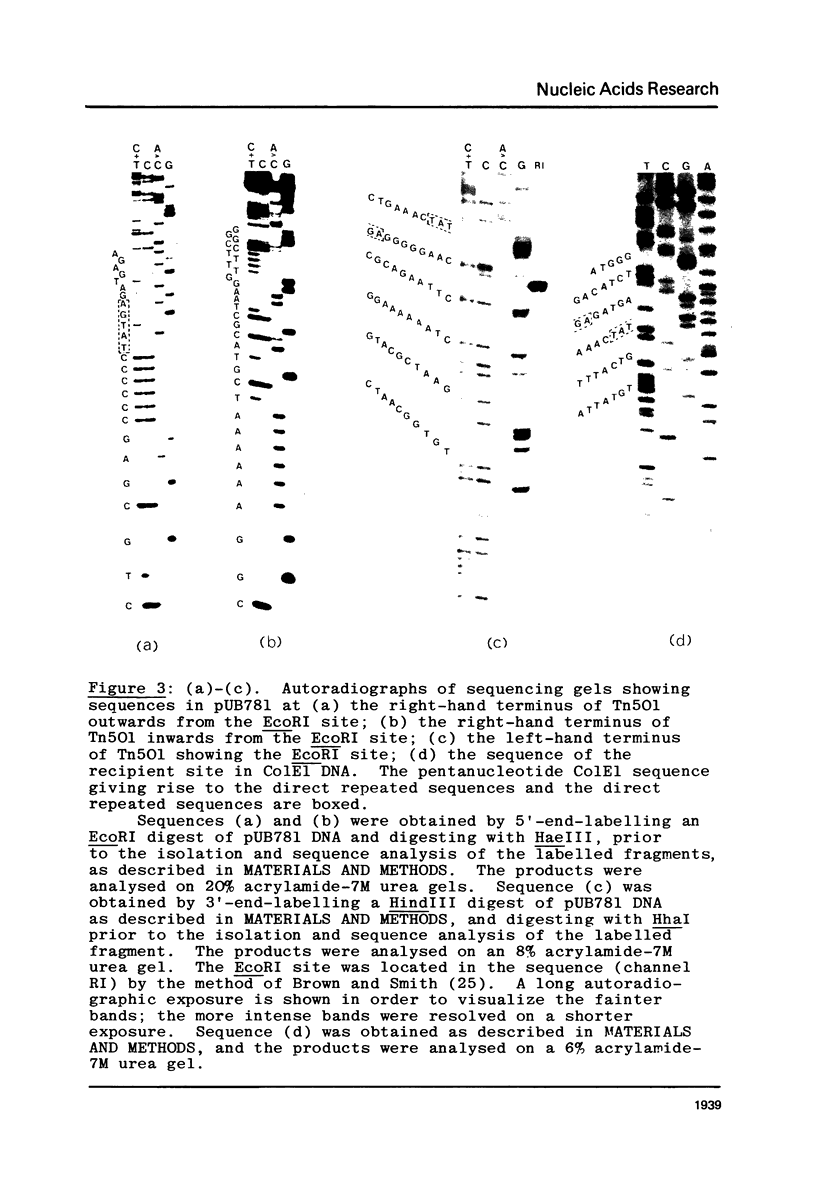
The nucleotide sequences at the ends of the mercury-resistance transposon, Tn501, have been determined. The terminal sequences are inverted repeated sequences 38 nucleotide pairs in length, which differ in 3 nucleotide pairs. The transposon is flanked by directly repeated sequences of 5 nucleotide pairs, originating from a single pentanucleotide sequence in the recipient replicon. There is no obvious homology between recipient replicons at the site of insertion of the transposon. The structures of the ends of Tn501 are compared with those of other transposons and insertion sequences. The use of Tn501 to locate an EcoRI site within a genetically defined sequence of interest is discussed.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B. Mu insertion duplicates a 5 base pair sequence at the host inserted site. Cell. 1979 Jan;16(1):123–129. doi: 10.1016/0092-8674(79)90193-4. [DOI] [PubMed] [Google Scholar]
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Campbell A., Berg D. E., Botstein D., Lederberg E. M., Novick R. P., Starlinger P., Szybalski W. Nomenclature of transposable elements in prokaryotes. Gene. 1979 Mar;5(3):197–206. doi: 10.1016/0378-1119(79)90078-7. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Sommer H., Saedler H. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 1979 Mar;6(3):1111–1122. doi: 10.1093/nar/6.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Habermann P., Klaer R., Kühn S., Starlinger P. IS4 is found between eleven or twelve base pair duplications. Mol Gen Genet. 1979 Oct 1;175(3):369–373. doi: 10.1007/BF00397237. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Johnsrud L., Calos M. P., Miller J. H. The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell. 1978 Dec;15(4):1209–1219. doi: 10.1016/0092-8674(78)90047-8. [DOI] [PubMed] [Google Scholar]
- Kleckner N. DNA sequence analysis of Tn10 insertions: origin and role of 9 bp flanking repetitions during Tn10 translocation. Cell. 1979 Apr;16(4):711–720. doi: 10.1016/0092-8674(79)90087-4. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Maat J., Smith A. J. A method for sequencing restriction fragments with dideoxynucleoside triphosphates. Nucleic Acids Res. 1978 Dec;5(12):4537–4545. doi: 10.1093/nar/5.12.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. R., Young R. A., Steitz J. A., Grindley N. D., Guyer M. S. Transposition of the Escherichia coli insertion element gamma generates a five-base-pair repeat. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4882–4886. doi: 10.1073/pnas.76.10.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier P. H., Cortese R. A fast and simple method for sequencing DNA cloned in the single-stranded bacteriophage M13. J Mol Biol. 1979 Mar 25;129(1):169–172. doi: 10.1016/0022-2836(79)90068-8. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M., Richmond M. H. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J Bacteriol. 1977 Mar;129(3):1227–1233. doi: 10.1128/jb.129.3.1227-1233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya T., Nomiyama H., Miyoshi J., Shimada K., Takagi Y. DNA sequences of the integration sites and inverted repeated structure of transposon Tn3. Nucleic Acids Res. 1979;6(5):1831–1841. doi: 10.1093/nar/6.5.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]