Abstract
Mitochondrial DNA (mtDNA) molecules from species of the genus Drosophila contain a region exceptionally rich in adenine + thymine (A+T). Using agarose gel electrophoresis and electron microscopy, we determined that in the mtDNA molecules of D. melanogaster, D. simulans, D. mauritiana, D. yakuba, D. takahashii, and D. virilis, the A+T-rich regions, which are 5.1, 4.8, 4.6, 1.1, 2.2, and 1.0 kilobase pairs in size, respectively, are at homologous locations relative to various common EcoRI and HindIII cleavage sites. Under conditions highly permissive for base pairing (35% formamide), heteroduplexes were constructed between EcoRI fragments and whole circular molecules of mtDNAs of the above mentioned six species in a variety of combinations. Complete pairing of molecules outside the A+T-rich region was found in all heteroduplexes examined. However, in contrast, A+T-rich regions of the different species failed to pair in all but those combinations of mtDNAs involving the three most closely related species. In heteroduplexes between D. melanogaster and D. simulans, and between D. melanogaster and D. mauritiana mtDNAs, up to 35% of the A+T-rich regions appeared double-stranded. These data indicate that much more extensive divergence of sequences has occurred in A+T-rich regions than in other regions of Drosophila mtDNA molecules.
Full text
PDF




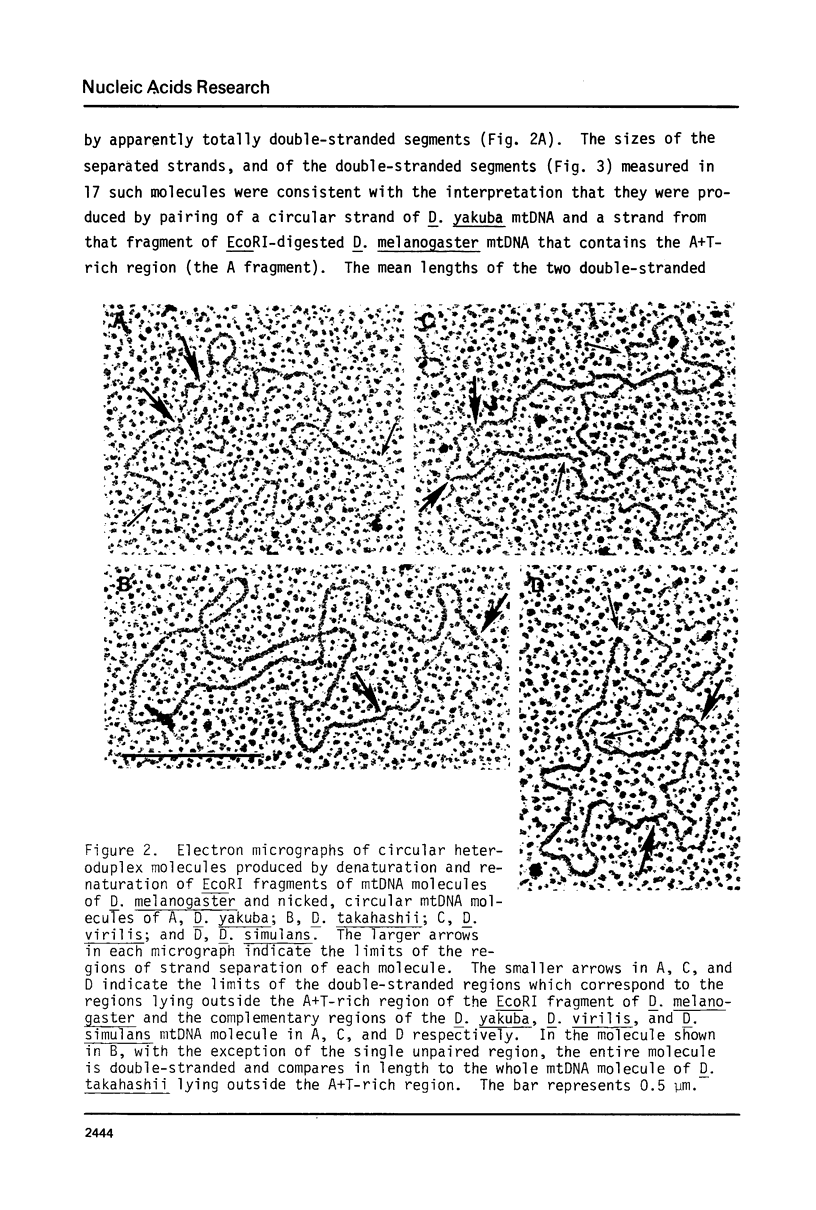
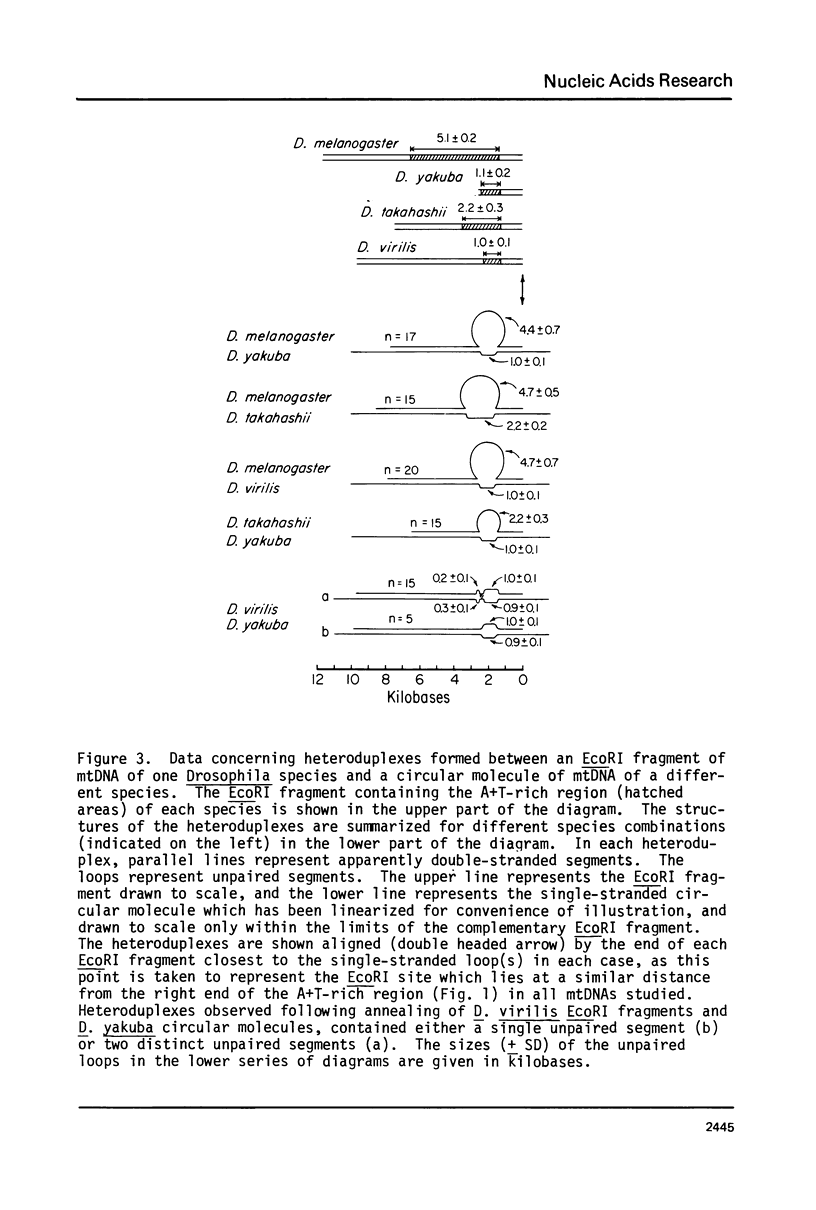


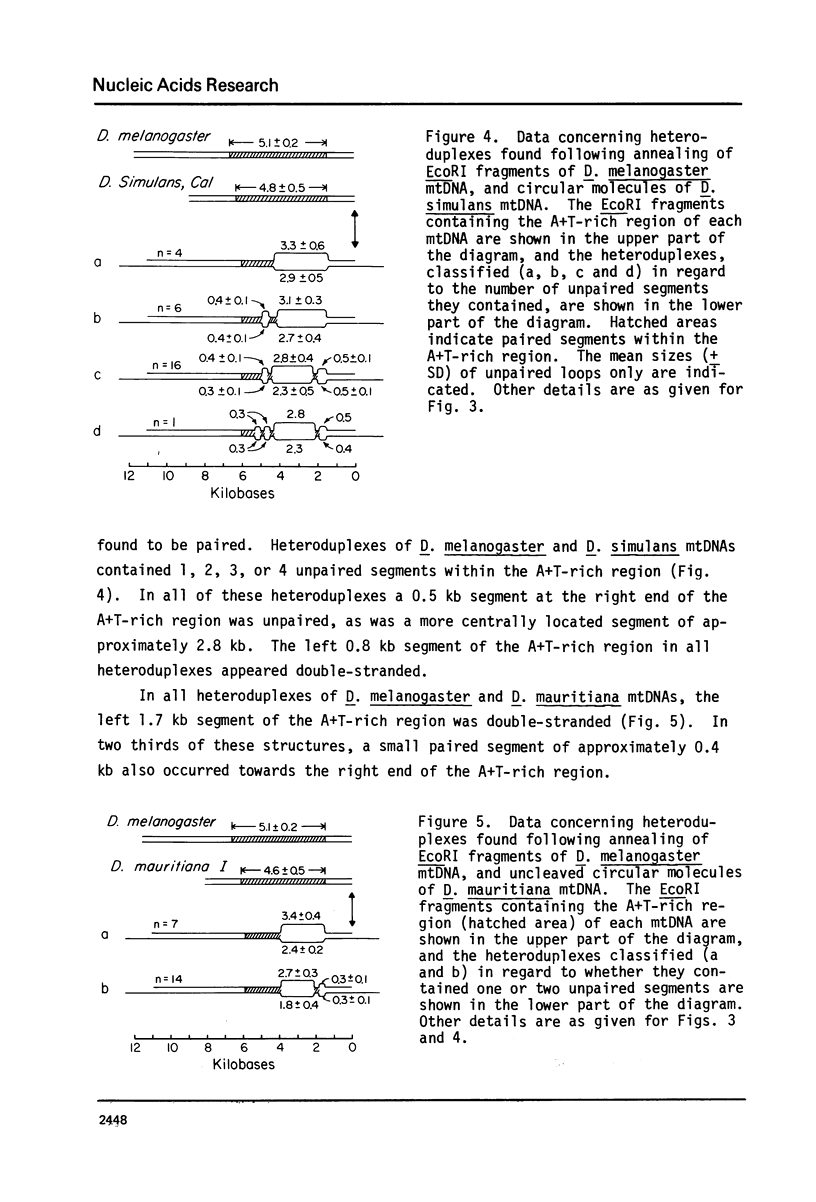




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultmann H., Laird C. D. Mitochondrial DNA from Drosophila melanogaster. Biochim Biophys Acta. 1973 Mar 19;299(2):196–209. doi: 10.1016/0005-2787(73)90342-0. [DOI] [PubMed] [Google Scholar]
- Buzzo K., Fouts D. L., Wolstenholme D. R. EcoRI cleavage site variants of mitochondrial DNA molecules from rats. Proc Natl Acad Sci U S A. 1978 Feb;75(2):909–913. doi: 10.1073/pnas.75.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauron C. M., Wolstenholme D. R. Structural heterogeneity of mitochondrial DNA molecules within the genus Drosophila. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3623–3627. doi: 10.1073/pnas.73.10.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D. L., Manning J. E., Wolstenholme D. R. Physicochemical properties of kinetoplast DNA from Crithidia acanthocephali. Crithidia luciliae, and Trypanosoma lewisi. J Cell Biol. 1975 Nov;67(2PT1):378–399. doi: 10.1083/jcb.67.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J. M., Wolstenholme D. R. Origin and direction of replication in mitochondrial DNA molecules from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3886–3890. doi: 10.1073/pnas.75.8.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E. S., Peacock W. J. Intramolecular heterogeneity of mitochondrial DNA of Drosophila melanogaster. J Cell Biol. 1977 May;73(2):279–286. doi: 10.1083/jcb.73.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klukas C. K., Dawid I. B. Characterization and mapping of mitochondrial ribosomal RNA and mitochondrial DNA in Drosophila melanogaster. Cell. 1976 Dec;9(4 Pt 1):615–625. doi: 10.1016/0092-8674(76)90044-1. [DOI] [PubMed] [Google Scholar]
- Lemeunier F., Ashburner M. A. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc R Soc Lond B Biol Sci. 1976 May 18;193(1112):275–294. doi: 10.1098/rspb.1976.0046. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Polan M. L., Friedman S., Gall J. G., Gehring W. Isolation and characterization of mitochondrial DNA from Drosophila melanogaster. J Cell Biol. 1973 Feb;56(2):580–589. doi: 10.1083/jcb.56.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. M., Langley C. H. Electron microscope heteroduplex study of Drosophila mitochondrial DNAs: evolution of A+T-rich region. Plasmid. 1979 Jan;2(1):69–78. doi: 10.1016/0147-619x(79)90007-6. [DOI] [PubMed] [Google Scholar]
- Shah D. M., Langley C. H. Inter- and intraspecific variation in restriction maps of Drosophila mitochondrial DNAs. Nature. 1979 Oct 25;281(5733):696–699. doi: 10.1038/281696a0. [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R., Fauron C. M. A partial map of the circular mitochondrial genome of Drosophila melanogaster. Location of EcoRI-sensitive sites and the adenine-thymine-rich region. J Cell Biol. 1976 Nov;71(2):434–448. doi: 10.1083/jcb.71.2.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakour R. A., Bultmann H. Evoluation of Drosophila mitochondrial DNAs. Analysis of heteroduplex molecules. Biochim Biophys Acta. 1979 Sep 27;564(2):342–351. doi: 10.1016/0005-2787(79)90231-4. [DOI] [PubMed] [Google Scholar]



