Abstract
The evolutionary origins of the multitude of duplicate genes in the plant genomes are still incompletely understood. To gain an appreciation of the potential selective forces acting on these duplicates, we phylogenetically inferred the set of metabolic gene families from 10 flowering plant (angiosperm) genomes. We then compared the metabolic fluxes for these families, predicted using the Arabidopsis thaliana and Sorghum bicolor metabolic networks, with the families' duplication propensities. For duplications produced by both small scale (small-scale duplications) and genome duplication (whole-genome duplications), there is a significant association between the flux and the tendency to duplicate. Following this global analysis, we made a more fine-scale study of the selective constraints observed on plant sodium and phosphate transporters. We find that the different duplication mechanisms give rise to differing selective constraints. However, the exact nature of this pattern varies between the gene families, and we argue that the duplication mechanism alone does not define a duplicated gene's subsequent evolutionary trajectory. Collectively, our results argue for the interplay of history, function, and selection in shaping the duplicate gene evolution in plants.
Keywords: dosage selection, genome duplication, gene duplication
Introduction
The contribution of gene duplication to evolution has long been a topic of interest (Taylor and Raes 2004), but in the last 10 years there has been a resurgence of interest in the varied fates of such duplications (Zhang et al. 2002; Kondrashov and Koonin 2004; Adams and Wendel 2005; Aury et al. 2006; Rodriguez et al. 2007; Barker et al. 2008; Liang et al. 2008; Ha et al. 2009; Innan and Kondrashov 2010; Ramsey 2011). Among those fates, the important roles played by genetic drift and simple changes in the gene “dosage” are increasingly appreciated. In several contributions, Lynch et al. have argued that the relatively small population sizes of multicellular eukaryotes could result in the fixation of many gene duplications through nonadaptive processes (Force et al. 1999; Lynch and Conery 2003; Lynch 2007). These processes, of course, still occur under the overall umbrella of natural selection. For instance, selection may act on gene dosage in one of two ways. First and most obviously, duplication of a gene may increase the rate of transcription and hence the translation of the encoded protein, increasing its abundance. We have previously referred to this possibility as a selection on “absolute” dosage (Bekaert et al. 2011). If a higher protein expression is selectively beneficial, we expect copy number polymorphisms will be fixed (Blanc and Wolfe 2004a; Kondrashov and Kondrashov 2006). The second possibility is that of a selection on the “relative dosage,” where an event affecting one of several genes that have coevolved together (i.e., a single gene duplication or differential paralog loss after polyploidy) introduces selective costs. This concept is known as the “dosage balance hypothesis” (Freeling 2009) and has been explored by a number of authors (Papp et al. 2003; Freeling and Thomas 2006; Birchler and Veitia 2007; Edger and Pires 2009). Here, we focus on the role of absolute dosage selection in determining duplicate fates.
As the first complete genome sequences became available, their patterns of gene duplication were explored to understand, among other questions, the role of natural selection in duplicate gene fixation (Lynch and Conery 2000; Gu et al. 2002; Wagner 2002; Gu et al. 2003). Those duplications had multiple origins, including whole-genome duplications (WGDs or polyploidy), as well as segmental, tandem, and retro-duplications (referred to here collectively as small-scale duplications or SSDs; Cannon et al. 2004; Thomas et al. 2006; Freeling 2009). The preponderance of polyploids among angiosperms (Wendel 2000) has led plant biologists to focus on understanding the patterns of the duplicate gene loss and the retention following WGD events (Bowers et al. 2003; Blanc and Wolfe 2004a; Blanc and Wolfe 2004b; De Bodt et al. 2005; Maere et al. 2005; Pfeil et al. 2005; Sterck et al. 2005; Cui et al. 2006; Freeling and Thomas 2006; Paterson et al. 2006; Schranz and Mitchell-Olds 2006; Town et al. 2006; Tuskan et al. 2006; Tang, Wang, et al. 2008; Barker et al. 2009; Edger and Pires 2009; Soltis et al. 2009; Wood et al. 2009; Duarte et al. 2010; Coate et al. 2011; Jiao et al. 2011; Schnable et al. 2011). In this work, we consider gene families with members derived from both SSD and WGD. These families are inferred from 10 angiosperm genomes: seven dicots (Arabidopsis, papaya, soybean, Medicago truncatula, poplar, peach, and grape) and three monocots (Brachypodium distachyon, rice, and sorghum).
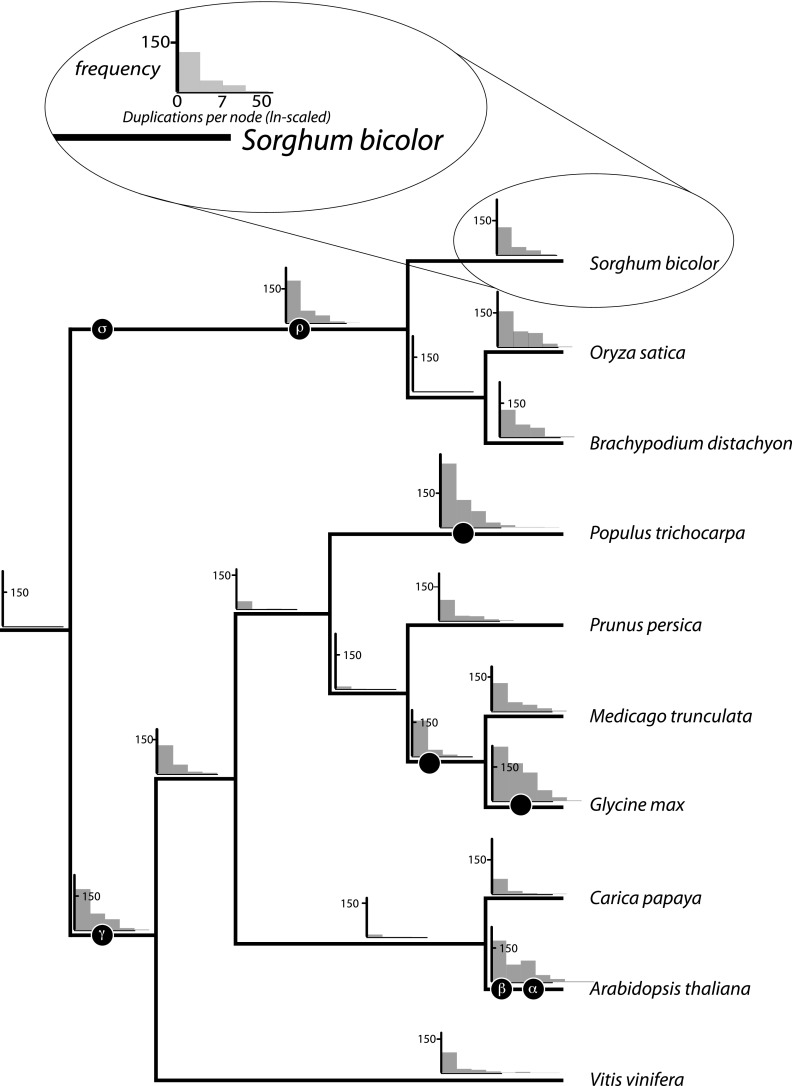
Of course, the taxa examined have a long history of polyploidy. Within the eudicots, the oldest genome duplication event, γ, was an ancient hexaploidy that characterizes the Rosidae (sensu Soltis et al. 2011), if not the core eudicots (Gunneridae sensu Jaillon et al. 2007; Lyons, Pedersen, Kane, Alam, et al. 2008; Lyons, Pedersen, Kane, Freeling, et al. 2008; Ming et al. 2008; Freeling 2009; Argout et al. 2011; Jiao et al. 2011; Shulaev et al. 2011; Soltis et al. 2011). Comparative genomics suggest that the lineage leading to poplar (Populus trichocarpa) underwent an additional WGD event, whereas that of the thale cress (Arabidopsis thaliana) had two: β and α. That these two duplications are independent is suggested by their absence in both grape (Vitis vinifera) and papaya (Carica papaya; fig. 1; Jaillon et al. 2007; Ming et al. 2008; Tang, Wang, et al. 2008; Freeling 2009). Analysis of the nonsynonymous substitution rates in the soybean (Glycine max) genome has revealed two WGD events post-WGD-γ: one shared with peanut (Arachis hypogaea), a basal legume, and a more recent soybean-specific duplication (Bertioli et al. 2009; Schmutz et al. 2010). The 3:1 ratio of grape to rice (Oryza sativa) genomic segments suggests that the γ paleohexaploidy is dicot-specific (Jaillon et al. 2007). However, cereal monocots also have a WGD event, ρ, basal to their radiation (Paterson et al. 2004); rice, sorghum (Sorghum bicolor) and purple false brome (B. distachyon) show no evidence of further WGD events (Throude et al. 2009; Vogel et al. 2010).
FIG. 1.—
Plant species used in reconciling gene trees. The phylogenetic relationships (branch lengths are arbitrary) among these species have been described previously (Moore et al. 2007; Paterson et al. 2009). The histograms depict the number of duplications per gene family. Thus, on the x-axes is the number of duplicates observed in a family at that node in the tree (on a natural log scale). The y-axes are then the frequency of families with that number of duplications. The scale is consistent across histograms. Black circles indicate whole-genome duplication events.
There is mounting evidence that the gene duplications created by WGD and by SSD differ in their ultimate fates (Seoighe and Wolfe 1999; Papp et al. 2003; Blanc and Wolfe 2004a, 2004b; Cannon et al. 2004; Aury et al. 2006; Thomas et al. 2006; Hakes et al. 2007; Conant and Wolfe 2008; Freeling 2008; Edger and Pires 2009; Freeling 2009; Coate et al. 2011). To cite just one example (relevant to this work), Maere et al. (2005) found that the ion transporters were overretained after WGD but underretained following SSD. The study of Arabidopsis WGDs by Blanc and Wolfe (2004a) reached similar conclusions but also found that genes involved in phosphate metabolism were significantly overretained following the recent WGD-α.
We are interested in whether the dosage effects are a strong predictor of duplicate retention, and here, we have taken both a “high level” phylogenomic approach and a “low-level” single-gene approach to look for evidence of such selection. Our first analysis extends our previous work in Arabidopsis, where we found an association between metabolic flux and some, but not all, of the Arabidopsis WGDs (Bekaert et al. 2011). Specifically, we hypothesize that genes in families with high flux will be, on average, over duplicated. Given that we have previously found significant differences in duplication propensity between cellular compartments (Bekaert et al. 2011; Hudson and Conant 2011), we also test for a relationship of duplicability and compartment. Additionally, we hypothesized that the WGD-produced and SSD-produced gene duplications will differ in their postduplication selective constraints. We evaluate this by narrowing our focus to a group of ion transporters. Such transporters have been found to have an outsized influence on the metabolic flux (Kacser and Burns 1981 notwithstanding; Brown et al. 1998; Pritchard and Kell 2002). Furthermore, their evolutionary behavior is distinct from other metabolic genes following both SSD and WGD (Lin and Li 2010; Bekaert and Conant 2011). Given the complexity of plant genome evolution, limiting our analysis to single gene families also has the advantage of allowing us to carefully distinguish WGD from SSD.
Materials and Methods
Estimation of Metabolic Flux
As previously described (Bekaert et al. 2011), we used the Systems Biology Research Tool v2.0.0 (Wright and Wagner 2008) to perform flux-balance analysis on the A. thaliana and S. bicolor metabolic networks (de Oliveira Dal'Molin et al. 2010a, 2010b). We estimated the maximal biomass production possible under photosynthetic conditions (a fixed level of photon import allowed, sugar imports forbidden) for both A. thaliana and S. bicolor networks (Pearson's correlation of flux r = 0.638, P < 10−15) and nonphotosynthetic conditions (photon import forbidden, fixed sugar imports allowed) for the A. thaliana network. Flux-balance analysis was also run by limiting the biomass and maximizing either photon import or sugar import (Bekaert et al. 2011), the results were similar and qualitatively the same. Because the distinctions between the two networks are in the photosynthetic reactions and because the sorghum metabolic network is derived from the Arabidopsis one, inclusion of the sorghum root data would be less informative and is hence omitted. In each case, we also made every possible reaction knockout whereby a given reaction's flux is constrained to zero and the remainder of the network is reoptimized. After knockout, all fluxes were normalized by the value of the biomass flux. Then, for each reaction, we selected the observed maximum flux, across all conditions. By doing so, we find what is essentially an upper bound on the flux of each reaction. It would obviously be desirable to also estimate the sensitivity of the network to changes in flux through each reaction. However, we do not have kinetic data for the entire network and these values cannot be estimated with flux-balance analysis. Instead, we compared this maximal flux to the duplication status of each reaction node. In cases where there was more than one flux value associated with a gene family, all possible flux values for that family were used in our association analyses, meaning that large gene families will not tend to be biased toward high flux because they encompass more reactions.
Gene Family Identification
We used the list of A. thaliana enzymes from the de Oliveira Dal'Molin et al. (2010a) metabolic network to identify enzyme gene families in the genomes of 10 flowering plants (fig. 1; A. thaliana, B. distachyon, C. papaya, G. max, M. trunculata, O. sativa, P. trichocarpa, Prunus persica, S. bicolor, and V. vinifera The Arabidopsis Genome Initiative 2000; Young et al. 2005; Ouyang et al. 2006; Tuskan et al. 2006; Jaillon et al. 2007; Ming et al. 2008; Paterson et al. 2009; Schmutz et al. 2010; The International Brachypodium Initiative 2010; Jung et al. 2009; Vogel et al. 2010). Homologous relationships were inferred using GenomeHistory (Conant and Wagner 2002), which calculated the nonsynonymous substitution rate (Ka) for all gene pairs with BLAST scores lower than 0.0001. Gene families were identified by single-linkage clustering with a cutoff in nonsynonymous divergence of Ka ≤ 0.20 for A. thaliana/A. thaliana comparisons and Ka ≤ 0.30 for all other comparisons (Powell et al. 2008). Gene pairs with Ka values below these thresholds were treated as nodes connected by an edge in the provisional gene family networks. These Ka parameters were selected after analyzing the results of using different Ka thresholds. For each threshold, we iteratively removed single edges from the provisional gene families. The chosen Ka thresholds were the largest values that did not cause a noticeable change in the constituency of the provisional gene families when any single edge was removed (data not shown). Families with fewer than four member genes were excluded. We used these gene families to associate a gene tree with each gene in the S. bicolor metabolic network.
Of the 138 pathways involved in Arabidopsis central metabolism, six contain no enzymes in the gene families we analyzed. Two of them (1,4-dichlorobenzene degradation and C21-steroid hormone metabolism) contain enzymes only present in Arabidopsis. The transport of α-d-glucose from the cytoplasm to the external cellular component contains a gene (AT5G18880), which is unclearly annotated. The transport of citrate and nitrate and the biosynthesis of monoterpenoid include four A. thaliana genes (citrate: AT1G02260, monoterpenoid: AT3G25830 and AT4G16730, and nitrate: AT5G14570) that our pipeline split into gene families that were too small to analyze phylogenomically.
Phylogenomics of Gene Families
Multiple sequence alignments of the protein sequences for each gene family were computed with MUSCLE v3.6 (Edgar 2004) using default parameters. Codon alignments were deduced from those alignments having 50 or more amino acids. We then inferred maximum likelihood gene trees using RAxML v7.0.4 (Stamatakis et al. 2008) with a general time-reversible model and discrete approximation of the gamma distribution (GTR + Γ). Confidence values were assigned to the gene trees from 100 bootstrap replicates. A relatively limited number of replicates were computed because we only wished to use these bootstrap statistics to identify nodes in the phylogeny with low support (<65%) prior to gene tree/species tree reconciliation. We thus reconciled all inferred gene trees with the species tree in figure 1 (Moore et al. 2007; Wang et al. 2009). To do so, we used NOTUNG v2.6 (Chen et al. 2000) to infer the most parsimonious pattern of the gene duplication and loss. Gene tree nodes with less than 65% bootstrap support were treated as polytomies and allowed to rearrange in order to minimize the number of duplications and/or losses (in practice, choosing support value thresholds between 50% and 80% produced similar results; data not shown). Using these parsimony reconstructions, we calculated the number of duplications (and number of duplications per species) for each gene tree.
Manual Annotation of Transporter Gene Trees
Coding sequences for the nine annotated PHT1s, one PHT2, three PHT3s, six PHT4s, and eight NHXs of A. thaliana were downloaded from TAIR (Swarbreck et al. 2008). A BLASTP search of the A. thaliana genome with these 19 and 8 sequences identified no further phosphate or sodium transporters in the genome. We then used BLASTP to search for ion transporter homologs in the genomes of papaya and poplar. We retained genes with BLAST E-values less than 10−20 as putative members of a given transporter family. Our homology estimation procedure always placed genes from C. papaya and P. trichocarpa into only a single A. thaliana transporter family. Gene trees were constructed as detailed above. In the case of the NHXs, one A. thaliana gene (At2g01980) aligned poorly with the other NHXs and was excluded from the alignment and gene tree.
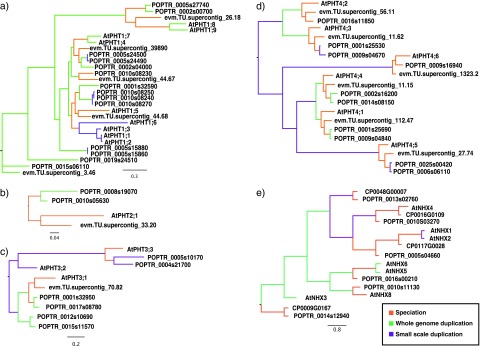
We manually assigned nodes in these phylogenies as either speciation or duplication events (fig. 2). Nodes connecting genes from the same species were labeled as duplication events where nodes connecting genes from different species were labeled speciation events. Because we were working with only a handful of genes, it was possible to make a more accurate distinction between SSD and WGD genes for these transporters than was possible for the genome-scale analyses. Thus, whole-genome duplicates were inferred in cases where the paralogs fit into distinct paralogous synteny blocks from the Plant Whole Genome Duplication Database (PGDD; Tang, Bowers, et al. 2008). Nodes connecting gene paralogs that could not be assigned using the PGDD were inferred to be SSDs (fig. 2). These manual duplication or speciation designations agreed with the automatic assessments of NOTUNG.
FIG. 2.—
Ion transporter gene trees used in this study. Branches demarcating speciation events are colored orange, whole-genome duplication events green, and non-WGDs purple. (a) High-affinity phosphate transporters AtPHT1; 1-AtPHT1; 9 with 16 P. trichocarpa and 7 C. papaya homologs. (b) Low-affinity phosphate transporters AtPHT2; 1 with 2 poplar and 1 papaya homologs. (c) Mitochondrial phosphate transporters AtPHT3; 1-ATPHT3; 3 with 6 poplar and 1 papaya homologs. (d) Chloroplast phosphate transporters AtPHT4; 1-AtPHT4; 6 with 10 poplar and 6 papaya homologs. (e) Sodium ion transporters AtNHX1-AtNHX6, AtNHX8 with 6 poplar and 4 papaya homologs.
Selective Constraint Following Speciation and Duplication Events in Five Families of Ion Transporters
The selective constraint (ratio of nonsynonymous substitutions to synonymous substitutions, i.e., Ka/Ks), for each gene tree was estimated by maximum likelihood under the MG/GY94 codon model (Goldman and Yang 1994; Muse and Gaut 1994): for details, see Conant et al. (2007). We tested three nested models of evolution: requiring all branches to have the same value of Ka/Ks (R_Null), allowing different values of Ka/Ks for branches following a speciation node from those following a duplication node (R_Dupl), and a model with differing values of Ka/Ks for branches following speciation, whole genome, and small-scale duplications (R_WGD). We compared these three models with nested likelihood ratio tests and evaluated statistical significance using the χ2 distribution, knowing that R_WGD has one more free parameter than R_Dupl, which in turn has one more parameter than R_Null.
Analysis of Constraints by Gene Ontology Slim Annotation
Gene ontology slim (GO Slim) annotations were obtained for each A. thaliana gene from TAIR. GO Slim categories were further condensed (supplementary table 1, Supplementary Material online) and transferred to our gene families. Spearman's rank correlations between the flux and number of duplications in each gene family were calculated in SAS (v9.2.1, Cary, NC) for all the cellular compartments and functions. Note that gene families could appear in more than one compartment or functional group. We applied a Bonferroni multiple-test correction equal to the number of either compartments or functional groups analyzed, resulting in the respective values of α, 0.0055 and 0.0042.
We also used the Wilcoxon rank test (SAS v9.2.1) to ask if the number of duplications per gene family differed for each cellular compartment or function as compared with the reminder of the genome. We used the same Bonferroni multiple-test corrections as previously.
Results
Computing Gene Families and Flux Values
We estimated the flux through each biochemical reaction in the Arabidopsis and sorghum metabolic networks using flux-balance analysis (Orth et al. 2010), maximizing the production of new cell mass for a fixed input of either light energy in both Arabidopsis and sorghum (in photosynthetic tissues) or carbohydrates for Arabidopsis (in nonphotosynthetic tissues, see Materials and Methods). We included the sorghum network to be sure that the differences in C3 and C4 photosynthesis were not greatly biasing our results.
Maximal flux values ranged from 0 to 3865120 (arbitrary flux-balance units) in the Arabidopsis leaf, from 0 to 6156740 in the Arabidopsis root, and from 0 to 2560860 in sorghum, when the biomass production is maximized and scaled to 1000 units. We then coupled those data to a set of cross-genome gene families identified from the 10 plant genomes (Materials and Methods). The result was a set of 735 gene families with associated metabolic fluxes. Of these 735 gene families, 463 have absolute flux values greater than zero. These families vary in size from 4 to 306 genes. The number of non–null-flux values associated with each family ranges from 1 to 13, with 90% having only one associated flux value and only three having 10 or more flux values. Those three families function as ATP synthases, phospholipid transporters, and cellulose synthases (functional Gene Ontology annotation from TAIR; Swarbreck et al. 2008). The number of gene duplications per family varies from 0 to 210, with a mean of 3.21 duplications per species. Reactions with no flux can result either from failure to include certain metabolites in the biomass reaction or from a reaction not being used in certain conditions. Because of the potential for error introduced by these two possibilities, we present our results both with and without null-flux reactions.
Correlation Between Number of Duplications and Maximum Metabolic Flux
The correlation between the number of duplications in a gene family and the maximal flux is positive and significant for both C3 and C4 model networks, whether or not null-flux reactions are included and whether duplications are calculated per species or per family (table 1).
Table 1.
Correlations Between Duplication and Flux by Gene Family
| All Flux Values | Excluding Null-Fluxa | |||
| rb | Pc | r | P | |
| Duplications per gene family | ||||
| All conditions | 0.245 | <10−15 | 0.336 | <10−15 |
| C3 leaves | 0.218 | <10−15 | 0.328 | <10−15 |
| C4 leaves | 0.176 | <10−8 | 0.218 | <10−4 |
| Roots | 0.223 | <10−15 | 0.359 | <10−15 |
| Duplications per species per gene familyd | ||||
| All conditions | 0.227 | <10−15 | 0.306 | <10−15 |
| C3 leaves | 0.203 | <10−15 | 0.272 | <10−14 |
| C4 leaves | 0.163 | <10−7 | 0.206 | <10−4 |
| Roots | 0.211 | <10−15 | 0.342 | <10−15 |
Flux values equaling 0 can have confounding biological and computational meanings.
Spearman's r.
Correlations and statistical significance calculated in R.
Number of duplication events per gene family divided by the number of species in that family.
Association of Flux and Duplication is neither Taxa nor Duplication-Mechanism Specific
As described, these species share a history of WGD (fig. 1). We summed the number of duplications on each branch in figure 1, separating those with lineage-specific WGDs from those without. Duplications in both groups are significantly and positively correlated with maximum flux (WGD: r = 0.111, P < 0.05; SSD: r = 0.094, P < 0.05). Of course, the branches containing WGDs will also have some background level of SSD, meaning that the duplications on these branches will not be exclusively due to WGD. However, the similarity in correlations seen between the two types of branch suggests that a more careful accounting of duplicates is unlikely to yield different results. Similarly, we found significant positive associations of duplication and flux for the monocot subtree as well as the eudicot tree with A. thaliana removed (P < 0.05). The similarity of the results for these subtrees implies that our results are not specific to Arabidopsis, even though one of the primary metabolic networks used is from this organism. Among the terminal nodes with rice and soybean show significant associations of flux and duplication after a Bonferroni multiple-testing correction (P < 0.00256). Unfortunately, for the remainder of the tip taxa, it is difficult to distinguish between the lack of an association and the lack of sufficient numbers of duplicates to discern if that association might exist. Similarly, the flux values inferred from the sorghum C4 leaves show a mixed pattern of associations and lack thereof depending on the precise data set used (0.1689 ≤ P ≤ 0.9653).
Association of Flux and Duplication Extends Across Compartments and Functional Annotations
Gene families were associated with GO Slim annotations (supplementary table 1, Supplementary Material online) for both cellular compartment and function. We found significant Spearman's correlations between the flux and duplication rate for the metabolic gene families from the chloroplast and mitochondria (table 2). Likewise, gene families that have a role in DNA or RNA binding or metabolism, hydrolase activity, and responses to stimuli or stress had significant correlations between the number of duplications and flux (table 3).
Table 2.
Duplication Status per Gene Family Split by Cellular Compartment
| Duplication versus Fluxa | Duplicationb | ||||
| Cellular Compartment | n | rc | P | Zd | P |
| Nucleus | 56 | 0.320 | 0.016 | 3.481 | 0.0005 |
| Cytosol | 74 | 0.133 | 0.258 | 4.910 | <0.0001 |
| Chloroplast and plastid | 273 | 0.275 | <0.0001 | −0.646 | 0.518 |
| Mitochondria | 134 | 0.434 | <0.0001 | 1.012 | 0.311 |
| Plasma membrane | 97 | 0.135 | 0.187 | 7.371 | <0.0001 |
| Endoplasmic reticulum | 44 | 0.304 | 0.045 | −1.161 | 0.246 |
| Golgi apparatus | 12 | 0.401 | 0.196 | 2.280 | 0.023 |
| Cell wall | 52 | 0.343 | 0.013 | 3.210 | 0.001 |
| Extracellular | 51 | 0.089 | 0.532 | 5.115 | <0.0001 |
Bold values are significant at a Bonferroni corrected α = 0.0055.
Duplications per gene family versus the maximum flux.
Wilcoxon rank test of difference across compartments (positive values: overduplication; negative values: underduplication).
Spearman's r, calculated in SAS (v9.2.2, Cary, NC).
Wilcoxon's Z, calculated in SAS (v9.2.2, Cary, NC).
Table 3.
Duplication Status per Gene Family Split by Functional Annotation
| Duplication versus Fluxa | Duplicationb | ||||
| Function | n | rc | P | Zd | P |
| Cell organization and biogenesis | 29 | 0.209 | 0.274 | 1.010 | 0.312 |
| Developmental processes | 20 | −0.132 | 0.578 | 1.693 | 0.090 |
| DNA or RNA binding or metabolism | 26 | 0.760 | <0.0001 | −2.284 | 0.022 |
| Electron transport | 7 | 0.860 | 0.013 | 0.460 | 0.645 |
| Hydrolase activity | 114 | 0.310 | <0.001 | −1.661 | 0.097 |
| Kinase activity | 62 | −0.031 | 0.817 | 1.167 | 0.243 |
| Nucleic acid or Nucleotide binding | 94 | 0.062 | 0.554 | 0.011 | 0.991 |
| Protein binding or metabolism | 121 | 0.139 | 0.128 | 1.463 | 0.143 |
| Signal transduction | 13 | 0.104 | 0.735 | 2.450 | 0.014 |
| Stimulus or stress response | 199 | 0.302 | <0.0001 | 2.650 | 0.008 |
| Transferase activity | 166 | 0.211 | 0.006 | −0.833 | 0.405 |
| Transporters or transport | 56 | −0.034 | 0.801 | 1.755 | 0.079 |
Bold values are significant at a Bonferroni corrected α = 0.0042.
Duplications per gene family versus the maximum flux.
Wilcoxon rank test of difference across compartments (positive values: overduplication; negative values: underduplication).
Spearman's r, calculated in SAS (v9.2.2, Cary, NC).
Wilcoxon's Z, calculated in SAS (v9.2.2, Cary, NC).
To determine whether duplication rates differed among compartments or classes, we used Wilcoxon rank-sum test (Z-scores in tables 2 and 3). Although gene families could appear in more than one annotation group, families located in the nucleus, cytosol, plasma membrane, cell wall, and extracellular space were significantly overduplicated compared with all other gene families (table 2). No functional categories were significantly overduplicated (table 3).
Selection on Sequence Evolution of Ion Transporters
We chose to analyze the ion transporters because of their interesting role as potential chokepoints. In the metabolic networks used in this analysis, the gene families representing transporters have significantly higher flux than nontransporter gene families (Mann–Whitney one-tailed P < 10−15). However, we found no significant correlation between the flux and duplicability among transporter gene families (P = 0.599). Therefore, we chose to look at the fine-scale differences in selection in two classes of ion transporters, phosphate and sodium. These elements have distinct roles in the growth and development of plants and hence potentially differing duplication dynamics. Phosphate transporters import an essential macronutrient, while sodium transporters primarily limit the import of potentially toxic sodium (Rausch and Bucher 2002; Kronzucker and Britto 2011). By narrowing our focus to just these 5 gene families and limiting ourselves to the three species (A. thaliana, P. trichocarpa, and C. papaya), it is possible to manually isolate SSD and WGD events. This inference in turn allows us to assess if the strength of selection differs following WGD, SSD, and speciation.
Phosphate Transporters
Phosphate transporters in A. thaliana are divided into four gene families. These families include the high-affinity transporters (PHT1; Mudge et al. 2002; Poirier and Bucher 2002), which import ions across the plasma membrane, and the mitochondrial (PHT3; Hamel et al. 2004) and chloroplast (PHT4; Guo et al. 2008) transporters, which act in their respective organelles. Finally, low-affinity (PHT2) phosphate transporters are also localized to the chloroplast (Versaw and Harrison 2002). We inferred gene phylogenies for the four phosphate transporter families and for one sodium transporter family (see Materials and Methods). Although the topology of phosphate transporter gene families is easily reconciled to the species tree, none of the clades contained the 4:2:1 ratio of A. thaliana to P. trichocarpa to C. papaya genes that would be expected if all transporters had been retained following the α, β, and P. trichocarpa–WGDs and no SSDs had been retained (fig. 2a and b). The average selective constraint (Ka/Ks) for PHT gene families varies considerably from 0.076 in high-affinity transporters to 0.207 in low-affinity transporters (table 4). The lowest Ka/Ks corresponds to the family with the largest observed number of duplications (high-affinity transporters: 19 duplications), whereas the highest Ka/Ks values correspond to the family with the fewest duplications (low-affinity transporters: 1 duplication). This observation is, however, without statistical significance. In all cases, the branches following gene duplications show significantly higher Ka/Ks than do those following speciation (table 4; but note that the small size of the low-affinity family limits the strength of our conclusion for that family). We also investigated selective constraints associated with duplication mechanism by dividing the branches following duplications into those due to WGD and to SSD. Here, the difference in selective constraint is less clear: for the high-affinity and chloroplast phosphate transporters, the Ka/Ks values for whole-genome duplicates are not significantly different than those for SSDs. Among the mitochondrial transporters, whole-genome duplicates have significantly higher Ka/Ks than small-scale duplicates, indicating a weaker selective constraint following WGD. The counts of WGDs versus SSDs per gene family are statistically uninformative (Fisher's Exact test: P = 0.75).
Table 4.
Selective Constraint Estimated with Three Models of Gene Evolution for Ion Transporters of A. thaliana, C. papaya, and P. trichocarpa
| PHT1–High-Affinity Phosphate Transporter | PHT2–Low-Affinity Phosphate Transporter | PHT3-Mitochondrial Phosphate Transporter | PHT4-Chloroplast Phosphate Transporter | NHX-Sodium Ion Transporter | |||||||
| Model | Branches | Ka/Ks | −lnL | Ka/Ks | −lnL | Ka/Ks | −lnL | Ka/Ks | −lnL | Ka/Ks | −lnL |
| R_Null | All | 0.076 | 0.207 | 0.114 | 0.148 | 0.049 | |||||
| 15379.5 | 3577.0 | 5898.3 | 24869.3 | 11210.1 | |||||||
| R_Dupl | Speciation | 0.063a | 0.156a | 0.080a | 0.123a | 0.062a | |||||
| Duplication | 0.082a | 0.415a | 0.133a | 0.249a | 0.031a | ||||||
| 15376.7 | 3570.2 | 5894.1 | 24847.0 | 11188.5 | |||||||
| R_WGD | Speciation | 0.063 | —b | 0.080a | 0.123 | 0.061a | |||||
| WGDc | 0.081 | — | 0.190a | 0.233 | 0.067a | ||||||
| SSDd | 0.085 | — | 0.112a | 0.265 | 0.018a | ||||||
| 15376.6 | — | 5891.2 | 24846.7 | 11175.3 | |||||||
Bold values indicate a significant improvement over the model immediately above at P < 0.05; nested likelihood ratio test (distributed χ2, P < 0.05, degrees of freedom = 1).
No small scale duplications in PHT2, so model R_Dupl is equivalent to model R_WGD.
WGD: determined by syntenic paralogy using the Plant Genome Duplication Database (Tang, Bowers, et al. 2008).
SSD: determined either by a lack of syntenic paralogy and/or by tandem duplication status.
Sodium Transporters
The angiosperm sodium ion transporters (NHX) are a single gene family responsible for keeping Na+ concentrations at nontoxic levels (Rodríguez-Rosales et al. 2008). The sodium ion transporters have a lower average Ka/Ks than do any of the phosphate transporter families (0.049 versus 0.076–0.207). Curiously, among these transporters, paralogs have significantly lower Ka/Ks values than do orthologs, indicating no release in a selective constraint after duplication (table 4). Genes duplicated by WGD seem to be under slightly less selective constraint than gene orthologs; however, SSDs seem to be under considerably higher selective constraint than either.
Discussion
Selection on Plant Gene Duplications
Although it has been hypothesized that a substantial fraction of the surviving duplicate genes in the genomes of multicellular eukaryotes might be due to the neutral fixation of duplicates (Lynch and Conery 2003), other potential forces could also be involved (Kondrashov and Kondrashov 2006; Innan and Kondrashov 2010). Here, we have taken both a low-level and a high-level approach to look for evidence of selection in the process of gene and genome duplications in the plants.
Selection, Sequence Evolution, and Ion Transporters
Part of our analysis focused on sequence evolution in two families of ion transporters. Transporters sometimes appear to be the limiting step in metabolic pathways (Brown et al. 1998; Pritchard and Kell 2002), a fact that may partly explain why their evolution after both SSD and WGD is distinct from other metabolic genes (Lin and Li 2010; Bekaert and Conant 2011). Limiting our analysis to single gene families also allows us to carefully distinguish WGD and SSD events and to model the selective constraints acting on these genes.
There are two primary hypotheses regarding the expected changes in selective constraint following gene duplication. Predominant and recent neo-functionalization would predict Ka/Ks> 1.0 (Zhang et al. 2003; Hahn 2009). On the other hand, subfunctionalization (and likely neutral retention by drift) would suggest that Ka/Ks is elevated after duplication but not above 1.0 (Hughes 1994; Zhang et al. 1998; Force et al. 1999; Lynch and Conery 2000). Importantly, both models predict an elevated value of Ka/Ks after duplication; however, evidence for such increases is mixed. Hughes and Hughes (1993) found no evidence for the relaxation of selective constraint among 17 genes in the tetraploid frog Xenopus laevis. Kondrashov et al. (2002) found that recent paralogs were under significantly lower selective constraints than orthologs, whereas others (Lynch and Conery 2000; Kondrashov et al. 2002; Zhang et al. 2003; Jordan et al. 2004) have found evidence for a decrease in selective constraint immediately following duplication. This relaxation appears to be temporary; Jordan et al. (2004) found that the average strength of purifying selection acting on old duplicates was higher than for nonduplicated genes. This observation presumably reflects the situation after the fate determining mutation, which breaks the selective symmetry of two duplicates and sends them down differing paths (Innan and Kondrashov 2010). Among PHTs, our results parallel those of Jordan et al. (2004) in finding a general relaxation of selective constraint after ion transporter duplication. This result is not supported among the NHX transporters. This difference may be due to the limited evolutionary paths opened by a duplication of sodium transporters compared with that of phosphate transporters (Kronzucker and Britto 2011).
We also extended our analysis to differences in constraint between SSD- and WGD-produced duplicates. We had no a priori hypothesis on which mechanism would impart higher selective constraint, and, in fact, we found both possible outcomes.
Associations Between Duplication Propensity and Metabolic Flux
We also made a large-scale analysis of the patterns of evolution in the metabolic network. To our knowledge, this analysis represents the first high-level phylogenomic–scale study of gene duplication and metabolism in angiosperms (for studies of metabolism following WGD in other organisms, see Gout et al. 2009; van Hoek and Hogeweg 2009). By focusing on metabolism, we can ask whether duplications are randomly distributed across the network (as might be expected if drift were the only force at work) or show biases in the patterns of fixation. Notably, we find that there is a statistically significant relationship between duplication propensity and each enzyme's predicted flux. This analysis follows our work on absolute and relative dosage among the Arabidopsis WGD duplicates (Bekaert et al. 2011), where we found that reactions with high flux were enriched for enzymes coded by duplicate genes produced by the ancient β event (but not the more recent α event). Here, we have shown that the relationship between the flux and duplication is not specific to Arabidopsis but a more general pattern in plants. Although it is certainly not the case that all gene duplications are associated with high-flux reactions (the association magnitudes found are small), selection for increased gene dosage (Kondrashov and Kondrashov 2006; Conant and Wolfe 2008) is an attractive explanation for the fixation of some of these duplicates. In fact, examples of plant duplications apparently fixed by such selection are well known (van Hoof et al. 2001; Widholm et al. 2001).
Because the association of flux with duplication holds for both SSD and WGD events, we propose that different types of selective environment favor dosage-based duplicates produced by the two mechanisms. Thus, SSD may be useful in situations where the increased dosage would be beneficial at the tips of a pathway or in secondary metabolism: this is likely the case for the copper tolerance duplication in bladder campion (Silene vulgaris; van Hoof et al. 2001). However, as Kacser and Burns (1981) pointed out, for most metabolic pathways, it is unlikely that a single reaction is flux limiting, meaning that a single gene duplication is unlikely to alter the flux in such a pathway. WGD is a potential route to increased flux in such situations, and it appears that such selection may have occurred after a WGD in the ancestor of bakers' yeast (Saccharomyces cerevisiae; Conant and Wolfe 2007; Merico et al. 2007; van Hoek and Hogeweg 2009).
Taking these analyses to the subcellular level, we find strong correlations between flux and duplication in the mitochondria and chloroplast, but not in the cytosol. This result suggests that the general association between flux and duplication is primarily driven by reactions in these compartments, an unsurprising conclusion given the roles of the chloroplast and the mitochondria as the plant cell's anabolic and energy-yielding centers. These patterns also accord well with our prior analyses of the compartmental evolution in the Arabidopsis and human metabolic networks (Bekaert et al. 2011; Hudson and Conant 2011).
Gene and Genome Duplication, Selection, and Contingency
Although a WGD that occurs in a particular individual is much less likely to be selectively neutral than an SSD (Vieta 2005), it does not follow that there should be strong selection at every locus duplicated in such an event. Although it might therefore appear that WGD produces a large class of duplicate genes that evolve more or less neutrally after WGD, this hypothesis is difficult to reconcile with observations such as the dosage balance hypothesis. To distinguish between these two hypotheses, one might consider what the sources of variation in the selective constraint are for the set of WGD-produced duplicate genes in a genome. In fact, the number of sources of variation in constraint among duplicates at large (Duret and Mouchiroud 2000; Pál et al. 2003; Drummond et al. 2006; Vitkup et al. 2006) suggests the importance of “contingency” in duplicate evolution. In other words, a duplicate's fate will depend on both its intrinsic properties (including factors studied here, such as function, cellular compartment, and duplication mechanism) as well as the environment in which it finds itself at birth.
Supplementary Material
Supplementary table 1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank Patrick Edger and our molecular evolution course for helpful discussions. This work was supported by the National Library of Medicine Biomedical and Health Informatics Training Fellowship (LM007089-19) to C.M.H., the University of Missouri Life Sciences Fellowship to E.E.P., the US National Science Foundation (DBI 0501712 and DBI 0638536) to J.C.P., and the Reproductive Biology Group of the Food for the 21st Century program at the University of Missouri to G.C.C.
References
- Adams K, Wendel J. Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics. 2005;171:2139–2142. doi: 10.1534/genetics.105.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. [DOI] [PubMed]
- Argout X, et al. The genome of Theobroma cacao. Nat Genet. 2011;43:101–108. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- Aury J-M, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Barker MS, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Conant GC. Copy number alterations among mammalian enzymes cluster in the metabolic network. Mol Biol Evol. 2011;28:1111–1121. doi: 10.1093/molbev/msq296. [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative followed by absolute dosage constraints. Plant Cell. 2011;23:1–10. doi: 10.1105/tpc.110.081281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli DJ, et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics. 2009;10:45. doi: 10.1186/1471-2164-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004a;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004b;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J, Chapman B, Rong J, Paterson A. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose-transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- Cannon S, Mitra A, Baumgarten A, Young N, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 2000;3:429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- Coate J, Schlueter J, Whaley A, Doyle J. Comparative evolution of photosynthetic genes in response to polyploid and nonpolyploid duplication. Plant Physiol. 2011;155:2081–2095. doi: 10.1104/pp.110.169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wagner A. GenomeHistory: a software tool and its application to fully sequenced genomes. Nucleic Acids Res. 2002;30:3378–3386. doi: 10.1093/nar/gkf449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wagner A, Stadler PF. Modeling amino acid substitution patterns in orthologous and paralogous genes. Mol Phylogenet Evol. 2007;42:298–307. doi: 10.1016/j.ympev.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Biol Evol. 2007;3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- de Oliveira Dal'Molin CG, Quek L-E, Palfreyman RW, Brumbley SM, Nielsen LK. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 2010a;152:579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal'Molin CG, Quek L-E, Palfreyman RW, Brumbley SM, Nielsen LK. C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol. 2010b;154:1871–1885. doi: 10.1104/pp.110.166488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D, Raval A, Wilke C. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- Duarte J, et al. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 2010;10:61. doi: 10.1186/1471-2148-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–85. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17:699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. The evolutionary position of subfunctionalization, downgraded. Genome Dyn. 2008;4:25–40. doi: 10.1159/000126004. [DOI] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Ann Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Gout J-F, Duret L, Kahn D. Differential retention of metabolic genes following whole-genome duplication. Mol Biol Evol. 2009;26:1067–1072. doi: 10.1093/molbev/msp026. [DOI] [PubMed] [Google Scholar]
- Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- Gu Z, et al. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Guo B, et al. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 2008;177:889–898. doi: 10.1111/j.1469-8137.2007.02331.x. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim E-D, Chen ZJ. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc Natl Acad Sci U S A. 2009;106:2295–2300. doi: 10.1073/pnas.0807350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Hakes L, Pinney J, Lovell S, Oliver S, Robertson D. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel P, et al. Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol Microbiol. 2004;51:307–317. doi: 10.1046/j.1365-2958.2003.03810.x. [DOI] [PubMed] [Google Scholar]
- Hudson C, Conant GC. Expression level, cellular compartment and metabolic network position all influence the average selective constraint on mammalian enzymes. BMC Evol Biol. 2011;11:89. doi: 10.1186/1471-2148-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. The evolution of functionally novel proteins after gene duplication. Proc R Soc B Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Hughes M, Hughes A. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol Biol Evol. 1993;10:1360–1369. doi: 10.1093/oxfordjournals.molbev.a040080. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768. [DOI] [PubMed]
- Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:U463–U465. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jordan I, Wolf Y, Koonin E. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, et al. 2009. Synteny of Prunus and other model plant species. BMC Genomic. 10:76. [DOI] [PMC free article] [PubMed]
- Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F, Rogozin I, Wolf Y, Koonin E. Selection on the evolution of gene duplicates. Genome Biol. 2002;3:research0008.1–research0008.9. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Kondrashov AS. Role of selection in fixation of gene duplications. J Theor Biol. 2006;239:141–151. doi: 10.1016/j.jtbi.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–290. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kronzucker HJ, Britto DT. 2011. Sodium transport in plants: a critical review. New Phytologist 189:54–81. [DOI] [PubMed]
- Liang H, Plazonic KR, Chen J, Li W-H, Fernandez A. Protein under-wrapping causes dosage sensitivity and decreases gene duplicability. PLoS Genet. 2008;4:e11. doi: 10.1371/journal.pgen.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Li WH. Expansion of hexose transporter genes was associated with the evolution of aerobic fermentation in yeasts. Mol Biol Evol. 2010;28:131–142. doi: 10.1093/molbev/msq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1154. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary demography of duplicate genes. J Struct Funct Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- Lyons E, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Freeling M. The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop Plant Biol. 2008;1:181–190. [Google Scholar]
- Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico A, Sulo P, Pi∫kur J, Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Ming R, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Bell C, Soltis P, Soltis D. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci U S A. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002;31:341–353. doi: 10.1046/j.1365-313x.2002.01356.x. [DOI] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Orth J, Thiele I, Palsson B. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2006;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst L. Rate of evolution and gene dispensability. Nature. 2003;421:496–497. doi: 10.1038/421496b. [DOI] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Paterson A, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, et al. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006;22:597–602. doi: 10.1016/j.tig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ. Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol. 2005;54:441–454. doi: 10.1080/10635150590945359. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Bucher M. Phosphate transport and homeostasis in Arabidopsis. In: Somerville C, Meyerowitz EM, editors. The Arabidopsis book. Rockville (MD): American Society of Plant Biologists; 2002. pp. 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A, Conant GC, Brown D, Carbone I, Dean R. Altered patterns of gene duplication and differential gene gain and loss in fungal pathogens. BMC Genomics. 2008;9:147. doi: 10.1186/1471-2164-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L, Kell DB. Schemes of flux control in a model of Saccharomyces cerevisiae glycolysis. Euro J Biochem. 2002;269:3894–3904. doi: 10.1046/j.1432-1033.2002.03055.x. [DOI] [PubMed] [Google Scholar]
- Ramsey J. Polyploidy and ecological adaptation in wild yarrow. Proc Natl Acad Sci U S A. 2011;108:7096–7101. doi: 10.1073/pnas.1016631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C, Bucher, M. 2002. Molecular mechanisms of phosphate transport in plants. Planta 216:23–37. [DOI] [PubMed]
- Rodriguez MA, Vermaak D, Bayes JJ, Malik HS. Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proc Natl Acad Sci U S A. 2007;104:15412–15417. doi: 10.1073/pnas.0707445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rosales MP, et al. Plant NHX cation/proton antiporters. Plant Signal Behav. 2008;4:265–276. doi: 10.4161/psb.4.4.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schnable J, Pedersen B, Subramaniam S, Freeling M. Dose–sensitivity, conserved non-coding sequences, and duplicate gene retention through multiple tetraploidies in the grasses. Frontiers Plant Sci. 2011;2:2. doi: 10.3389/fpls.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH. Yeast genome evolution in the post-genome era. Curr Opin Microbiol. 1999;2:548–554. doi: 10.1016/s1369-5274(99)00015-6. [DOI] [PubMed] [Google Scholar]
- Shulaev V, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soltis DE, et al. Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A fast bootstrapping algorithm for the RAxML web-servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Sterck L, et al. EST data suggest that poplar is an ancient polyploid. New Phytol. 2005;167:165–170. doi: 10.1111/j.1469-8137.2005.01378.x. [DOI] [PubMed] [Google Scholar]
- Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tang H, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Ann Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Thomas B, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throude M, et al. Structure and expression analysis of rice paleo duplications. Nucleic Acids Res. 2009;37:1248–1259. doi: 10.1093/nar/gkn1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town C, et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- van Hoek MJ, Hogeweg P. Metabolic adaptation after whole genome duplication. Mol Biol Evol. 2009;26:2441–2453. doi: 10.1093/molbev/msp160. [DOI] [PubMed] [Google Scholar]
- van Hoof NA, et al. Enhanced copper tolerance in Silene vulgaris (Moench) Garcke populations from copper mines is associated with increased transcript levels of a 2b-type metallothionein gene. Plant Physiol. 2001;126:1519–1526. doi: 10.1104/pp.126.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ. A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell. 2002;14:1751–1766. doi: 10.1105/tpc.002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieta R. Paralogs in polypoids: one for all and all for one? Plant Cell. 2005;17:4–11. doi: 10.1105/tpc.104.170130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitkup D, Kharchenko P, Wagner A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 2006;7:R39. doi: 10.1186/gb-2006-7-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Wagner A. Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol. 2002;19:1760–1768. doi: 10.1093/oxfordjournals.molbev.a003998. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci U S A. 2009;10:3853–3858. doi: 10.1073/pnas.0813376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- Widholm JM, et al. Glyphosate selection of gene amplification in suspension cultures of 3 plant species. Physiol Plant. 2001;112:540–545. doi: 10.1034/j.1399-3054.2001.1120411.x. [DOI] [PubMed] [Google Scholar]
- Wood T, et al. The frequency of polyploid speciation in vascular plants. Mol Biol Evol. 2009;19:1464–1473. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J, Wagner A. The Systems Biology Research Tool: evolvable open-source software. BMC Syst Biol. 2008;2:55. doi: 10.1186/1752-0509-2-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, et al. Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol. 2005;137:1174–1181. doi: 10.1104/pp.104.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gu Z, Li WH. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 2003;4:R56. doi: 10.1186/gb-2003-4-9-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenberg H, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Vision TJ, Gaut BS. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol Biol Evol. 2002;19:1464–1473. doi: 10.1093/oxfordjournals.molbev.a004209. [DOI] [PubMed] [Google Scholar]