Abstract
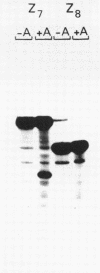
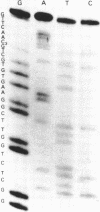
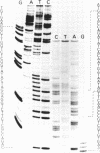
It is already known that phi X gene A protein converts besides phi X RFI DNA also the RFI DNAs of the single-stranded bacteriophages G4, St-1, alpha 3 and phi K into RFII DNA. We have extended this observations for bacteriophages G14 and U3. Restriction enzyme analysis placed the phi X gene A protein cleavage site in St-1 RF DNA in the HinfI restriction DNA fragment F10 and in the overlapping HaeIII restriction DNA fragment Z7. The exact position and the nucleotide sequence at the 3'-OH end of the nick were determined by DNA sequence analysis of the single-stranded DNA subfragment of the nicked DNA fragment F10 obtained by gelelectrophoresis in denaturing conditions. A stretch of 85 nucleotides of St-1 DNA around the position of the phi X gene A protein cleavage site was established by DNA sequence analysis of the restriction DNA fragment Z7F1. Comparison of this nucleotide sequence with the previously determined nucleotide sequence around the cleavage site of phi X gene A protein in phi X174 RF DNA and G4 RF DNA revealed an identical sequence of only 10 nucleotides. The results suggest that the recognition sequence of the phi X174 gene A protein lies within these 10 nucleotides.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Jansz H. S. Bacteriophage phiX174 DNA synthesis in a replication-deficient host: determination of the origin of phiX DNA replication. J Mol Biol. 1976 Apr 15;102(3):633–656. doi: 10.1016/0022-2836(76)90339-9. [DOI] [PubMed] [Google Scholar]
- Duguet M., Yarranton G., Gefter M. The rep protein of Escherichia coli: interaction with DNA and other proteins. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):335–343. doi: 10.1101/sqb.1979.043.01.040. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Kornberg A. Purification and characterization of phiX174 gene A protein. A multifunctional enzyme of duplex DNA replication. J Biol Chem. 1979 Jun 25;254(12):5328–5332. [PubMed] [Google Scholar]
- Fiddes J. C., Barrell B. G., Godson G. N. Nucleotide sequences of the separate origins of synthesis of bacteriophage G4 viral and complementary DNA strands. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1081–1085. doi: 10.1073/pnas.75.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke B., Ray D. S. Formation of the parental replicative form DNA of bacteriophage phi-X174 and initial events in its replication. J Mol Biol. 1971 Nov 14;61(3):565–586. doi: 10.1016/0022-2836(71)90065-9. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Barrell B. G., Staden R., Fiddes J. C. Nucleotide sequence of bacteriophage G4 DNA. Nature. 1978 Nov 16;276(5685):236–247. doi: 10.1038/276236a0. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Roberts R. J. dna, single stranded/*metab. Virology. 1976 Sep;73(2):561–567. doi: 10.1016/0042-6822(76)90421-9. [DOI] [PubMed] [Google Scholar]
- Grindley J. N., Godson G. N. Evolution of bacteriophage phi X174. IV. Restriction enzyme cleavage map of St-1. J Virol. 1978 Sep;27(3):738–744. doi: 10.1128/jvi.27.3.738-744.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda J. E., Yudelevich A., Hurwitz J. Isolation and characterization of the protein coded by gene A of bacteriophage phiX174 DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2669–2673. doi: 10.1073/pnas.73.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda J. E., Yudelevich A., Shimamoto N., Hurwitz J. Role of polymeric forms of the bacteriophage phi X174 coded gene A protein in phi XRFI DNA cleavage. J Biol Chem. 1979 Oct 10;254(19):9416–9428. [PubMed] [Google Scholar]
- Jansz H. S., Pouwels P. H., Schiphorst J. Preparation of double-stranded DNA (replicative form) of bacteriophage phi-X174: a simplified method. Biochim Biophys Acta. 1966 Sep;123(3):626–627. doi: 10.1016/0005-2787(66)90233-4. [DOI] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., Baas P. D., Jansz H. S., van Arkel G. A., Weisbeek P. J. Nucleotide sequence of the origin of replication in bacteriophage phiX174 RF DNA. Nature. 1978 Feb 2;271(5644):417–420. doi: 10.1038/271417a0. [DOI] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., de Winter J. M., Weisbeek P. J. Cleavage of single-stranded DNA by the A and A* proteins of bacteriophage phi X174. Nucleic Acids Res. 1979 Dec 20;7(8):2177–2188. doi: 10.1093/nar/7.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Marians K. J., Ikeda J. E., Schlagman S., Hurwitz J. Role of DNA gyrase in phiX replicative-form replication in vitro. Proc Natl Acad Sci U S A. 1977 May;74(5):1965–1968. doi: 10.1073/pnas.74.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Takanami M., Kojo H. Cleavage site specificity of an endonuclease prepared from Heamophilus influenzae strain H-I. FEBS Lett. 1973 Feb 1;29(3):267–270. doi: 10.1016/0014-5793(73)80035-3. [DOI] [PubMed] [Google Scholar]
- Takanami M. Specific cleavage of coliphage fd DNA by five different restriction endonucleases from Haemophilus genus. FEBS Lett. 1973 Aug 15;34(2):318–322. doi: 10.1016/0014-5793(73)80821-x. [DOI] [PubMed] [Google Scholar]
- Tessman E. S. Mutants of bacteriophage S13 blocked in infectious DNA synthesis. J Mol Biol. 1966 May;17(1):218–236. doi: 10.1016/s0022-2836(66)80104-3. [DOI] [PubMed] [Google Scholar]
- van Mansfeld A. D., Langeveld S. A., Weisbeek P. J., Baas P. D., van Arkel G. A., Jansz H. S. Cleavage site of phiX174 gene-A protein in phiX and G4 RFI DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):331–334. doi: 10.1101/sqb.1979.043.01.039. [DOI] [PubMed] [Google Scholar]