Summary
Resistance in tomato to infection by Pseudomonas syringae involves both detection of PAMPs and recognition by the host Pto kinase of pathogen effector AvrPtoB which is translocated into the host cell and interferes with PAMP-triggered immunity (PTI). The N-terminal portion of AvrPtoB is sufficient for its virulence activity and for recognition by Pto. An amino acid substitution in this protein, F173A, abolishes these activities. To investigate the mechanisms of AvrPtoB virulence, we screened for tomato proteins that interact with AvrPtoB and identified Bti9, a LysM receptor-like kinase. Bti9 has the highest amino acid similarity to Arabidopsis CERK1 among the tomato LysM-RLKs and belongs to a clade containing three other tomato proteins, SlLyk11, SlLyk12, and SlLyk13 all of which interact with AvrPtoB. The F173A substitution disrupts the interaction of AvrPtoB with Bti9 and SlLyk13 suggesting these LysM-RLKs are its virulence targets. Two independent tomato lines with RNAi-mediated reduced expression of Bti9 and SlLyk13 were more susceptible to P. syringae. Bti9 kinase activity was inhibited in vitro by the N-terminal domain of AvrPtoB in an F173-dependent manner. These results indicate Bti9/SlLyk13 play a role in plant immunity and the N-terminal domain of AvrPtoB may have evolved to interfere with their kinase activity. Finally, we found that Bti9 and Pto interact with AvrPtoB in a structurally similar although not identical fashion suggesting Pto may have evolved as a molecular mimic of LysM-RLK kinase domains.
Keywords: Pseudomonas syringae, Solanum lycopersicum, tomato, PAMP-triggered immunity, Pto kinase, AvrPtoB, Disease
Introduction
An initial layer of induced plant defense against potential pathogens involves host perception of pathogen-associated molecular patterns (PAMPs), typically by plasma membrane-localized pattern recognition receptors (PRRs) (Gomez-Gomez and Boller 2002, Zipfel 2008). PRRs, in concert with associated proteins, then activate signaling pathways leading to PAMP-triggered immunity (PTI) (Chinchilla et al. 2007, Chisholm et al. 2006, Jones and Dangl 2006). To date, diverse PAMPs including flagellin, elongation factor Tu (EF-Tu), chitin, and peptidoglycan (PGN) are known to be recognized by plants (Boller 1995, Zipfel 2008). However, the PRRs that play a role in recognition of the majority of PAMPs remain unknown.
The two best characterized plant PRRs are from Arabidopsis and both recognize bacterial PAMPs: FLS2 detects a 22-amino acid epitope (flg22) in flagellin and EFR detects an 18-amino acid epitope in EF-Tu (Chinchilla et al. 2006, Gómez-Gómez and Boller 2000, Zipfel et al. 2006). Both of these proteins are leucine-rich repeat (LRR) receptor-like kinases (LRR-RLKs) and FLS2, in particular, has been studied intensively. FLS2 binds the flg22 peptide after which it associates with another LRR-RLK, the BRI1-associated kinase BAK1 (Chinchilla, et al. 2006, Chinchilla, et al. 2007). Plants with mutations in the FLS2 gene are compromised for PTI and are more susceptible to certain bacterial pathogens (Hann and Rathjen 2007, Zipfel et al. 2004). Recently FLS2 has been shown to also recognize Ax21, a PAMP from Xanthomonas campestris pv. campestris and, surprisingly, the Clavata3 peptide which is expressed exclusively in the shoot apical meristem (Danna et al. 2011, Lee et al. 2011).
Pathogens have evolved virulence proteins (‘effectors’) which they deliver into the plant cell to suppress PTI (Hogenhout et al. 2009). Gram-negative bacterial pathogens, for example, use a type III secretion (TTS) system to inject effector proteins directly into the plant cell cytoplasm (Cornelis 2006, Grant et al. 2006, Guo et al. 2009, Mudgett 2005). Strain DC3000 of Pseudomonas syringae pv. tomato (Pst), the causal pathogen of tomato bacterial speck disease, expresses and delivers ~30 type III effectors into host cell (Lindeberg et al. 2006).
On plants that express the resistance (R) protein Pto and the nucleotide-binding-leucine rich repeat (NB-LRR) protein Prf, either of two Pst effectors, AvrPto or AvrPtoB (HopAB2) is recognized by the Pto kinase via a physical interaction, leading to activation of effector-triggered immunity (ETI) (Pedley and Martin 2003). On plants that lack a functional Pto/Prf pathway, however, AvrPto and AvrPtoB promote virulence of Pst in an additive manner (Cunnac et al. 2011, Kvitko et al. 2009, Lin and Martin 2005). Both effectors suppress PTI signaling in Arabidopsis and Nicotiana benthamiana plants (Hann et al. 2010, He et al. 2006).
The PTI-suppressing function of AvrPto and AvrPtoB occurs upstream of MAPK signaling and AvrPto is known to target the cytoplasmic (kinase) domains of PRR complexes to suppress PTI (He, et al. 2006, Shan et al. 2008, Xiang et al. 2011, Xiang et al. 2008). Unlike AvrPto, which encodes a small (18 kDa) protein, AvrPtoB is a 59 kD modular protein (Kim et al. 2002, Salmeron and Staskawicz 1993). The C-terminal domain (CTD) of AvrPtoB is a structural mimic of a eukaryotic E3 ubiquitin ligase that facilitates degradation of another tomato R protein, Fen, to suppress ETI, allowing Pst to retain the virulence activities associated with the AvrPtoB N-terminal region (Abramovitch et al. 2006, Janjusevic et al. 2006, Rosebrock et al. 2007).
The N-terminal portion of AvrPtoB has two distinct domains that contribute to its virulence activity. The region consisting of amino acids 1–387 of AvrPtoB (AvrPtoB1-387) but not a shorter fragment, AvrPtoB1-307, is able to suppress FLS2-mediated PTI (He, et al. 2006, Shan, et al. 2008). On tomato plants lacking Prf (RG-prf3), the other domain, AvrPtoB1-307, is sufficient to confer virulence comparable to AvrPtoB in DC3000ΔavrPto/ΔavrPtoB (a strain with avrPto and avrPtoB deleted) (Xiao et al. 2007).
AvrPtoB1-307 is also able to elicit Pto-mediated ETI on tomato plants having a functional Pto/Prf pathway. Interestingly, substitution in AvrPtoB1-307 of a key amino acid, phenylalanine-173 to alanine (F173A) abolishes both its virulence and avirulence activity. The F173A mutation, however, has no apparent effect on AvrPtoB PTI suppression in Arabidopsis (Xiao, et al. 2007). As with AvrPto, the molecular mechanisms used by AvrPtoB to suppress PTI are not fully understood. Previous studies showed the N-terminal region of AvrPtoB (AvrPtoB1-387 in Arabidopsis and AvrPtoB1-307 in tomato) to be sufficient for virulence, probably due to targeting of PRRs or BAK1 (He, et al. 2006, Shan, et al. 2008, Xiao, et al. 2007). However, two recent studies reported the E3 ligase activity of AvrPtoB CTD to be required for suppression of PTI while promoting virulence of Pst in Arabidopsis (Gimenez-Ibanez et al. 2009a, Gohre et al. 2008). Despite this discrepancy, it is clear AvrPtoB targets other PRRs or adaptor proteins in addition to FLS2/EFR/BAK1, as AvrPtoB suppresses PTI triggered by PAMPs in addition to flagellin and EF-Tu (He, et al. 2006, Shan, et al. 2008, Zipfel, et al. 2004). In this regard, AvrPtoB has recently been shown to target the Arabidopsis LysM-RLK, CERK1, which itself was shown to play a role in the immune response against Pst (Gimenez-Ibanez, et al. 2009a).
To identify possible virulence target(s) of AvrPtoB in tomato, we performed a yeast two-hybrid screen using the effector as a bait against a tomato leaf cDNA library. One AvrPtoB tomato-interacting (Bti) protein identified in the screen, Bti9, was found to encode a lysin motif (LysM) receptor-like kinase (LysM-RLK) whose closest amino acid match in Arabidopsis is CERK1. Here we characterize the role of Bti9 and a closely-related tomato protein, SlLyk13, in plant immunity and investigate the mechanism by which AvrPtoB may interfere with the activity of these proteins.
Results
The N-terminal region of AvrPtoB is sufficient to suppress immunity in tomato triggered by bacterial PAMPs other than flagellin
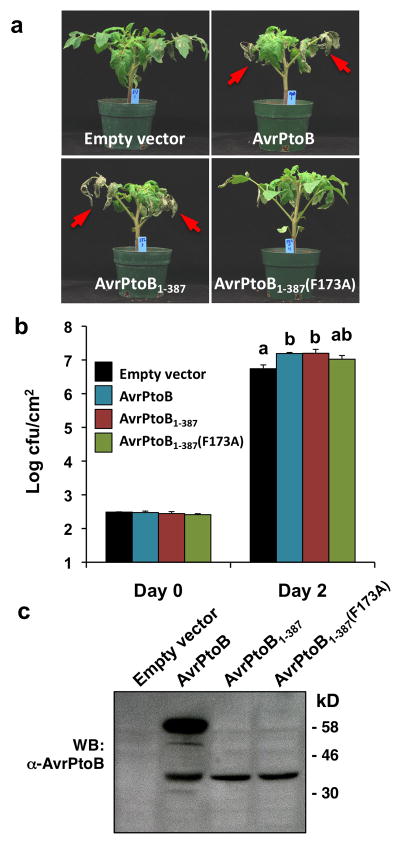
We reported previously that either of the N-terminal fragments, AvrPtoB1-307 or AvrPtoB1-387, was able to enhance Pst virulence on tomato RG-prf3 plants to the same extent as full-length AvrPtoB when expressed in DC3000ΔavrPto/ΔavrPtoB (Xiao, et al. 2007). To determine if the AvrPtoB N-terminal domain is capable of suppressing PTI triggered by Pst PAMPs other than flagellin, we inoculated RG-prf3 plants with DC3000Δ avrPtoΔavrPtoBΔhopQ1-1 ΔfliC strains (DC3000 with deletion of genes avrPto, avrPtoB, hopQ1-1, and fliC) carrying empty vector, full-length AvrPtoB, AvrPtoB1-387 or AvrPtoB1-387(F173A). The F173A substitution also abolishes AvrPtoB1-307 virulence, however, we used AvrPtoB1-387 instead of AvrPtoB1-307 in these experiments because its expression in Pst is more similar to that of full-length AvrPtoB (Xiao, et al. 2007). The fliC gene encodes flagellin and this strain therefore lacks this PAMP (Kvitko, et al. 2009). The hopQ1-1 gene is irrelevant to this experiment as it confers avirulence of Pst on N. benthamiana plants, but not on tomato (Wei et al. 2007). To distinguish between subtle differences in disease symptoms, we used a low titer of 3 × 104 cfu/mL bacteria for inoculation. Consistent with our previous findings (Xiao, et al. 2007), we found that DC3000ΔavrPtoΔ avrPtoB ΔhopQ1-1ΔfliC strains expressing AvrPtoB or AvrPtoB1-387 caused more severe disease symptoms on tomato RG-prf3 plants compared to strains carrying an empty vector or AvrPtoB1-387(F173A) (Figure 1A).
Figure 1. The N-terminal region of AvrPtoB is sufficient to suppress immunity in tomato triggered by PAMPs of Pseudomonas syringae other than flagellin.
(A) Disease symptoms of RG-prf3 plants 4 days after inoculation with Pst strains DC3000ΔavrPtoΔavrPtoBΔhopQ1-1ΔfliC carrying either empty vector (pCPP45) or plasmids expressing AvrPtoB, AvrPtoB1-387, or AvrPtoB1-387(F173A). Red arrows point to enhanced necrosis observed on lower leaves, a characteristic of AvrPtoB virulence (Xiao, et al. 2007). (B) Bacterial populations in leaves of plants as shown in (A). Pst strains were infiltrated into RG-prf3 plants using an inoculum of 3 × 104 cfu/mL. Each treatment represents the mean of four plants and bars show the standard error. Experiments were repeated three times with similar results. Means with different letters were significantly different based on a Tukey-Kramer HSD test (α= 0.05). (C) Expression of AvrPtoB, AvrPtoB1-387, or AvrPtoB1-387(F173A) in DC3000ΔavrPtoΔavrPtoBΔhopQ1-1ΔfliC carrying corresponding plasmids grown in minimal medium under hrp induction conditions (Lin, et al. 2006) confirmed using a polyclonal anti-AvrPtoB antibody (Lin et al., 2006). The lower band in the AvrPtoB lane is likely a degradation product.
Assessment of bacterial growth in these plants revealed that two days after inoculation, the Pst strains expressing AvrPtoB or AvrPtoB1-387 grew to a statistically significant higher level than the strain carrying an empty vector (Figure 1B). The growth of Pst expressing AvrPtoB1-387(F173A) was not significantly different from any of the other three strains (including the empty vector control; Figure 1B) although this strain caused less lower leaf necrosis. Each of the AvrPtoB proteins was expressed in Pst (Figure 1C). Importantly, the Pst strain expressing AvrPtoB1-387 had the same virulence activity as the strain expressing AvrPtoB, indicating the N-terminal domain (lacking the CTD E3 ligase) is sufficient to suppress PTI triggered by PAMPs other than flagellin of Pst in tomato. Note also that although the abundance of AvrPtoB1-387 protein was less than AvrPtoB, this amount appeared to be sufficient for full virulence as no additional growth was observed with the strain expressing full-length AvrPtoB (Figure 1BC). Together, these observations indicate the N-terminal domain is sufficient for full AvrPtoB virulence on tomato and that F173 plays an important role in disease symptom formation (especially necrosis of the lower leaves).
A LysM-RLK, Bti9, interacts with AvrPtoB1-307
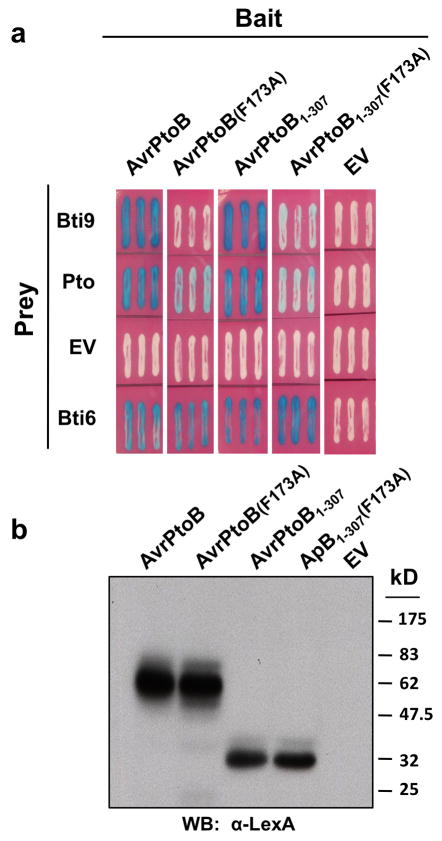
To identify host targets of AvrPtoB we performed a yeast two-hybrid screen using the effector AvrPtoB as bait with a tomato leaf cDNA library. One interactor, Bti9, was found to interact strongly with AvrPtoB and AvrPtoB1-307 but significantly less so with virulence- attenuated proteins AvrPtoB(F173A) and AvrPtoB1-307(F173A) (Figure 2A). Bti9 did not interact with AvrPto (not shown). All AvrPtoB proteins were expressed in yeast as confirmed by Western blotting and by their interaction with Bti6, a non-specific AvrPtoB interactor (Figure 2AB).
Figure 2. Bti9 interacts with AvrPtoB1-307 in an F173-dependent manner.
(A) Interaction in a yeast two-hybrid assay between Bti9 or Pto and AvrPtoB or its variants. A non-specific AvrPtoB tomato-interacting protein, Bti6, encoding a putative lactate dehydrogenase was used as a positive control (Xiao, et al. 2007). Blue patches indicate positive interactions. Photographs were taken 24 hours after yeast transformants were streaked on –Ura –His –Trp plates (Xiao, et al. 2007). (B) Expression of AvrPtoB and its variants as bait proteins in yeast cells that also express Bti9 was confirmed by Western blotting using an anti-LexA antibody.
The Bti9 region that interacted with AvrPtoB1-307 encodes a serine/threonine protein kinase domain with 76% amino acid identity to Arabidopsis Chitin Elicitor Receptor Kinase 1 (CERK1) (AtLysM-RLK1, AtLyk1) (Miya et al. 2007, Wan et al. 2008). Generation of a full-length cDNA of Bti9 revealed it encodes a protein with a predicted extracellular domain containing three lysin motifs (LysM) and the C-terminal kinase that interacts with AvrPtoB1-307 (Figure S1). The presence of a putative signal sequence and transmembrane region in Bti9 (Figure S1) suggested the protein is localized to the plasma membrane. Indeed, we detected Bti9 at the periphery of the cell when it was expressed transiently as a GFP fusion in tomato protoplasts (Figure S2), supporting a possible role for the protein in extracellular pathogen recognition.
LysM domains are known to bind N-acetylglucosamine (GlcNAc)-containing glycan molecules including peptidoglycan (PGN) from several bacteria and chitin from fungi (Bateman and Bycroft 2000, Bielnicki et al. 2006, Buist et al. 2008, Iizasa et al. 2010, Petutschnig et al. 2010, Steen et al. 2003). LysM-RLKs play an important role in Lotus japonicus and Medicago truncatula in their association with Rhizobium bacteria (Limpens et al. 2003, Radutoiu et al. 2003, Radutoiu et al. 2007, Smit et al. 2007). Furthermore, CERK1 was recently shown to be involved in immunity against Pst (Gimenez-Ibanez, et al. 2009a, Gohre, et al. 2008). These observations raised the possibility Bti9 plays a role in recognition of a bacterial PAMP and, as a result, has become a host target of the AvrPtoB N-terminal domain. We therefore tested whether the expression of Bti9, like FLS2 and other immunity-associated kinase genes (Zipfel, et al. 2004), is induced in leaves upon challenge by PAMPs. We observed Bti9 transcript abundance increased 4 hours after exposure to DC3000 as compared to control leaves. Interestingly, at 8 hours after exposure to wild-type DC3000, but not to a type III secretion mutant, this increase was no longer observed (Figure S3). These results suggest Bti9 gene expression is induced by PAMPs and that type III effectors interfere with its expression.
Two LysM-RLKs are implicated in AvrPtoB-suppressed immunity against P. syringae
The tomato genome sequence became available during the course of this work and allowed us to search for homologs of Bti9 (http://solgenomics.net/). This search revealed tomato has 13 LysM-RLK genes that we have named in accordance with previous recommendations and, in part, their relationship to the five Arabidopsis LysM-RLKs (Figure S4; (Zhang et al. 2007)). Three of these genes, SlLyk11, SlLyk12, and SlLyk13 reside in the same clade as Bti9 (also referred to as SlLyk1) and share extensive sequence similarity (Figure S5).
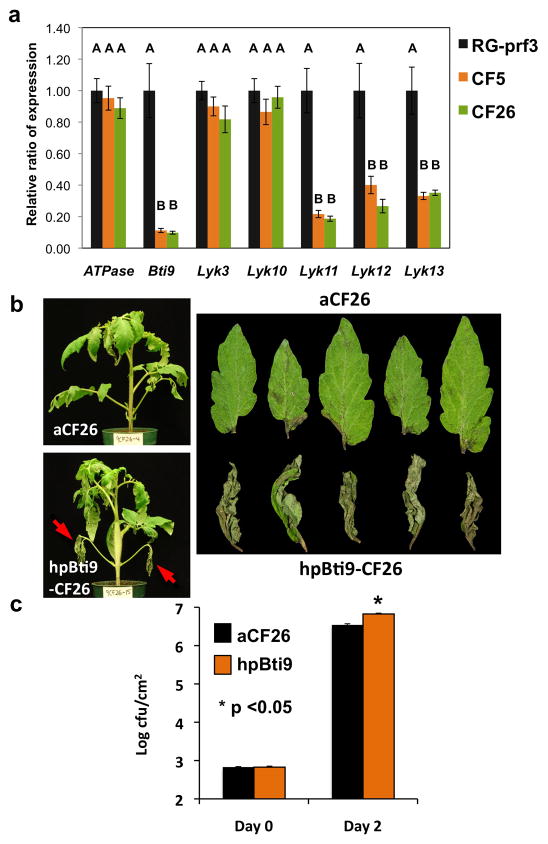
To test whether Bti9 and possibly other members of the ‘Bti9 clade’ are involved in the tomato immune response we developed stable transgenic plants that carry a ‘hairpin’ Bti9 (hpBti9) construct which could potentially silence all four genes (Helliwell et al. 2002, Wesley et al. 2001). The presence and copy number of the transgene in plants of T2 transgenic lines was examined (Figure S6). Two lines, CF5 and CF26, from independent transformation events were chosen for further characterization. Quantitative RT-PCR revealed that both lines had reduced transcript abundance of Bti9, SlLyk11, SlLyk12, and SlLyk13 (Figure 3).
Figure 3. Bti9 and SlLyk13 are implicated in tomato immunity against P. syringae pv. tomato.
(A) Transcript abundance of Bti9 and its three closest tomato homologs was reduced in CF5 and CF26 transgenic lines carrying the hpBti9 construct. Expression was analyzed by qRT-PCR using SlEF1α as a normalization control. Similar results were obtained using SlATPase normalization. The relative ratio of expression sets 1.0 as the normalized expression of the gene in wild-type plants. Each bar represents the mean of 8 plants and the bars show the standard error of the mean (σE). Means with different letters were significantly different based on a Tukey-Kramer HSD test (α = 0.05). (B) Disease symptoms of plants as shown in (A) where either hpBti9 was lost by segregation (termed ‘azygous’ CF26; aCF26) or where hpBti9 was present (hpBti9-CF26) thus transcript abundance of Bti9 was reduced. The photos were taken 4 days after vacuum infiltration of plants with DC3000ΔavrPtoΔ avrPtoBΔ hopQ1-1ΔfliC (3 × 104 cfu/mL). Red arrows in the lower panel (left) point to necrosis of lower leaves, which is a characteristic symptom of AvrPtoB virulence. Right panel shows close-up of lower leaves from aCF26 or from hpBti9-CF26. (C) Bacterial populations in leaves of aCF26 or hpBti9-CF26 tomato plants after inoculation of DC3000ΔavrPtoΔ avrPtoB ΔhopQ1-1Δ fliC as described in (B). Each treatment represents the mean of 5 plants and the bar shows the standard error. The asterisk denotes the difference of bacterial populations in leaves of aCF26 and hpBti9-CF26 was significant as tested using JMP software package (version 7, http://www.jmp.com/; Cary, NC) by a Student’s T test (α = 0.05). This experiment was performed three times with similar results.
The hpBti9 plants and appropriate controls were inoculated with Pst strain DC3000ΔavrPtoΔavrPtoBΔhopQ1-1ΔfliC and monitored for disease symptoms. We observed the hpBti9 plants displayed more severe disease symptoms than control plants (Figure 3B and Figure S7A). Interestingly, the hpBti9 plants developed extensive necrosis on their lower leaves, which is reminiscent of the phenotype associated with virulence activity of the AvrPtoB N-terminal region (Figure 1A and 3B, and (Xiao, et al. 2007)) Bacterial population assays showed there was a slight, but statistically significant, increase in Pst growth in leaves of the hpBti9 plants as compared with control leaves two days after inoculation (Figure 3C and Figure S7B). These disease assays on two independent hpBti9 transgenic lines support a role for one or more members of the Bti9 clade in tomato immunity against Pst.
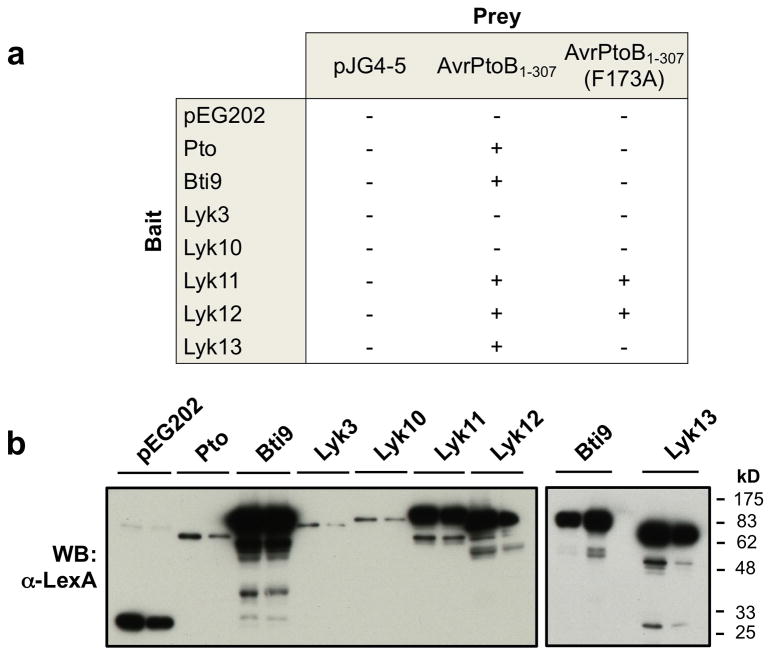
As one way of assessing which of the four Bti9-clade genes may contribute to the plant immune response, we examined whether the kinase domain encoded by SlLyk11, SlLyk12, or SlLyk13 interacts with AvrPtoB in an F173-dependent manner. Our rationale was that the F173A substitution abolishes AvrPtoB1-307 virulence activity and therefore if a SlLyk protein plays a role in the immune response suppressed by AvrPtoB then its interaction with this effector would be expected to require F173. Yeast two-hybrid analyses showed both SlLyk11 and SlLyk12 interacted with AvrPtoB1-307 but this occurred even when the F173A substitution was present (Figure 4A). SlLyk13, however, displayed an interaction profile identical to Bti9. The less-related proteins, SlLyk3 and SlLyk10, did not interact with AvrPtoB1-307. Each of the Lyk proteins was expressed in yeast and, while SlLyk3 and SlLyk10 were expressed poorly, their abundance was the same as Pto which interacts strongly with AvrPtoB1-307 in this assay (Figure 4B). We conclude from these experiments that Bti9 and SlLyk13 are the best candidates for contributing to the immunity against Pst which is suppressed by AvrPtoB1-307.
Figure 4. SlLyk13 but not SlLyk11 and SyLyk12 interacts with AvrPtoB1-307 in an F173-dependent manner.
(A) Interaction in a yeast-two hybrid assay between proteins in the Bti9 clade with AvrPtoB or its variants. +, a positive interaction (blue yeast patches observed in the assay); −, weak or no interaction (white or faint blue yeast patches). (B) Expression of Bti9 and its homologs in yeast was confirmed by protein blotting using an anti-LexA antibody.
AvrPtoB inhibits Bti9 kinase activity
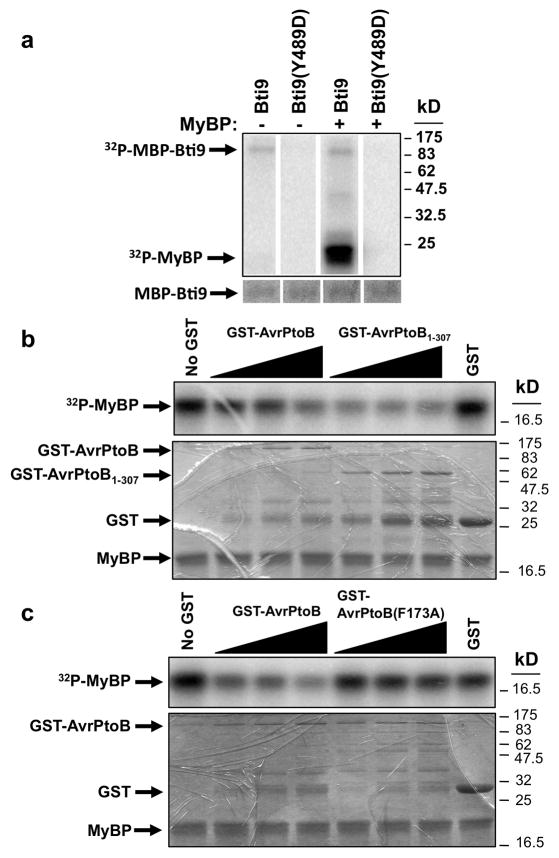
We focused on Bti9 as representative of both Bti9 and SlLyk13 in order to test if it is an active protein kinase. An in vitro kinase assay showed Bti9 weakly autophosphorylates and strongly transphosphorylates myelin basic protein, an artificial substrate (Figure 5A). In common with the Pto variant Y207D (Rathjen et al. 1999), substitution at the corresponding Bti9 amino acid, Y489D, caused a complete loss of Bti9 kinase activity (autophosphorylation and transphosphorylation). Bti9 is therefore an active protein kinase.
Figure 5. Bti9 is an active kinase whose activity is inhibited by AvrPtoB in vitro.
The top panel in each part shows a phosphor-image of the kinase assay. The bottom panel in parts B and C shows the Coomassie-stained gel that was used for phosphor-imaging of the kinase assay. (A) Bti9 encodes an active kinase. A maltose-binding protein (MBP) fusion to Bti9 weakly autophosphorylated and strongly trans-phosphorylated myelin basic protein (MyBP). Substitution of Y to D at amino acid 489 abolished Bti9 kinase activity. Reactions without MyBP were used as controls. The Coomassie-stained gel shows equal abundance of Bti9 or Bti9(Y489D) protein (barely visible because only 200 ng was used per lane). (B) AvrPtoB and AvrPtoB1-307 inhibited Bti9 kinase activity. Addition of GST-AvrPtoB or GST-AvrPtoB1-307 fusion proteins to the Bti9 kinase assays reduced the ability of Bti9 to phosphorylate MyBP in a dose-dependent manner. Top panel: first lane shows phosphorylation of MyBP by Bti9 without addition of AvrPtoB or AvrPtoB1-307. The next six lanes show the assay with addition of 2.5, 5.0, or 7.5 μg of GST-AvrPtoB or GST-AvrPtoB1-307, respectively. The last lane shows addition of GST alone did not inhibit Bti9 kinase activity. (C) The F173A substitution abolished the ability of AvrPtoB to inhibit Bti9 kinase activity. Top panel: first lane shows phosphorylation of MyBP by Bti9 without addition of AvrPtoB. The next six lanes show the assay with addition of 2.5, 5.0, or 7.5 μg of GST-AvrPtoB or GST-AvrPtoB(F173A), respectively. The last lane shows addition of GST alone did not inhibit Bti9 kinase activity.
The kinase activity of FLS2 is required for its role in innate immunity (Gómez-Gómez, et al. 2001). In addition, recent work on PTI suppression by AvrPto showed this effector inhibits kinase activity of PRR complexes (Shan, et al. 2008, Xiang, et al. 2011, Xiang, et al. 2008). We therefore examined whether the AvrPtoB N-terminal region affects Bti9 kinase activity. Standard in vitro kinase activity assays for Bti9 were conducted in the presence of increasing amounts of GST-AvrPtoB, GST-AvrPtoB1-307 or GST alone. A Bti9 reaction without of any of these proteins was used as a control. Both GST-AvrPtoB and GST-AvrPtoB1-307 inhibited Bti9 kinase activity in a dose-dependent manner (Figure 5B). In contrast, GST alone had no effect on Bti9 kinase activity. GST-AvrPtoB1-307 inhibited Bti9 kinase activity even more effectively than AvrPtoB, but this may be due to the higher relative molarity of this smaller protein (Figure 5B). We next examined the ability of AvrPtoB(F173A) to inhibit Bti9 kinase activity (Figure 5C). We found AvrPtoB(F173A) was unable to inhibit Bti9 kinase activity, even though the abundance of this variant was the same as wild-type AvrPtoB (Figure 5C). Collectively, these results provide additional evidence for interaction between AvrPto and Bti9 and also support a model in which AvrPtoB inhibits kinase activity of Bti9, and possibly that of SlLyk13, to block PTI signaling.
Bti9/SlLyk13 are probably not the only virulence targets of AvrPtoB1-307 in tomato
A single pathogen effector can have multiple host targets for promoting pathogen virulence (Hogenhout, et al. 2009). In the case of AvrPtoB, the region between amino acids 307–387 has been reported to be required for interference with FLS2/BAK1-mediated PTI (Xiao, et al. 2007, Shan, et al. 2008). It is therefore possible the AvrPtoB1-307 domain has targets in addition to Bti9/SlLyk13. To test this, we monitored bacterial growth of DC3000ΔavrPtoΔavrPtoBΔhopQ1-1ΔfliC either expressing or not expressing AvrPtoB1-387 on RG-prf3 and hpBti9 plants. Consistent with our earlier observations, AvrPtoB1-387 enhanced virulence of this DC3000 strain on RG-prf3 plants and silencing of Bti9/SlLyk13 rendered tomato plants more susceptible (Figure S8). Importantly, when this DC3000 strain was expressing AvrPtoB1-387 it was still more virulent on hpBti9 plants than the same strain carrying an empty vector. This suggests the AvrPtoB N-terminal region has other host targets in addition to Bti9. However, we cannot rule out the possibility this enhanced virulence is due to AvrPtoB suppression of residual Bti9/SlLyk13 accumulation in the cell in spite of the hpBti9-mediated gene silencing.
Bti9 and Pto interact with AvrPtoB1-307 in a structurally similar but not identical way
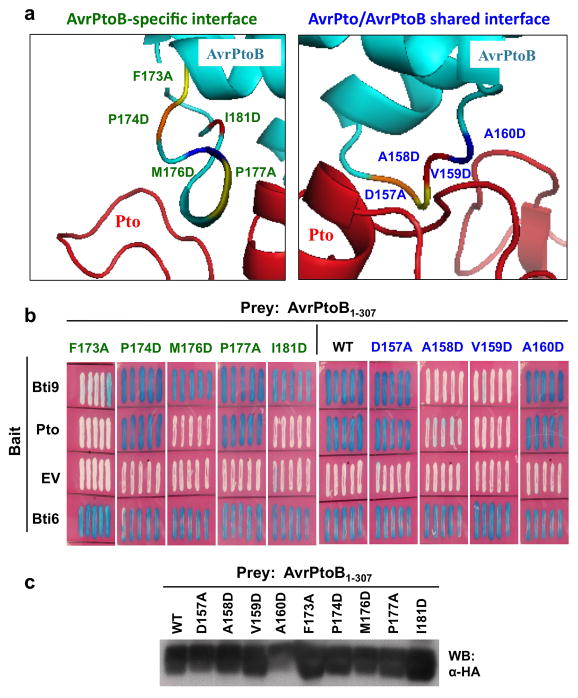
The crystal structure of the subdomain in AvrPtoB1-307 that is sufficient for its interaction with Pto has been solved (AvrPtoB121-200; (Dong et al. 2009)). This structure revealed Pto has one interface that it uses for interaction with both AvrPto and AvrPtoB and another that is unique for AvrPtoB (Figure 6A). Notably, the F173 residue, which is required for AvrPtoB1-307 virulence, is also required for recognition by Pto indicating the tomato immune system has evolved to specifically detect this virulence determinant of AvrPtoB.
Figure 6. Bti9 and Pto interact with AvrPtoB1-307 in a similar but not identical way.
(A) Close-up of the crystal structure of the complex between AvrPtoB (in turquoise) and Pto (in red) (Dong, et al. 2009). The interface with Pto that is specific to AvrPtoB is shown in the left panel and the interface shared by both AvrPtoB and AvrPto is shown in the right panel. Residues at which amino acid substitutions were made are shown and their positions indicated in different colors (Dong, et al. 2009). (B) Interactions in a yeast two-hybrid assay of Bti9 or Pto with AvrPtoB1-307 and variants. Blue patches indicate a positive interaction. A non-specific AvrPtoB tomato-interacting protein, Bti6, encoding a putative lactate dehydrogenase was used as a positive control (Xiao, et al. 2007). (C) Immunoblot analysis with anti-HA antibody in the bottom panel shows similar expression in yeast of wild-type AvrPtoB1-307 and the variant proteins from the prey vector.
To further examine the possible similarity between the interaction of Bti9 and Pto with AvrPtoB1-307, we took advantage of a series of substitutions in AvrPtoB that were developed for analysis of the AvrPtoB121-200-Pto complex (Dong, et al. 2009). The expression of each of these variants was verified and their interactions with Bti9 and Pto were tested in the yeast two-hybrid system (Figure 6BC). The interactions of these two proteins with AvrPtoB1-307 were largely similar with the only differences being M176D and I181D which disrupted the interaction with Pto but not with Bti9. These observations support a model in which Pto evolved as a structural mimic, albeit not a perfect one, of Bti9/SlLyk13.
Discussion
By searching for host targets of AvrPtoB virulence activity we isolated a LysM-RLK gene, Bti9, and later identified three Bti9-related genes in the tomato genome. Our rationale for initially focusing on Bti9 was that it interacts with a minimal fragment of AvrPtoB (amino acids 1–307) that enhances virulence of Pst on tomato to the same degree as full-length AvrPtoB (Xiao, et al. 2007). Furthermore, this interaction was dependent on phenylalanine-173, a residue that is required for AvrPtoB1-307 virulence activity. Similar to many kinase genes involved in PTI, Bti9 gene expression was induced by PAMPs and appeared to be suppressed by type III effectors. The fact that four genes exist in the Bti9 clade raised the possibility they may have some redundancy in function. Our hpBti9 construct successfully knocked down expression of all four genes and resulted in increased susceptibility to Pst. However, only Bti9 and SlLyk13 interacted with AvrPtoB in an F173-dependent manner suggesting they are the most likely virulence targets of this effector protein. AvrPtoB also interacted with Bti9 in vitro as inferred from its ability to inhibit Bti9 kinase activity. This inhibition was dependent on F173 which supports kinase inhibition as the primary mechanism for interference in the function of Bti9, and possibly SlLyk13, and thus PTI suppression. Finally, based on the similar interaction of Bti9 and Pto with AvrPtoB our data support the possibility Pto evolved as a molecular mimic (a decoy) of Bti9, although it does not appear to be ‘perfect’ structural mimic as some interaction differences were observed. Together, our data support a model in which Bti9 and/or SlLyk13 play a role in perception of a bacterial PAMP and in PTI signaling and have therefore been targeted for interference by AvrPtoB.
We have shown here and previously that the N-terminal portion of AvrPtoB is sufficient to enhance Pst virulence in tomato to the same degree as full-length AvrPtoB (Xiao, et al. 2007). We have also reported that a naturally-occurring truncated AvrPtoB homolog (HopPmaL) which lacks the E3 ligase domain retains full virulence in tomato (Lin et al. 2006). These observations are in contrast to two recent papers both of which conclude the E3 ubiquitin ligase of AvrPtoB is required for virulence on Arabidopsis. It is possible there are actual differences between tomato and Arabidopsis in this regard. However, an alternative explanation is that certain experimental conditions differed between our work and these other studies. For example, the expression levels in Pst were very low of the E3 ligase-deficient versions of AvrPtoB in one of the studies and may have underestimated the virulence activity of these proteins (Gimenez-Ibanez, et al. 2009a). In the other study, the AvrPtoB1-387 fragment was tested and shown to have less virulence activity but its activity was not compared with full-length AvrPtoB expressed from the same plasmid (Gohre, et al. 2008). Consistent with our observations, the AvrPtoB1-387 N-terminal domain is sufficient to suppress molecular readouts associated with PTI in Arabidopsis also indicating the E3 ligase is dispensable for virulence in this species (Shan, et al. 2008). In the cases where AvrPtoB has been compared with similarly-expressed truncated versions the results indicate the E3 ligase is not required for AvrPtoB virulence activity. To date, the only well-substantiated role reported for the AvrPtoB E3 ligase is to facilitate ubiquitination-mediated degradation of the Fen kinase to overcome ETI (Rosebrock, et al. 2007). The presence of Fen-like kinases in diverse plant species suggests this mechanism has been evolutionarily important in defeating this conserved immune response.
Expression of a hairpin Bti9 construct in tomato resulted in increased susceptibility to Pst infection with no other morphological or developmental defects being observed in these plants. We used a DC3000 strain that lacks flagellin in order to exclude the strong FLS2-mediated immune response and uncover other potential PTI responses and observed enhanced disease symptoms in the hpBti9 plants particularly on the lower leaves. This lower-leaf necrosis is remarkably similar to the phenotype associated with AvrPtoB virulence activity and, along with our yeast two-hybrid data, further support Bti9/SlLyk13 as targets of AvrPtoB that may play an especially important role in older leaves. Despite the effective silencing of Bti9/SlLyk13 we still observed AvrPtoB virulence activity on hpBti9 plants. The most likely explanation for this observation is probably that AvrPtoB1-307 has additional virulence targets in tomato. However, it is also possible that AvrPtoB1-307 interferes with residual Bti9/SlLyk13 protein still present in our hpBti9 plants allowing enhanced disease and bacterial growth.
The Bti9 clade contains four closely related LysM-RLK genes all of which were silenced to some degree in our hpBti9 plants. However, we present data that suggests Bti9 and/or SlLyk13 are the most likely targets of AvrPtoB as these proteins interact in a biologically relevant way with AvrPtoB. SlLyk11 and SlLyk12 interact even with virulence-attenuated AvrPtoB(F173A). They are likely not responsible for the PTI response that we see suppressed by AvrPtoB, but our data cannot exclude the possibility they may contribute to the tomato immune response.
We found both full-length AvrPtoB and AvrPtoB1-307 inhibited Bti9 kinase activity in a dose-dependent manner and this inhibition was dependent on the virulence-associated F173 residue. Although we have not demonstrated kinase activity is required for Bti9 signaling, that is the case for other immunity-associated kinases such as FLS2 and TPK1b and it seems likely it is the case for Bti9 (Abuqamar et al. 2008, Gomez-Gomez et al. 2001). The fact that AvrPtoB inhibits this activity therefore would support a model in which this is the primary mechanism of AvrPtoB activity directed against Bti9/SlLyk13. In the future, it will be important to identify substrates of these proteins that act in the tomato immune response as that will allow further testing of the effectiveness of AvrPtoB to interfere with natural Bti9 phosphorylation events.
Bti9 and Pto interacted with AvrPtoB1-307 using mostly the same effector residues thereby supporting the hypothesis that Pto might have evolved as a molecular mimic of the Bti9 kinase domain. However, two aspartate substitutions, at AvrPtoB residues M176 and I181, disrupted the interaction with Pto but not with Bti9. These residues lie in a loop of AvrPtoB that interacts with the AvrPtoB-specific contact site of Pto. These observations raise the possibility that AvrPtoB might be able to acquire mutations that would alter M176 and I181 allowing the effector to escape Pto detection while at the same time retaining its ability to interfere with Bti9. The fact such mutations are not present in over 20 AvrPtoB (HopAB) homologs suggests such mutations might have other detrimental effects on AvrPtoB structure or function (http://pseudomonas-syringae.org/).
CERK1, which based on amino acid sequence similarity is the closest Arabidopsis protein to Bti9, has also been reported to play a role in immunity against Pst and to be targeted by AvrPtoB (Gimenez-Ibanez, et al. 2009a). Our results are largely consistent with this previous work while providing additional insights into the AvrPtoB-Bti9 interaction and also supporting an alternative model for how AvrPtoB interferes with Bti9 function. In common with this earlier paper, we discovered the AvrPtoB1-307 domain is sufficient for interaction with Bti9 and we extended this to show the virulence-associated residue F173 is required for this interaction. The compromised immunity to Pst observed with a cerk1 Arabidopsis mutant and our hpBti9 tomato plants indicate that, in addition to playing a role in chitin perception (Miya, et al. 2007, Wan, et al. 2008), these kinases also may play a role in detection of a bacterial PAMP. LysM domains are known to bind certain N-acetylglucosamine (GlcNAc)-containing glycan molecules including peptidoglycan (PGN) from bacteria (Petutschnig, et al. 2010) and it is an attractive hypothesis that Bti9/SlLyk13 and CERK1 may bind PGN from Pst to activate PTI. However, preliminary experiments indicate this is not the case (Gimenez-Ibanez et al. 2009b) and the identity of the putative cognate PAMP remains unknown.
The previous paper on CERK1 presented evidence that the AvrPtoB E3 ligase is required for effector virulence and that it played a role in proteasome-independent degradation of CERK1 via a vacuolar-mediated process (Gimenez-Ibanez, et al. 2009a). In contrast, we observed the E3 ligase domain to be dispensable for full AvrPtoB virulence in tomato. Instead, we discovered AvrPtoB inhibits Bti9 kinase activity supporting this inhibition as the primary mechanism of AvrPtoB virulence. A possible synthesis of these results would be that Bti9 kinase activity is inhibited and the inactivated Bti9 is turned over by a vacuolar-dependent receptor recycling pathway. Down-regulation of receptors by internalization and subsequent lysosomal degradation typically involves monoubiquitination or K63-linked polyubiquitination of the substrates (Lauwers et al. 2009, Mukhopadhyay and Riezman 2007) and the AvrPtoB E3 ligase would likely not play a biologically significant role in this process.
We propose a model in which Bti9/SlLyk13 are localized to the plasma membrane and play a direct or indirect role in recognizing a bacterial PAMP upon pathogen infection (Figure S9). This recognition activates Bti9/SlLyk13 kinase activity that in turn phosphorylates an unknown substrate(s) leading to downstream signaling culminating in PTI. To subvert this PTI response, the N-terminal region of AvrPtoB has evolved to inhibit Bti9/SlLyk13 kinase activity, which blocks PTI signaling and signals the cell to recycle these proteins via a vacuolar-mediated process (Gimenez-Ibanez, et al. 2009a). To counter this interference, tomato has evolved Pto, likely as a molecular mimic of the Bti9/SlLyk13 kinase domain. Pto interacts with the AvrPtoB N-terminal region, which elicits ETI against the pathogen. Examination of this model in the future will benefit from a crystal structure of the AvrPtoB1-307-Bti9 (or AvrPtoB1-307-SlLyk13) complex, identification of the cognate PAMP and possible Bti9/SlLyk13 adaptor proteins, and importantly an understanding of substrates and the pathways these LysM-RLK activate.
Experimental Methods
Generation of full-length cDNA for the Bti9 gene
The C-terminal domain of Bti9 (nucleotide 814 to the end) in prey vector pJG4-5 was recovered from yeast and sequenced. 5′-rapid amplification of cDNA ends (RACE) using gene-specific primer LRZ01 (Table S1) and the ′-RACE System (Invitrogen, CA) was performed to obtain the full-length sequence of the gene.
Generation of hpBti9 transgenic plants
Gateway cloning (Hartley et al. 2000) was used to generate a Bti9 ‘hairpin’ construct (hpBti9) consisting of the C-terminal region of the Bti9 gene (nucleotides 1206 to 1807). A primer pair LRZ02 and LRZ03 (Table S1) was used to amplify the C-terminal fragment with Easy-A High-Fidelity PCR Cloning Enzyme (Stratagene, CA) and cloned into pCR8/GW/TOPO vector (Invitrogen, CA). The fragment was transferred into the pHellsgate8 binary vector (Helliwell, et al. 2002, Wesley, et al. 2001) by Gateway LR reactions. The pHellsgate8 plasmid containing the C-terminal region of Bti9 was transformed into tomato RG-prf3 (Pto/Pto, prf/prf) using Agrobacterium-mediated gene transfer technique (Hoekema et al. 1983). The expression level of Bti9 in T1 transformants was initially assessed by reverse transcriptase-PCR using primer pair LRZ04 and LRZ05 (Table S1). T1 transformants that showed reduced Bti9 expression were advanced to the T2 generation. The copy number of the hpBti9 construct in plants of T2 lines was examined by DNA gel blots as shown in Figure S4. T2 plants derived from T1 lines 9CF-5 and 9CF-26, both of which had a single copy of the hpBti9 transgene in RG-prf3 background, were used for pathogen assays.
Assessment of pathogen virulence in tomato leaves
RG-prf3 (Pto/Pto, prf/prf) and hpBti9 plants were grown in the greenhouse. Plasmids carrying avrPtoB or its variants expressed from a hrp promoter were developed using vector pCPP45 and transformed into Pst DC3000ΔavrPtoΔavrPtoBΔhopQ1-1ΔfliC (Lin and Martin 2005). Inoculation of tomato plants was performed as described (Anderson et al. 2006). In brief, three- to four-week old plants were vacuum-infiltrated with the Pst strains using an inoculum of 3 x104 cfu/mL. The inoculated plants were maintained in a growth chamber at 24/20 °C day/night temperature, 50% humidity, and 16-/8-hour light/dark cycle. Bacterial populations were recovered and measured at 0 and 2 days after inoculation. Photos of the plants were taken at 4 days after inoculation. Plants of the same size were used for each experiment, and lower leaves from approximately the same position on each plant were sampled. As described (Anderson, et al. 2006), consistency in handling of bacterial strains and plants is critical for reproducible virulence assays. The presence of the hpBti9 transgene in each T2 plant was examined by PCR of genomic DNA using primers LRZ06 and LRZ07 (Table S1) (T-DNA right border) and primers LRZ08 and LRZ09 (T-DNA left border). Expression of Bti9 was analyzed by semi-quantitative PCR as described above.
Quantitative real time reverse transcription PCR (qRT-PCR)
qRT-PCR was performed as described (Nguyen et al. 2010) but with qRT-PCR cycling conditions as follows: 50 °C for 2 minutes, 95 °C for 10 minutes and 40 cycles of 95 °C for 30 seconds, 55 °C for 30 seconds and 72 °C for 30 seconds. qRT-PCR expression data were analyzed using a Tukey-Kramer HSD test (α = 0.05). Primers and their concentrations are in Table S1.
Yeast two-hybrid assay
A LexA yeast two-hybrid system was used to test protein interactions (Golemis et al. 2008). The vector pEG202 was used for baits and pJG4-5 for preys. Pto in bait and prey vector, AvrPtoB and its variants in bait vector, and AvrPtoB1-307 and its variants in prey vector were described previously (Lin, et al. 2006, Xiao, et al. 2007) while SlLyks were in bait vector. Screening for AvrPtoB tomato-interacting (Bti) proteins was described previously (Abramovitch, et al. 2006) using a tomato cDNA prey library (Zhou et al. 1995).
Protein expression and in vitro kinase assay
Mutation of Bti9 Y489D was generated by site-directed mutagenesis using primers LRZ10 and LRZ11 (Table S1). Bti9 and Bti9(Y489D) were expressed as fusions with MBP (pMAL-c2; New England Biolabs, MA) in BL21 (DE3) E. coli (Invitrogen, Carlsbad) and purified using amylose resin (New England Biolabs). AvrPtoB(F173A) was prepared using site-directed mutagenesis (Xiao, et al. 2007). AvrPtoB, AvrPtoB(F173A) and AvrPtoB1-307 were expressed as N-terminal GST-fused proteins (pDEST15, Invitrogen) in BL21 (DE3) E. coli cells and purified using glutathione Sepharose 4 fast flow resin (GE Health, WI). GST-fused proteins were eluted from Sepharose 4 resin and concentrated using an Amicon Ultra Centrifugal Filter Unit (Millipore, MA). In vitro kinase assays were done in 30 μl reactions containing 200 nanograms of Bti9 or Bti9(Y489D), 7.5 micrograms myelin basic protein (M1891; Sigma-Aldrich, St. Louis, MO), 5 μCi of [γ-32P]-ATP, 20 μl kinase buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM NaCl, 1 mM DTT, 5 uM ATP, and1 mM EDTA) at room temperature for 30 minutes. The reactions were stopped with SDS-loading buffer and heated at 100°C for 5 min before electrophoresis on 12.5% SDS polyacrylamide gel. The gel was dried and radioactivity visualized using a PhosphorImager (Molecular Dynamics, CA). For kinase inhibition assays, purified proteins of AvrPtoB or its derivatives were added to the reactions before they were incubated at room temperature. For visualization of protein loading, the dried gel was stained with Coomassie Brilliant Blue R250 and photographed after de-staining.
Western blotting
Standard methods were used for Western blotting. Rat anti-HA primary antibody (clone 3F10) was from Roche Applied Science (Indianapolis, IN). Rabbit anti-LexA primary antibody was from Invitrogen. HRP-conjugated secondary antibodies and ECL Plus detection system (GE Healthcare) were used for detection. For detection of AvrPtoB, the rabbit polyclonal anti-AvrPtoB antibody was used with a similar dilution as described (Lin et al. 2006).
Supplementary Material
Figure S1. Domain organization and amino acid sequence of the Bti9 protein.
Figure S2. Subcellular localization of Bti9 in tomato protoplasts revealed by confocal fluorescence microscopy.
Figure S3. Bti9 transcript abundance.
Figure S4. Comparison of the Bti9 kinase domain with that of LysM RLK proteins in tomato and other plant species.
Figure S5. Comparison of the DNA sequence spanning the hpBti9 fragment with the corresponding region in SlLyk11, SlLyk12, and SlLyk13.
Figure S6. Characterization of hpBti9 T2 transformants.
Figure S7. Characterization of the involvement of Bti9 in tomato innate immunity using plants from hpBti9 T2 transgenic line CF-5
Figure S8. AvrPtoB still exhibits virulence activity on hpBti9 tomato plants.
Figure S9. Model for suppression of Bti9/SlLyk13-mediated immunity by AvrPtoB.
Primer sequences used for RT-PCR.
Genbank accession and ID numbers for the genes and proteins used in this study.
Detection of Bti9 subcellular localization.
Acknowledgments
We thank Kent Loeffler from the Department of Plant Pathology and Plant-Microbe Biology, Cornell University for photography, Mamta Srivastava at the Plant Cell Imaging Center of the Boyce Thompson Institute for assistance with confocal microscopy, and Heather McLane for Figure S3. This work was supported, in part, by the National Science Foundation (IOB-0841807) and the National Institutes of Health (R01-GM078021) to GBM.
Contributor Information
Lirong Zeng, Email: lxzeng@ualr.edu.
André C. Velásquez, Email: acv24@cornell.edu.
Kathy R. Munkvold, Email: kar47@cornell.edu.
Jingwei Zhang, Email: jz293@cornell.edu.
References
- Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci U S A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuqamar S, Chai MF, Luo H, Song F, Mengiste T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. The Plant cell. 2008;20:1964–1983. doi: 10.1105/tpc.108.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Pascuzzi PE, Xiao F, Sessa G, Martin GB. Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell. 2006;18:502–514. doi: 10.1105/tpc.105.036590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Bielnicki J, Devedjiev Y, Derewenda U, Dauter Z, Joachimiak A, Derewenda ZS. B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins. 2006;62:144–151. doi: 10.1002/prot.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T. Chemoperception of microbial signals in plant cells. Annual Reviews of Plant Physiology and Plant Molecular Biology. 1995;46:189–214. [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci U S A. 2011;108:2975–2980. doi: 10.1073/pnas.1013031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna CH, Millet YA, Koller T, Han SW, Bent AF, Ronald PC, Ausubel FM. The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9286–9291. doi: 10.1073/pnas.1106366108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Xiao F, Fan F, Gu L, Cang H, Martin GB, Chai J. Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell. 2009;21:1846–1859. doi: 10.1105/tpc.109.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009a;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Ntoukakis V, Rathjen JP. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal Behav. 2009b;4:539–541. doi: 10.4161/psb.4.6.8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohre V, Spallek T, Haweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol. 2008;18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- Golemis EA, Serebriiskii I, Finley RL, Jr, Kolonin MG, Gyuris J, Brent R. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 2008. Interaction trap/two-hybrid system to identify interacting proteins; pp. 20.21.21–20.21.35. [Google Scholar]
- Gomez-Gomez L, Bauer Z, Boller T. Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. The Plant cell. 2001;13:1155–1163. [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 2002;7:251–256. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR. The Majority of the Type III Effector Inventory of Pseudomonas syringae pv. tomato DC3000 Can Suppress Plant Immunity. Mol Plant Microbe Interact. 2009;22:1069–1080. doi: 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann DR, Gimenez-Ibanez S, Rathjen JP. Bacterial virulence effectors and their activities. Curr Opin Plant Biol. 2010;13:388–393. doi: 10.1016/j.pbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Hann DR, Rathjen JP. Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 2007;49:607–618. doi: 10.1111/j.1365-313X.2006.02981.x. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM. High-throughput vectors for efficient gene silencing in plants. Functional Plant Biology. 2002;29:1217–1225. doi: 10.1071/FP02033. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. A binary plant vector strategy based on separation of vir and Tregion of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact. 2009;22:115–122. doi: 10.1094/MPMI-22-2-0115. [DOI] [PubMed] [Google Scholar]
- Iizasa Ei, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Kvitko BH, Park DH, Velasquez AC, Wei CF, Russell AB, Martin GB, Schneider DJ, Collmer A. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E, Jacob C, André B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol. 2009;185:493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chah OK, Sheen J. Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature. 2011;473:376–379. doi: 10.1038/nature09958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- Lin NC, Martin GB. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe Interact. 2005;18:43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- Lin NC, Abramovitch RB, Kim YJ, Martin GB. Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl Environ Microbiol. 2006;72:702–712. doi: 10.1128/AEM.72.1.702-712.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact. 2006;19:1151–1158. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Chakravarthy S, Velásquez AC, McLane HS, Zeng L, Park DW, Collmer A, Martin GB. Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Molecular Plant-Microbe Interactions. 2010;23:991–999. doi: 10.1094/MPMI-23-8-0991. [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease. Ann Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. The Journal of biological chemistry. 2010;285:28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EMH, Albrektsen AS, James EK, Thirup S, Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen JP, Chang JH, Staskawicz BJ, Michelmore RW. Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 1999;18:3232–3240. doi: 10.1093/emboj/18.12.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Staskawicz BJ. Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239:6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, Martin GB, Sheen J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A, Buist G, Leenhouts KJ, El Khattabi M, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem. 2003;278:23874–23881. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1–1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007;51:32–46. doi: 10.1111/j.1365-313X.2007.03126.x. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zhang J, Chen J, Chen M, Zhou JM. BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Molecular plant-microbe interactions: MPMI. 2011;24:100–107. doi: 10.1094/MPMI-04-10-0096. [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou JM. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Xiao F, He P, Abramovitch RB, Dawson JE, Nicholson LK, Sheen J, Martin GB. The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 2007;52:595–614. doi: 10.1111/j.1365-313X.2007.03259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, Cannon SB, Stacey G. Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007;144:623–636. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Loh YT, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine-threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 2008;20:10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Domain organization and amino acid sequence of the Bti9 protein.
Figure S2. Subcellular localization of Bti9 in tomato protoplasts revealed by confocal fluorescence microscopy.
Figure S3. Bti9 transcript abundance.
Figure S4. Comparison of the Bti9 kinase domain with that of LysM RLK proteins in tomato and other plant species.
Figure S5. Comparison of the DNA sequence spanning the hpBti9 fragment with the corresponding region in SlLyk11, SlLyk12, and SlLyk13.
Figure S6. Characterization of hpBti9 T2 transformants.
Figure S7. Characterization of the involvement of Bti9 in tomato innate immunity using plants from hpBti9 T2 transgenic line CF-5
Figure S8. AvrPtoB still exhibits virulence activity on hpBti9 tomato plants.
Figure S9. Model for suppression of Bti9/SlLyk13-mediated immunity by AvrPtoB.
Primer sequences used for RT-PCR.
Genbank accession and ID numbers for the genes and proteins used in this study.
Detection of Bti9 subcellular localization.