Abstract
Seed dormancy has been associated with red grain color in cereal crops for a century. The association was linked to qSD7-1/qPC7, a cluster of quantitative trait loci for seed dormancy/pericarp color in weedy red rice. This research delimited qSD7-1/qPC7 to the Os07g11020 or Rc locus encoding a basic helix-loop-helix family transcription factor by intragenic recombinants and provided unambiguous evidence that the association arises from pleiotropy. The pleiotropic gene expressed in early developing seeds promoted expression of key genes for biosynthesis of abscisic acid (ABA), resulting in an increase in accumulation of the dormancy-inducing hormone; activated a conserved network of eight genes for flavonoid biosynthesis to produce the pigments in the lower epidermal cells of the pericarp tissue; and enhanced seed weight. Thus, the pleiotropic locus most likely controls the dormancy and pigment traits by regulating ABA and flavonoid biosynthetic pathways, respectively. The dormancy effect could be eliminated by a heat treatment, but could not be completely overcome by gibberellic acid or physical removal of the seed maternal tissues. The dormancy-enhancing alleles differentiated into two groups basically associated with tropical and temperate ecotypes of weedy rice. Of the pleiotropic effects, seed dormancy could contribute most to the weed adaptation. Pleiotropy prevents the use of the dormancy gene to improve resistance of white pericarp cultivars against pre-harvest sprouting through conventional breeding approaches.
SEEDS acquire primary dormancy during development to enhance adaptation of wild species to diverse environments by distributing germination over time and space. Domestication tends to reduce dormancy by selection for rapid, uniform germination (Harlan et al. 1973). Differentiation in seed dormancy between cereal crops and wild relatives has been associated with seed morphologies (Nilsson-Ehle 1914; Johnson 1935) and quantitative trait loci (QTL). Cloning of validated dormancy loci provides in-depth insights into regulatory mechanisms underlying natural variation in this adaptive or domestication-related trait (Bentsink et al. 2006; Sugimoto et al. 2010).
Weedy rice refers to Oryza spp., which competes with cultivated rice (Oryza sativa L. and O. glaberrima Steud.) from tropical to temperate areas (Oka 1988; Delouche et al. 2007). The most persistent type of weedy rice is red rice, which is characterized by a red pericarp color. Red rice has strong seed dormancy (Cohn and Hughes 1981; Noldin et al. 2006). Genetic analysis has associated pericarp color with seed dormancy in red rice (Gu et al. 2005a).This association was first reported for wheat (Triticum aestivum L.), where red grain genotypes were more dormant than the white ones, and this morphology has been used to select cultivars for resistance to pre-harvest sprouting (Nilsson-Ehle 1914; Flintham 2000). However, it remains unknown if the association in rice, wheat, and other crops arises from a tight linkage between genes for these two traits or from pleiotropy.
Genetic control of red grain color involves the homoelogous R1–R3 loci in wheat (Flintham 2000) and the Rc and Rd loci in rice (Kinoshita 1984). Rc on chromosome 7 and Rd on chromosome 1 encode a basic helix-loop-helix (bHLH) family transcription factor and a dehydroflavonol-4 reductase (DFR), respectively (Sweeney et al. 2006; Furukawa et al. 2007). However, several groups failed to detect a dormancy locus from the Rc or Rd region in wild or cultivated rice (Lin et al. 1998; Cai and Morishima 2000; Miura et al. 2002; Thomson et al. 2003; Lee et al. 2005).We mapped a cluster of QTL for seed dormancy (qSD7-1) and pericarp color (qPC7) on the short arm of chromosome 7 in weedy red rice (Gu et al. 2005b). The QTL-containing genomic segment was introduced from weedy into cultivated rice to facilitate cloning and characterization of the dormancy gene (Gu et al. 2006). Here we delimit the clustered QTL to a single locus and characterize the dormancy gene for additional effects, downstream gene networks for abscisic acid (ABA) and flavonoid biosynthesis, and allelic differentiation in weedy rice.
Materials and Methods
Plant materials and cultivation
An introgression line, ILSD7-1/PC7, was selected from the advanced backcross population segregating only for a qSD7-1/qPC7-containing segment introduced from SS18-2 in the EM93-1 genetic background (Gu et al. 2006). The donor parent SS18-2 is a line of weedy red rice, and the recipient parent EM93-1 is a white pericarp line of cultivated rice (O. sativa subsp. indica). The EM93-1 background for the QTL-containing region delimited by RM6338 and RM8006 (Figure 1A) was determined with 140 markers relatively evenly distributed on the framework linkage map (Gu et al. 2004). A cross was made between ILSD7-1/PC7 and EM93-1 to genetically dissect qSD7-1 from qPC7 using the breeding scheme step by step from the F2 to F6 generation (see supporting information, Figure S1 for breeding scheme and population sizes). About 6300 F2 plants were grown in an isolated field plot to harvest fully mature seeds. A single seed (F3) was pooled from each of the F2 plants to form F3 subpopulations of red or white pericarp seeds. The F3–F6 populations were grown in a greenhouse to identify recombinants or isogenic lines for the dormancy-enhancing (SD7-1D) or -reducing (SD7-1d) allele of the qSD7-1 underlying gene SD7-1.
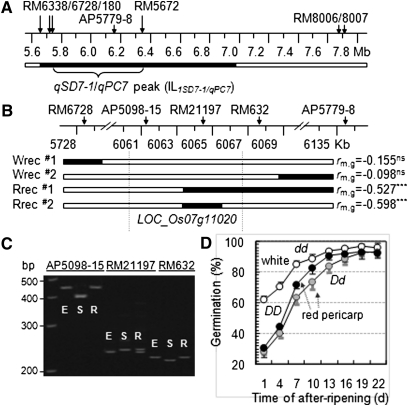
Figure 1 .
Fine mapping of the qSD7-1/qPC7 region. (A) Physical map of the QTL peak-containing region. ILqSD7-1/qPC7 is the introgression line with only one chromosomal segment from the weedy rice SS18-2 (solid bar) in the background of the cultivated rice EM93-1 (open bars). (B) Graphic representation of recombinants selected for progeny testing. Each recombinant was identified as a plant heterozygous for an SS18-2-derived subsegment (solid bar) and produced white (Wrec) or red (Rrec) pericarp-colored seeds. rm,g is the correlation coefficient between genotypes of the marker on the subsegment and germination values in the recombinant-derived progeny population (see Figure S1 for population pedigree and sizes), with a negative value indicating that the SS18-2-derived allele reduces germination rate and the superscript indicating that the correlation was not significant (ns) or significant at P < 0.0001 (***). Vertical lines delimit the Os07g11020 locus underlying the QTL cluster. (C) Gel image showing marker (AP5098-15, RM21197, and RM632) genotypes of the parental lines EM93-1 (E) and SS18-2 (S) and the recombinant Rrec#2 (R). (D) Genotypic difference in seed dormancy and pericarp color. The duration of dormancy was evaluated by germination of seeds after-ripened for 1–22 d. Data shown are genotypic means (circles) and SD of 15 plants selected from the (B) intragenic recombinant Rrec#2-derived progeny population. The genotypes homozygous for the dormancy-enhancing allele (DD, solid circles) or heterozygous (Dd, shaded circles) at SD7-1 displayed red pericarp color, and the genotype homozygous for the dormancy-reducing allele at SD7-1 exhibited white pericarp color (dd, open circles). Germination profiles for individual after-ripening periods are presented in Figure S2.
Marker genotyping and recombinant identification
New markers (Table S1) were developed on the basis of the genome sequence for Nipponbare (O. sativa subsp. japonica; International Rice Genome Sequencing Project 2005) for the contig corresponding to the QTL peak-containing region. F3–F6 seedlings were determined for marker genotypes using previously described methods (Gu et al. 2004). Recombinant genotypes between the markers were transplanted into pots to harvest seeds for dormancy assay by marker-assisted progeny testing. Seeds were air-dried for 3 d and then stored at −20° before dormancy assay by germination.
Phenotypic identification
Seed dormancy:
The degree of dormancy was measured by the percentage of germination. Prior to germination, seed samples from individual plants were after-ripened (warm-dried at 24°–25°) for 1–21 d, depending on experiments or populations. Three replicates of ∼50 seeds each were germinated at 30° using the previously described methods (Gu et al. 2004). For progeny testing, ∼100 marker-genotyped plants from a recombinant-derived population were evaluated, and a significant marker–germination correlation was used to indicate the presence of a dormancy-enhancing allele in the recombinant. The SD7-1D and SD7-1d isogenic lines were used to estimate genotypic responses to dormancy-breaking treatments: intact seeds heating at 45° for 7 d or incubated with buffer containing 1 or 3 mM gibberellic acid [GA3 (Acros Organics), a germination-promoting hormone commonly used in seed biology experiments], hull-removed seeds (i.e., caryopses), and pericarp/testa-scraped caryopses prepared using the method described in Gu et al. (2003).
Pericarp color:
Pericarp color for individual plants was visually scored as red (brown for transgenic lines) or white. Red and white pericarp genotypes were coded as 1 and 0, respectively, to correlate the pigment with the dormancy trait in a segregating population.
Seed moisture:
Difference in moisture content between red and white pericarp genotypes was determined for freshly harvested and drying seeds. Ten plants that flowered on the same day from each of SD7-1D and SD7-1d were harvested at 40 d after flowering. One hundred fully developed seeds from each plant were weighed immediately after harvesting, air-dried at 24°, and weighed every 24 h. After 11 d, these samples were dried at 105° for 72 h to measure dry weight to calculate moisture contents. This initial experiment detected a significant difference in 100-seed weight between the isogenic lines. Therefore, these lines were grown in the following two seasons to confirm the genotypic effect on seed weight, which was estimated as the percentage of (SD7-1D-SD7-1d)/SD7-1d in 100-seed weight.
Abscisic acid:
The plant hormone ABA is a key signaling molecule for dormancy induction (Finkelstein et al. 2008). Thus, ABA content was measured for SD7-1D and SD7-1d developing seeds sampled (∼200 spikelets each) at 0 (control), 10, 20, and 30 d post anthesis (DPA). Sample preparation and ABA detection were conducted using the methods described in Destefano-Beltran et al. (2006). 2H6-ABA (50 ng) was used as the internal standard. ABA was quantified using a high performance liquid chromatography-mass spectrometer system. Ions monitored were 263 for ABA and 269 for the internal standard. This experiment was replicated three times.
Genomic DNA isolation and sequence analysis
DNA segments covering SD7-1 and its flanking regions were amplified from SS18-2, EM93-1, and SD7-1D by PCR. Primers targeting 10 overlapping fragments were designed on the basis of the reference genome (International Rice Genome Sequencing Project 2005). PCR products were cloned into pGEM T-Vector (Promega) and sequenced from both directions. Sequences were aligned and analyzed using the Lasergene 7 Software Suite.
cDNA isolation and sequence analysis
RNA samples of SD7-1D and SD7-1d were prepared from 10-d developing seeds or expanding leaf tissue using the method of Chang et al. (1993) and reverse-transcribed using the SuperScript First-Strand Synthesis Kit (Invitrogen). Primers (Table S1) for the reverse transcription and subsequent PCR were designed on the basis of the SD7-1 genomic DNA sequence from SD7-1D to amplify the two overlapping cDNA fragments and full-length cDNAs (Figure 2A). PCR products were cloned as described above and sequenced. Protein sequences deduced on the basis of the full-length cDNAs were annotated with the Pfam protein family database (Finn et al. 2010).
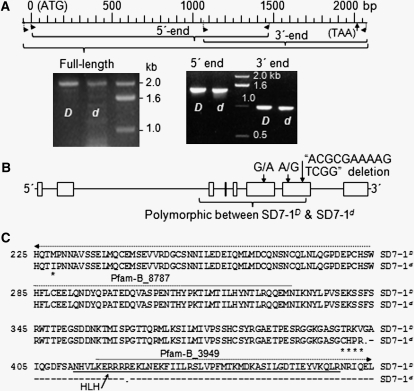
Figure 2 .
cDNAs and allelic variation of the SD7-1 locus. (A) cDNA fragments from SD7-1D and SD7-1d. Images show cDNA clones for the dormancy-enhancing (D) and -reducing (d) alleles. The scale shows positions of the PCR primers and the start (ATG) and stop (TAA) codons on the cDNA sequence of the D allele. (B) Gene structure. Exons (boxes) and introns (line fragments) were predicted by aligning the genomic DNA against the full-length cDNA sequence for the D allele. Arrows indicate variations in cDNA sequence between D and d. (C) Protein sequences. Amino acid sequences were deduced on the basis of the D and d cDNA sequences. Solid/dotted lines indicate the Pfam-A (HLH)/-B matches, respectively, predicted with the Pfam protein family database (Finn et al. 2010) for allele D. Asterisks/dashes indicate changed amino acid residuals located before/after, respectively, the first predicted stop codon (dot) in allele d.
Complementary test of Rc for seed dormancy
The Rck transgenic T1 line Rc3-#16, developed by transformation of Nipponbare with the Rc genomic DNA from the indica cultivar Kasalath (Furukawa et al. 2007), was advanced to the T3 generation to improve seed set and to select stable transgenic lines. Selected T3 lines differed in pericarp color (brown vs. white). However, the difference in mean germination rate between the T3 brown (64%) and white (72%) pericarp lines was not significant (P > 0.05). Thus, the Rck transgene was introduced into the EM93-1 background by three generations of recurrent backcrossing combined with phenotypic selection. Seeds from a selected BC3F1 plant were developed into the BC3F2 population to test for association between the pigment and dormancy traits.
Phylogenetic analysis of SD7-1/Rc alleles
The alleles from additional red pericarp (RcRc) genotypes were sequenced to determine if their differentiation affects seed dormancy. Seven lines of red weedy or cultivated (landrace) rice, which were evaluated for seed dormancy in the previous research (Gu et al. 2005a), and two lines of wild rice (O. rufipogon) were selected to represent different geographic origins. Genomic DNAs for the SD7-1/Rc alleles isolated by PCR were directly sequenced, and sequences were analyzed using the methods described above. Phylogenetic analyses for these and previously sequenced alleles were conducted using the MEGA4 software (Tamura et al. 2007).
Real-time quantitative reverse transcription PCR
Transcriptional levels of SD7-1 and genes predicted for ABA and flavonoid biosynthesis were quantified in SD7-1D and SD7-1d developing caryopses to infer regulatory mechanisms of seed dormancy and red pericarp color. Individual spikelets from SD7-1D and SD7-1d plants were marked for the date of anthesis at the peak time of flowering. About 60 spikelets harvested at 0 (control), 5, 10, 15, 20, 25, and 30 DPA were used to prepare caryopses for RNA extraction. Each sample of 4 μg RNA was used to prepare cDNA. PCR primers were designed on the basis of the reference genome (International Rice Genome Sequencing Project 2005) to target open reading frames of predicted ABA and flavonoid biosynthesis genes (Table S1). The genes encoding RNA II polymerase and GTP-binding protein were selected as controls on the basis of a transcriptomic analysis, in which their expression levels were not different in the tested tissue. Real-time reactions were performed using SYBER Green PCR Master Mix (Applied Biosystems). Primer validation and differential expression analysis were conducted using the method described in Chao (2008).
Histological analysis of pericarp color
Developing seeds sampled at 5, 10, 15, 20, and 30 DPA were fixed with a formalin aceto alcohol solution for 3 h, dehydrated, and embedded in Paraplast Plus (Sigma) using methods described in Luna (1968). Longitudinal microtome sections of 8–10 μm thick were stained with hematoxylin and eosin (Sigma) and imaged using an Olympus AX70 Upright Compound Microscope.
Results
Fine mapping of qSD7-1/qPC7
An F2 population from the EM93-1/ILqSD7-1/qPC7 cross consisted of 4711 red and 1568 white pericarp-colored plants. The red:white ratio fits the 3:1 expectation (χ2 = 0.003; P = 0.95) for a dominant gene, which should be qPC7. There were no plants with a brown or other pericarp color in the F2 population, indicating that EM93-1 carries functional alleles at the Rd and other loci required for expressing the red phenotype.
A total of 1383 F3 seedlings from the white F2’s were genotyped with the RM6728 and AP5779-8 markers (Figure 1A). Of the 74 recombinant events identified, 66 and 8 gametes retain an SS18-2 allele at RM6728 and AP5779-8, respectively. The white recombinants Wrec#1 and #2, heterozygous at RM6728 or AP5779-8 (Figure 1B), were advanced to the F4 generation to precisely determine if qSD7-1 was separated from qPC7. There was no significant correlation between the marker genotypes and germination rates in these two white progeny populations (Figure 1B), indicating that qSD7-1/1PC7 does not locate on either heterozygous region.
Genotyping 1238 F3 seedlings from the red F2’s identified 58 recombinant events between RM6728 and AP5779-8. One (i.e., Rrec#1) of the recombinants was derived from the crossover occurring between AP5098-15 and RM21197, both of which are located within the The Institute for Genome Research locus Os07g11020 (Figure 1B). Rrec#1 was heterozygous for the RM21197–AP5779-8 region and produced red pericarp seeds (F4). Progeny testing detected a significant marker–germination correlation (R2 = 0.28) in the Rrec#1-derived F4 population, indicating that qSD7-1 and qPC7 co-locate on the heterozygous interval of <70 kb (Figure 1B).
Genotyping of the Rrec#1-derived 2295 F5 plants identified 12 recombinant events between RM21197 and AP5779-8. Recombinant Rrec#2, which is heterozygous at RM21197 but homozygous for the EM93-1 alleles at flanking markers RM632 and AP5098-15 located within Os07g11020 (Figure 1C), also produced red pericarp seeds. Progeny testing of this intragenic recombinant detected a significant marker–germination correlation (R2 = 0.36), with red pericarp genotypes having more dormant seeds than the white ones (Figure 1D). Co-location unambiguously demonstrated that Os07g11020 underlies both qSD7-1 and qPC7. Os07g11020 was cloned as Rc (Sweeney et al. 2006; Furukawa et al. 2007). On the basis of the progeny population (F6) segregating only for the intragenic segment between AP5098-15 and RM632, we estimated that the dormancy gene at qSD7-1, or SD7-1, has both additive (a) and dominance (d, d/a varying from 1.1 to 2.3) effects on germination (Figure S2).Thus, the isogenic lines SD7-1D and SD7-1d were selected from the F6 homozygous genotypes to represent dormancy-enhancing and -reducing alleles, respectively, for the following experiments.
Gene structure and functional mutation for seed dormancy
Alignment of SD7-1 genomic DNA sequences from SS18-2 and EM93-1 detected 32 mutations, including 24 single nucleotide polymorphisms (SNPs) and 8 short (1–34 bp) insertion/deletions (Table S2). The sequence alignment between the dormancy-enhancing and -reducing alleles from SD7-1D and SD7-1d, respectively, revealed that the isogenic lines differ in the intragenic region of 3206–5190 bp from the start codon (Table S2). Thus, Rrec#2 must be derived from the first crossover between 3109 and 3206 bp in the F1 or F2 and from the second crossover between 5190 and 5401 bp in the F4 (Figure S1).
Partial and full-length cDNAs for the SD7-1 locus were isolated from SD7-1D and SD7-1d 10-d developing seeds (Figure 2A). Alignment of the full-length cDNA against the genomic DNA sequences from SD7-1D demonstrated that SD7-1 consists of eight exons and seven introns (Figure 2B). Comparison between the SD7-1D and SD7-1d full-length cDNAs confirmed three mutations in the coding region of the dormancy-reducing allele, including a G/A transition in exon 6 and an A/G transition and a 14-bp deletion in exon 7 (Figure 2B).
The deduced protein sequence (Figure 2C) from the SD7-1D full-length cDNA contains a major (Pfam-A match HLH) and two minor (Pfam-B match) domains (Finn et al. 2010). Of the two SNPs, only the G/A transition in exon 6 changed a codon from methionine to isoleucine in the Pfam-B_8787 domain. This transition could be excluded as a functional mutation, as it is also present in the dormant red pericarp lines C9541, LD, Kebra60, Pokkalli, and W1713 (Figure 4). The 14-bp deletion is predicted to generate a premature stop codon located before the overlapped HLH and Pfam-B_3949 domains, resulting in a truncated protein in SD7-1d (Figure 2C). Annotation of this truncated protein did not find any significant Pfam-A or -B match, suggesting that the dormancy-reducing allele is a loss-of-function mutation. Therefore, this deletion is the only molecular lesion accounting for the phenotypic variation in seed dormancy and pericarp color between the isogenic lines.
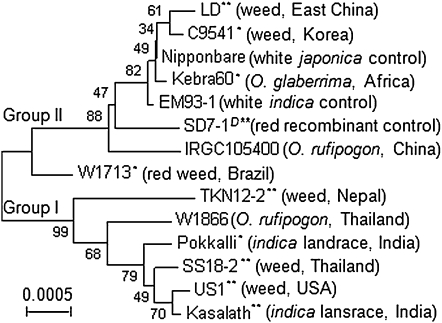
Figure 4 .
Evolutionary relationship of Rc alleles from a set of red pericarp lines. Phylogenetic tree was developed on the basis of genomic DNA sequences in the final data set of 6303 positions from 14 [12 red and 2 white (control)] genotypes of diverse origins. Numbers next to the branches are bootstrap values of 1000 replicates. Superscripts indicate dormant genotypes (*) or that Rc in the genotype (**) was associated with seed dormancy (Gu et al. 2005a).
Effect of a transgenic Rc on seed dormancy
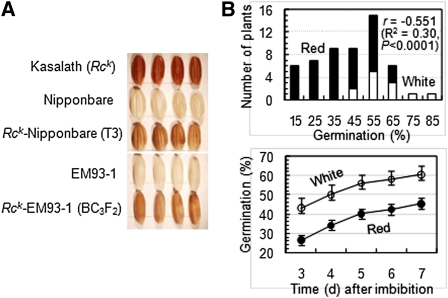
The Rck transgene changed pericarp color from brown in the Nipponbare to light red in the EM93-1 background (Figure 3A). In a BC3F2 population, the red:white segregation ratio of 42:12 fits the 3:1 Mendelian expectation (χ2 = 0.22; P = 0.63) for a dominant gene, and the pericarp color trait correlated with seed dormancy (Figure 3B). The correlation accounted for 30% of the phenotypic variance, and average germination was 16% lower for red than for white pericarp genotypes in two independent germination tests, indicating that Rck is also functional for seed dormancy. The failure to detect this function in the Nipponbare background (Lin et al. 1998; Miura et al. 2002) could result from other factors that interfere with Rck expression and/or phenotypic assessment.
Figure 3 .
Effects of the Rck transgene. (A) Images of caryopsis morphologies. The Kasalath-derived Rck transgene in the Nipponbare background (Furukawa et al. 2007) was introduced from a transgenic T3 plant into the EM93-1 background by three generations of recurrent backcrossing. Caryopses shown are from the donor (red), recipients (white), T3 (brown), and BC3F2 (light red) plants. (B) Germination profile of a BC3F2 population. The population consisted of 42 red and 12 white pericarp-colored plants and was evaluated for seed dormancy in two germination experiments. The red and white pericarp genotypes were correlated with germination rates at 7 d after imbibition in the first experiment (top). Circles/bars indicate means/SD of daily cumulative germination in the second experiment (bottom).
Differentiation of functional Rc alleles in seed dormancy
A total of 45 variable sites detected from 12 Rc alleles distinguish the donors from one another (Figure 4). All these alleles retain the 14-bp sequence deleted in the mutant rc alleles from EM93-1 and Nipponbare. Fifteen of the 45 sites locate in the exon regions outside the sites reported for the mutant Rc-s (Sweeney et al. 2006) and the reverting mutants Rc-g and Rcr derived from the rc alleles in white pericarp cultivars (Brooks et al. 2008; Lee et al. 2009). Seven of the 15 sites, including the G/A transition in exon 6, changed amino acid residues in deduced proteins.
The 12 Rc alleles are equally clustered into two distinct clades or groups, each consisting of donors from weedy, wild (O. rufipogon), and cultivated rice (Figure 4). Five of the six Rc alleles in group I distributed in tropical areas of Asia, where wild rice co-existed with weedy and indica types of rice (Oka 1988). Rc in US1, a red rice line collected from the U.S. temperate area (Suh et al. 1997), is an exception in group I. Rc alleles in group II are closest to the rc alleles and distribute in diverse donors, including the intragenic recombinant SD7-1D and the weedy rice from east China (LD) and Korea (C9541) where there was no O. rufipogon. Rc alleles from the temperate ecotype of weedy lines LD and C9541 could represent the functional alleles most distant to those in group I (Figure 4). All the donor lines of weedy and cultivated rice in both groups were identified as dormant genotypes, or dormancy was associated with the Rc locus in our previous (Gu et al. 2005a) or present research. Thus, phylogenetic and dormancy data suggest that Rc alleles functional for pericarp color most likely also function for seed dormancy.
Pleiotropic effects and responses to dormancy-breaking treatments
Independent experiments also detected differences in ABA accumulation and seed weight between the isogenic lines. ABA content increased during embryogenesis and reached a peak at 10 DPA in SD7-1D and SD7-1d. However, the hormone level was about two times higher in the dormant than in the nondormant line at peak time (Figure 5A). The genotypic difference suggests that SD7-1 may control seed dormancy by regulating ABA biosynthesis and/or metabolism in developing seeds.
Figure 5 .
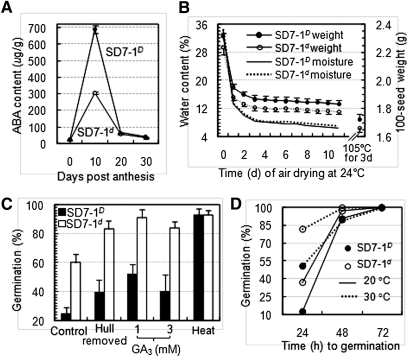
Phenotypic differences between SD7-1D and SD7-1d. (A) Abscisic acid (ABA) accumulation in developing seeds. Bars represent SD of three replicates. (B) Seed moisture and weight. Seeds in amounts of 100 were sampled from a plant; bars represent SD of 10 plants. (C) Germination of intact seeds (control), hull-removed seeds, and seeds treated with gibberellic acid (GA3) and heated at 45° for 7 d. Germination was evaluated with three samples (50 seeds/sample) from a plant; bars represent SD of 15 plants. (D) Germination of pericarp/testa-removed caryopses at two different temperatures. Germination was evaluated with 100 caryopses.
The experiment shown in Figure 5B was conducted to determine if pigmented pericarp tissue has an effect on seed moisture content, which is a physiological factor directly affecting dormancy release and germination capability in red rice (Leopold et al. 1988). The initial experiment did not detect a significant difference for moisture content in either freshly harvested or air-drying seeds from the isogenic lines, but revealed a greater (3%) weight for SD7-1D than for SD7-1d seeds (Figure 5B). This observation was confirmed in other two experiments that showed increasing seed weight by 3.0–3.6% (Table S3).
Several dormancy-breaking treatments were used to evaluate genotypic difference in germination response between SD7-1D and SD7-1d. One week of heating at 45° almost completely removed dormancy from both lines. Removal of the hull (lemma and palea) tissue from intact seeds or application of 1 or 3 mM GA3 increased germination but did not eliminate the genotypic difference (Figure 5C), suggesting that the SD7-1-controlled dormancy is not imposed through the hull or a GA-related pathway. Removal of the pericarp and testa tissues from the caryopses still allowed detection of the difference in germination velocity before 72 hr of incubation at 30° or 20° (Figure 5D). Thus, this difference could attribute to the residual effect of SD7-1 on germination after the maternal tissues are physically removed.
Downstream gene networks regulated by SD7-1/Rc and tissue-specific expression
Both SD7-1 functional and mutant alleles were transcribed at a relatively high level before 20 DPA (Figure 6A). Compared to the mutant allele, the functional allele displayed a higher level of expression from 5 to 15 DPA. The fold change in transcript level between SD7-1D and SD7-1d was 230 at 5 DPA and 85 at 10 DPA, suggesting that the dormancy gene is self-upregulated. The temporal expression pattern indicates that SD7-1 plays its regulatory roles during early seed development.
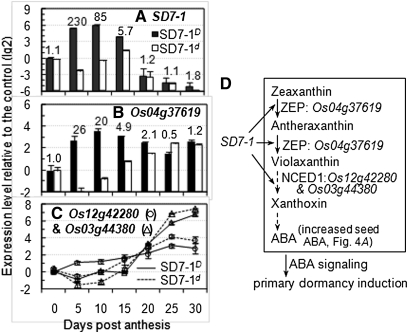
Figure 6 .
Transcription profile of selected genes in SD7-1D and SD7-1d. (A) The seed dormancy gene SD7-1. RNAs prepared from spikelet at 0 (control) and caryopses at 5–30 d post anthesis were quantified by quantitative RT-PCR (qRT-PCR), with the RNA II polymerase and GTP-binding protein genes as the control in this and the following analyses. (B and C) Predicted genes for ABA biosynthesis. Bars in B indicate means (SD) of three replicates for transcripts quantified by qRT-PCR. The number over each pair of bars is the fold change in transcription level between SD7-1D and SD7-1d. (D) A model for a regulatory mechanism of seed dormancy. SD7-1 upregulates expression of key genes on the ABA biosynthetic pathway, resulting in increased ABA accumulation in early developing seeds, which induces the development of primary dormancy. The pathway was adopted from Nambara and Marion-Poll (2005). ABA, abscisic acid; ZEP, zeaxanthin epoxidase; NCED1, nine-cis-epoxycarotenoid dioxygenase 1.
Expression of SD7-1 coincided with early ABA accumulation (Figure 5A) and with transcription of the predicted gene Os04g37619 or OsABA1 (Agrawal et al. 2001) encoding zeaxanthin epoxidase (ZEP) in SD7-1D (Figure 6, A and B). Transcript levels of ZEP were 20 to 26 times higher in SD7-1D than in SD7-1d from 5 to 10 DPA. In addition, the Os12g42280 and Os03g44380 paralogs encoding 9-cis-epoxycarotenoid dioxygenase 1 (NCED1) were also differentially expressed between the isogenic lines from 5 to 10 DPA (Figure 6C). However, the transcription patterns of NCED1s and ZEP were different. For example, the transcription levels of Os12g42280 and Os03g44380 were only a few times higher in SD7-1D than in SD7-1d from 5 to 10 DPA and kept increasing in both dormant and nondormant lines after 15 DPA (Figure 6C). ZEP and NCED are key enzymes of the ABA biosynthetic pathway (Nambara and Marion-Poll 2005). Thus, we propose that SD7-1 enhances ABA synthesis and subsequent dormancy induction in SD7-1D (Figure 6B).
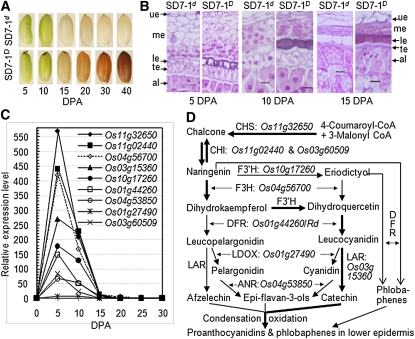
The difference in pericarp color between SD7-1D and SD7-1d became visible after 15 DPA (Figure 7A). However, a histological analysis indicated that pigments accumulated in the lower epidermal cell layer of the pericarp tissue in SD7-1D by 5 DPA (Figure 7B). Of the genes predicted to encode enzymes of the flavonoid biosynthetic pathway reported for other species (Lepiniec et al. 2006), nine were coordinately upregulated in SD7-1D before 15 DPA. The largest differences occurred at 5 DPA, with changes varying from 6- to 569-fold (Figure 7C). The more upregulated genes—Os11g32650 for chalcone synthase (569×), Os11g02440 for chalcone isomerase (442×), Os04g56700 for flavanone 3-hydroxylase (420×), Os10g17260 for flavonoid 3′-hydroxylase (179×), Os01g44260 or Rd for DFR (149×), and Os03g15360 for leucoanthocyanidin reductase (270×)—are thought to compose the main pathway for synthesis of the brown pigment proanthocyanidins (Figure 7D). Differential expression between SD7-1D and SD7-1d was not detected for predicted genes encoding enzymes (e.g., Os10g02880 for O-methyltransferase) required for biosynthesis of the red pigment anthocyanidins. However, it is known that DFR is also involved in the biosynthetic pathway from flavanones to the red compounds phlobaphenes (Lepiniec et al. 2006). It is possible that phlobaphenes may also contribute to the pericarp pigmentation in rice.
Figure 7 .
Differential expression of pericarp color and flavonoid biosynthesis genes between SD7-1D and SD7-1d. (A) Images of developing caryopses. Red pericarp color became visible at ∼15 d post anthesis (DPA) when chlorophyll disappeared. (B) Microscopic images of developing caryopses. Longitudinal sections for the dorsal side show the pigment accumulation only in the lower epidermal cell layer of the pericarp tissue in the dormant line, starting at 5 DPA, and filled the cells at ∼10 DPA. al, aleuronic layer; te, testa; ue, me, or le, upper, middle, or lower epidermal layers of the pericarp tissue. Bar, 10 μm. (C) Expression profile of predicted genes for seed flavonoid biosynthesis. mRNA samples prepared from spikelet at 0 and caryopses at 5–30 DPA were quantified by qRT-PCR and expressed as the ratio of SD7-1D to SD7-1d. The RNA II polymerase and GTP-binding protein genes were used as the control in the qRT-PCR analysis. (D) Flavonoid biosynthetic pathways activated by SD7-1/Rc in rice seeds. The pathway was modified from Lepiniec et al. (2006) on the basis of expression levels of the detected genes (C) encoding enzymes: ANR, anthocyanidin reductase; CHI, chalconeisomerase; CHS, chalcone synthase; DFR, dihydroflavonol reductase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; LAR, leucoanthocyanidin reductase; LDOX, leucoanthocyanidin dioxygenase. The thick arrow-headed lines in D are the proposed main paths as their underlying genes were more upregulated by SD7-1 than the genes on the other paths (C).
Discussion
Association between seed dormancy and pericarp color
Map-based cloning delimited qSD7-1/qPC7 to a single locus and provided unambiguous evidence that the association between seed dormancy and pericarp color arises from the pleiotropic effects of Os07g11020. Os07g11020 was identified as Rc for the qualitative trait red pericarp color (Kinoshita 1984; Sweeney et al. 2006; Furukawa et al. 2007) and also serves as one (SD7-1) of the polygenes controlling the quantitative trait seed dormancy. Due to pleiotropy, SD7-1 cannot be used to improve white pericarp cultivars for resistance to pre-harvest sprouting by conventional breeding approaches.
The pleiotropic gene controls the qualitative and quantitative traits most likely through different physiological pathways. SD7-1/Rc activates a conserved gene network for flavonoid biosynthesis only in the lower epidermal cells of the pericarp tissue, instead of in the testa tissue as in Arabidopsis (Debeaujon et al. 2000), to produce the pigments leading to a red or brown appearance. Although the pigmented pericarp, which is tightly enclosed by the hull in rice, could regulate some germination events, it is more likely that enhanced ABA biosynthesis is the primary reason for dormancy induction on the basis of well-documented information (Kermode 2005; Finkelstein et al. 2008). Of the three genes in the proposed ABA-mediated dormancy induction pathway (Figure 6D), OsABA1 appears to be the major target because its expression was upregulated by SD7-1 at a much higher level and the gene product ZEP catalyzes two steps from zeaxanthin to violaxanthin (Nambara and Marion-Poll 2005). There could be undetected gene networks downstream of SD7-1 involved in regulation of primary dormancy in rice.
Available information does not infer an orthologous relation between Rc and the wheat R genes. For example, the Os07g11020 orthologs are predicted for Arabidopsis thaliana (TRANSPARENT TESTA8), Brachypodium distachyon (BRADI1G54070), Zea mays (GRMZM2G042733), and Sorghum bicolor (Sb02g006390), but not for barley and wheat (http://www.gramene.org/Oryza_sativa); wheat ESTs from the R-containing deletion bins were aligned to the rice genome outside Os07g11020 (Sorrells et al. 2003; Kuraparthy et al. 2008). Instead of a conserved orthologous gene, wheat and rice may share similar regulatory mechanisms for the dormancy–grain-color association. The wheat sequence Tamyb10, amplified using primers designed on the basis of the maize P gene encoding a Myb transcription factor, could be an R candidate because it was coordinately expressed with flavonoid biosynthesis genes and was located in the R-containing deletion bins (Himi and Noda 2005). bHLH, Myb, and WD40 repeat proteins are components of the protein complex required to initiate flavonoid biosynthesis in the other plants (Ramsay and Glover 2005). Research is being conducted to identify the SD7-1 partners and other downstream genes in the rice system.
Red rice—beyond a pigment issue
Of the pleiotropic effects of SD7-1 detected in this research, seed dormancy must contribute most to the adaptation of red rice. There are two independent digenic systems controlling red (Rc/Rd) or purple (Prp-b/Prp-a) pericarp color in rice, and both require functional flavonoid biosynthesis (Kinoshita 1984; Abdel-Aal et al. 2006). There is no report on a selective advantage of red over purple pericarp color in rice. Thus, natural selection has favored red rice most likely because of seed dormancy instead of pericarp color per se. In addition to the main effect, SD7-1 also regulates seed dormancy by two or higher orders of epistasis with other dormancy loci (Gu et al. 2004).
The two groups of Rc alleles in weedy rice basically associate with geographic origins of the donor lines from tropical (group I) or temperate (group II) areas. A similar pattern was reported for Rc alleles from indica and japonica landraces (Sweeney et al. 2007). This similarity suggests that Rc alleles from tropical (e.g., SS18-2 and TKN12-2) and temperate (e.g., LD and C9541) weed ecotypes share some evolutionary mechanisms with those from indica and japonica subspecies, respectively, such as selection pressures from temperatures and cropping systems or common gene donors. Exceptions exist for Rc alleles from weedy genotypes in new rice-growing areas, such as US1 (group I) and other U.S. weedy rice lines discussed in detail by Gross et al. (2010). However, our research suggests that the dormancy function of SD7-1 alleles in red rice may not vary with their phylogenetic distances but that detection of the dormancy effect can be influenced by the genetic backgrounds and/or experimental methods. For example, the dormancy effect of SD7-1 from SS18-2 or Kasalath was significant in the EM93-1, but not in the CO39 (Gu et al. 2003) or Nipponbare background (Lin et al. 1998; Miura et al. 2002).
In conclusion, natural selection retained the red pericarp color characteristic because a functional SD7-1 allele can regulate multiple physiological pathways, which enhance survival of seeds in the soil seed bank and distribute germination over time. Domestication favored white percarp color because the mutant characteristic associates with the pleasant appearance and flavor/taste of grains or the absence of proanthocyanidins in the percarp tissue (Sweeney et al. 2007), which is caused by the inactivation of a functional flavonoid biosynthetic pathway downstream of the pleiotropic gene.
Acknowledgments
We thank B. Carsrud, Y. Wang, T. Nelson, C. Huckle, L. Kelley, B. Bigger, and L. L. Huckle for technical assistance. Wild rice lines were obtained from the National Small Grain Collection of the U.S. Department of Agriculture (USDA)-Agricultural Research Service or National Institute of Genetics, Japan. Funding for this research was supported by USDA-National Research Initiative (NRI) grant #2008-35301-19058 and in part by USDA-NRI grant #0200668, National Science Foundation grant #0641376, and the South Dakota Agricultural Extension Station.
Literature Cited
- Abdel-Aal E. S. M., Young J. C., Rabalski I., 2006. Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food Chem. 54: 4696–4704 [DOI] [PubMed] [Google Scholar]
- Agrawal G. K., Yamazaki M., Kobayashi M., Hirochika R., Miyao A., et al. , 2001. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 125: 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C. J., Koornneef M., 2006. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S. A., Yan W., Jackson A. K., Deren C. W., 2008. A natural mutation in rc reverts white-rice-pericarp to red and results in a new, dominant, wild-type allele: Rc-g. Theor. Appl. Genet. 117: 575–580 [DOI] [PubMed] [Google Scholar]
- Cai H. W., Morishima H., 2000. Genomic regions affecting seed shattering and seed dormancy in rice. Theor. Appl. Genet. 100: 840–846 [Google Scholar]
- Chang S., Puryear J., Cairney J., 1993. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11: 113–116 [Google Scholar]
- Chao W. S., 2008. Real-time PCR as a tool to study weed biology. Weed Sci. 56: 290–296 [Google Scholar]
- Cohn M. A., Hughes J., 1981. Seed dormancy in red rice (Oryza sativa) I. Effect of temperature on dry-afterripening. Weed Sci. 29: 402–404 [Google Scholar]
- Debeaujon I., Léon-Kloosterziel K. M., Koornneef M., 2000. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delouche J. C., Burgos N. R., Gealy D. R., De San Martin G. Z., Labrada R., et al. , 2007. Weedy Rices: Origin, Biology, Ecology, and Control. FAO Plant Production and Protection Paper 188. Food and Agricultural Organization, Rome [Google Scholar]
- Destefano-Beltrán L., Knauber D., Huckle L., Suttle J. C., 2006. Effects of postharvest storage and dormancy status on ABA content, metabolism, and expression of genes involved in ABA biosynthesis and metabolism in potato tuber tissues. Plant Mol. Biol. 61: 687–697 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C., 2008. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Tate J., Coggill P., Heger A., et al. , 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham J. E., 2000. Different genetic components control coat-imposed and embryo-imposed dormancy in wheat. Seed Sci. Res. 10: 43–50 [Google Scholar]
- Furukawa T., Maekawa M., Oki T., Suda I., Iida S., et al. , 2007. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 49: 91–102 [DOI] [PubMed] [Google Scholar]
- Gross B. L., Reagon M., Hsu S. C., Caicedo A. L., Jia Y., et al. , 2010. Seeing red: the origin of grain pigmentation in US weedy rice. Mol. Ecol. 19: 3380–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X.-Y., Chen Z.-X., Foley M. E., 2003. Inheritance of seed dormancy in weedy rice (Oryza sativa L.). Crop Sci. 43: 835–843 [Google Scholar]
- Gu X.-Y., Kianian S. F., Foley M. E., 2004. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166: 1503–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X.-Y., Kianian S. F., Foley M. E., 2005a Dormancy imposed by covering tissues interrelated with seed shattering and morphological characteristics in weedy rice (Oryza sativa L.). Crop Sci. 45: 948–955 [Google Scholar]
- Gu X.-Y., Kianian S. F., Hareland G. A., Hoffer B. L., Foley M. E., 2005b Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa L.). Theor. Appl. Genet. 110: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Gu X.-Y., Kianian S. F., Foley M. E., 2006. Isolation of three dormancy QTLs from weedy rice as Mendelian factors. Heredity 96: 93–99 [DOI] [PubMed] [Google Scholar]
- Harlan J. R., de Wet J. M. J., Price E. G., 1973. Comparative evolution of cereals. Evolution 27: 311–325 [DOI] [PubMed] [Google Scholar]
- Himi E., Noda K., 2005. Red grain colour gene (R) of wheat is a Myb-type transcription factor. Euphytica 143: 239–242 [Google Scholar]
- International Rice Genome Sequencing Project, 2005. The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Johnson L. P. V., 1935. The inheritance of delayed germination in hybrids of Avena fatua and A. sativa. Can. J. Res. 13: 367–387 [Google Scholar]
- Kermode A. R., 2005. Role of abscisic acid in seed dormancy. J. Plant Growth Regul. 24: 314–334 [Google Scholar]
- Kinoshita T., 1984. Genetics, pp. 187–274 Biology of Rice, edited by Tsunoda S., Takahashi N. JSSP/Elsevier, Tokyo [Google Scholar]
- Kuraparthy V., Sood S., Gill B. S., 2008. Targeted genomic mapping of a red seed color gene (R-A1) in wheat. Crop Sci. 48: S37–S48 [Google Scholar]
- Lee D., Lupotto E., Powell W., 2009. G-string slippage turns white rice red. Genome 52: 490–493 [DOI] [PubMed] [Google Scholar]
- Lee S. J., Oh C. S., Suh J. P., McCouch S. R., Ahn S. N., 2005. Identification of QTLs for domestication-related and agronomic traits in an Oryza sativa×O. rufipogon BC1F7 population. Plant Breed. 124: 209–219 [Google Scholar]
- Leopold A. C., Glenister R., Cohn M. A., 1988. Relationship between water content and afterripening in red rice. Physiol. Plant. 74: 659–662 [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J. M., Baudry A., Pourcel L., et al. , 2006. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Sasaki T., Yano M., 1998. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor. Appl. Genet. 96: 997–1003 [Google Scholar]
- Luna L. G., 1968. Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology. McGraw-Hill Book Co., New York [Google Scholar]
- Miura K., Lin S. Y., Yano M., Nagamine T., 2002. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 104: 981–986 [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A., 2005. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nilsson-Ehle H., 1914. Zur Kenntnis der mit der Keimungsphysiologie des Weizens in Zusammenhang stehenden inneren Faktoren. Zeitschrift für Pflanzenzüchtung 2: 153–187 [Google Scholar]
- Noldin J. A., Chandler J. M., McCauley G. N., 2006. Seed longevity of red rice ecotypes buried in soil. Planta Daninha 24: 611–620 [Google Scholar]
- Oka H. I., 1988. Origin of Cultivated Rice. Japan Scientific Society Press, Tokyo [Google Scholar]
- Ramsay N. A., Glover B. J., 2005. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Sorrells M. E., La Rota M., Bermudez-Kandianis C. E., Greene R. A., Kantety R., et al. , 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., et al. , 2010. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 107: 5792–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H.-S., Sato Y. I., Morishima H., 1997. Genetic characterization of weedy rice (Oryza sativa L.) based on morphophysiology, isozymes and RAPD markers. Theor. Appl. Genet. 94: 316–321 [Google Scholar]
- Sweeney M. T., Thomson M. J., Pfeil B. E., McCouch S. R., 2006. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M. T., Thomson M. J., Cho Y. G., Park Y. J., Williamson S. H., et al. , 2007. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 3: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S., 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thomson M. J., Tai T. H., McClung A. M., Lai X. H., Hinga M. E., et al. , 2003. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493 [DOI] [PubMed] [Google Scholar]