Abstract
The effective population size (Ne) is one of the most fundamental parameters in population genetics. It is thought to vary across the genome as a consequence of differences in the rate of recombination and the density of selected sites due to the processes of genetic hitchhiking and background selection. Although it is known that there is intragenomic variation in the effective population size in some species, it is not known whether this is widespread or how much variation in the effective population size there is. Here, we test whether the effective population size varies across the genome, between protein-coding genes, in 10 eukaryotic species by considering whether there is significant variation in neutral diversity, taking into account differences in the mutation rate between loci by using the divergence between species. In most species we find significant evidence of variation. We investigate whether the variation in Ne is correlated to recombination rate and the density of selected sites in four species, for which these data are available. We find that Ne is positively correlated to recombination rate in one species, Drosophila melanogaster, and negatively correlated to a measure of the density of selected sites in two others, humans and Arabidopsis thaliana. However, much of the variation remains unexplained. We use a hierarchical Bayesian analysis to quantify the amount of variation in the effective population size and show that it is quite modest in all species—most genes have an Ne that is within a few fold of all other genes. Nonetheless we show that this modest variation in Ne is sufficient to cause significant differences in the efficiency of natural selection across the genome, by demonstrating that the ratio of the number of nonsynonymous to synonymous polymorphisms is significantly correlated to synonymous diversity and estimates of Ne, even taking into account the obvious nonindependence between these measures.
THE effective population size (Ne) is one of the most fundamental quantities in population genetics, evolutionary biology, and molecular ecology, since it determines the effectiveness of natural selection and the level of neutral genetic diversity that a population contains (Charlesworth 2009). Populations and regions of the genome with small Ne tend to have low levels of genetic diversity, to be susceptible to the accumulation of deleterious mutations through genetic drift, and to have potentially low rates of adaptive evolution (Charlesworth 2009).
The effective population size is expected to vary across the genome as a consequence of genetic hitchhiking (Smith and Haigh 1974) and background selection (Charlesworth et al. 1993). The action of both positive and negative natural selection, particularly in regions of the genome with low rates of recombination, is expected to reduce the effective population size leading to lower levels of genetic diversity and reduced effectiveness of selection. Hence variation in the rate of recombination and the density of selected sites is expected to generate variation in Ne.
The evidence that there is variation in Ne within a genome comes from three sources. First, it has been shown that levels of neutral genetic diversity are correlated to rates of recombination in Drosophila (Begun and Aquadro 1992), humans (Hellmann et al. 2003), and some plant species (Tenaillon et al. 2004; Roselius et al. 2005). This could be due to variation in the mutation rate since neutral genetic diversity is proportional to the effective population size multiplied by the mutation rate. However, the level of neutral sequence divergence between species, which should be proportional to the mutation rate, is not correlated to the rate of recombination in Drosophila (Begun and Aquadro 1992) and the plant species (Roselius et al. 2005) that have been investigated. Furthermore, although there is a correlation between neutral sequence divergence and recombination rate in humans, this correlation is not sufficient to explain the correlation between diversity and the recombination rate (Hellmann et al. 2005). It is has also been shown that the Y and W chromosomes, which have no recombination over most of their length, have substantially lower diversity than other chromosomes and that this cannot be attributed to differences in the mutation rate or the fact that there are fewer Y and W chromosomes than autosomes (Filatov et al. 2001; Montell et al. 2001; Bachtrog and Charlesworth 2002; Hellborg and Ellegren 2004). It thus seems that the effective population size varies across genomes and is positively correlated to the rate of recombination.
Second, under the neutral theory of molecular evolution it is expected that levels of diversity and divergence should be proportional to each other, since both depend on the neutral mutation rate. Deviations from this hypothesis, caused by variation in Ne, can be tested using the HKA test and derivatives of it (Hudson et al. 1987; Ingvarsson 2004; Wright and Charlesworth 2004; Innan 2006). Evidence for departures from the neutral hypothesis, based on the HKA test, comes from multiple multilocus surveys in plants (Roselius et al. 2005; Schmid et al. 2005), the chicken Z chromosome (Sundström et al. 2004), humans (Zhang et al. 2002), and Drosophila (Moriyama and Powell 1996; Machado et al. 2002).
Third, variation in the effective population size should manifest itself as variation in the effectiveness of selection and this has also been observed. In Drosophila it has been shown that codon usage bias is lower in the regions of the genome with very low rates of recombination (Hey and Kliman 2002; Kliman and Hey 2003; Marais et al. 2003). It has also been shown that the number of nonsynonymous polymorphisms (Pn) relative to the number of synonymous polymorphisms (Ps) is higher in the low-recombining parts of the Drosophila melanogaster genome (Presgraves 2005), that the rate of nonsynonymous (dN) relative to the rate of synonymous (dS) substitution is positively correlated to the frequency of recombination (Betancourt and Presgraves 2002), and that the overall efficiency of selection appears to be lower in the regions of the genome with low rates of recombination (Presgraves 2005; Larracuente et al. 2008). Likewise it has been shown that dN/dS is higher on the Y or W chromosome than on the other chromosomes in humans (Wyckoff et al. 2002) and birds (Berlin and Ellegren 2006) and on the fourth chromosome of Drosophila species (Arguello et al. 2010). In contrast, Bullaughey et al. (2008) found no correlation between dN/dS and the rate of recombination in primates.
It is thought that the correlation between dN/dS or Pn/Ps and the rate of recombination is due to regions of the genome with little or no recombination having low effective population size and hence reduced effectiveness of natural selection (Betancourt et al. 2009). Pn/Ps is negatively correlated to the rate of recombination because regions with low effective population size allow more slightly deleterious mutations to segregate for a longer time. In contrast, dN/dS can be either positively or negatively correlated to the rate of recombination depending on the prevalence of advantageous mutations. If advantageous mutations are common, then regions of the genome with high rates of recombination are expected to evolve faster because they have a higher effective mutation rate and because selection is effective on a greater proportion of mutations. In contrast, if advantageous mutations are rare, then regions of the genome with high rates of recombination may have low values of dN/dS because selection against slightly deleterious mutations is more effective.
Although it is well established that Ne varies across the genome in a few species, it is unclear whether this is true of all species and, more importantly, how much variation in Ne there is and whether this variation results in differences in the effectiveness of selection. Here we test whether there is variation in the effective population size by considering whether there is significant variation in neutral diversity, taking into account that this might be due to variation in the mutation rate by using the divergence between species to control for differences in the mutation rate. We also quantify the variation in Ne. We estimate Ne from the nucleotide diversity at putatively neutral sites, since this is expected to be equal to 4Neμ in a diploid organism, where Ne is the effective population size and μ is the mutation rate per generation. We use the divergence between two species at neutral sites as an estimate of the mutation rate per generation. Note that since we are comparing loci within a genome, they all share the same generation time (unless they are on the sex chromosomes or in the mitochondrial DNA) and so this does not have to be explicitly taken into account. We can therefore estimate the effective population size for each locus. However, although each individual estimate is unbiased, the distribution of these values has a variance that is greater than the true variance because of sampling error; a locus might have a particularly low diversity just by chance and not because its effective population size is particularly low. To get around this problem we use a hierachical Bayesian framework to estimate the distribution of Ne across genes, taking into account the sampling error associated with both the polymorphism and the divergence data.
We test for and investigate the variation in the effective population size in 10 eukaryotic species including humans, D. melanogaster, A. thaliana, and Saccharomyces paradoxus (Table 1). We find that there is statistically significant variation in Ne across genes, but that it is rather modest in most of the organisms. We also investigate whether variation in Ne within a genome leads to variation in the proportion of effectively neutral mutations, by testing whether the ratio of the number of nonsynonymous to synonymous polymorphisms is correlated to the effective population size, in a way that circumnavigates the obvious nonindependence between the two variables. We find overall evidence for a correlation between these two parameters and hence conclude that even modest variation in the effective population size is sufficient to generate variation in the effectiveness of natural selection.
Table 1 . Summary of data sets used for the analyses.
| Species | Outgroup | Loci | Sites | Alleles | θs | ds | Data set |
|---|---|---|---|---|---|---|---|
| Drosophila melanogaster | D. simulans | 302 | 40,920 | 8 | 0.019 | 0.13 | Shapiro et al. (2007) |
| Homo sapiens | Macaca mulatta | 434 | 170,441 | 32 | 0.001 | 0.08 | EGP/PGAa |
| Mus musculus castaneus | Rattus norvegicus | 66 | 5,127 | 20 | 0.010 | 0.21 | Halligan et al. (2010) |
| Arabidopsis thaliana | A. lyrata | 918 | 64,927 | 24 | 0.008 | 0.14 | Nordborg et al. (2005) |
| Capsella grandiflora | Neslia paniculata | 251 | 31,273 | 8 | 0.019 | 0.16 | Slotte et al. (2010) |
| Sorghum bicolor | S. propinquum | 134 | 6,799 | 14 | 0.004 | 0.02 | Hamblin et al. (2006) |
| Boechera stricta | A. thaliana | 129 | 10,048 | 40 | 0.003 | 0.21 | Song et al. (2009); Gossmann et al. (2010) |
| Arabidopsis lyrata | A. thaliana | 66 | 5,260 | 24 | 0.018 | 0.15 | Ross-Ibarra et al. (2008); Foxe et al. (2008) |
| Capsella rubella | A. thaliana | 49 | 5,014 | 16 | 0.004 | 0.29 | Foxe et al. (2009); Guo et al. (2009) |
| Saccharomyces paradoxus | S. cerevisiae | 94 | 28,019 | 8 | 0.002 | 0.36 | Tsai et al. (2008) |
Number of synonymous sites (Sites) and nucleotide diversity (θs) are from the polymorphism data. ds, average divergence between the species pairs at silent sites.
EGP, http://egp.gs.washington.edu; and PGA, http://pga.gs.washington.edu, August 2010.
Materials and Methods
Sequence data
We obtained data from different plant species, mouse, fruitfly, and yeast using publicly available data from GenBank (http://www.ncbi.nlm.nih.gov/Genbank). Polymorphism data for Homo sapiens were downloaded from Enivironmental Genome Project (egp.gs.washington.edu) and Seattle SNPs (pga.gs.washington.edu) Web sites and for Arabidopsis thaliana from http://walnut.usc.edu/2010. The annotated protein-coding genome of A. thaliana was obtained from TAIR 8 (ftp://ftp.arabidopsis.org), and the annotated A. lyrata genome was obtained from JGI http://genome.jgi-psf.org/. The annotated protein-coding genomes of Pan troglodytes, Macaca mulatta, and Rattus norvegicus were obtained from Ensembl (http://www.ensembl.org/info/data/ftp/index.html). The S. cerevisae genome chromosome III was obtained from http://www.yeastgenome.org. We restricted our analysis of D. melanogaster to data from the Zimbabwe population, of the S. paradoxus data set to the European population, and of the human data set to African populations, since all of these represent the ancestral populations of the three species (Garrigan and Hammer 2006; Stephan and Li 2007; Liti et al. 2009). Qualitatively similar results were obtained in the three cases when using global data.
Preparation of the data
The analysis was performed using protein-coding sequences. Coding regions were assigned using protein-coding genomic data or, if given, were taken from the GenBank input files. Sequences were aligned using Clustalw, using default parameter values (Thompson et al. 1994). The outgroup ortholog was assigned using the best BLAST (Altschul et al. 1990) hit or, if given, was taken from the polymorphism data set. We used only polymorphism data for which we could assign an outgroup sequence. For all analyses the number of synonymous substitutions and polymorphisms served as the neutral standard. For computational reasons all sites had to have been sampled in the same number of chromosomes within each species; because some loci had been sampled in more individuals than others and other loci had missing data, we reduced the data set to a common number of chromosomes by randomly sampling the polymorphisms at each site without replacement. The numbers of synonymous and nonsynonymous sites and substitutions were estimated by randomly selecting one allele from the polymorphism data and comparing it against the outgroup using the F3x4 model implemented in PAML (Yang 1997) in which codon frequencies are estimated from the nucleotide frequencies at the three codon positions. The proportion of sites estimated by PAML was also used to compute the numbers of synonymous and nonsynonymous sites for the polymorphism data. Although how we choose to define a site can be important in some circumstances (Bierne and Eyre-Walker 2003), this is not likely to be a problem in the current context because we use the same definition for both the divergence and the polymorphism data; as such, the number of sites effectively cancels out in most of our analyses (however, see discussion of selection on synonymous codon bias below). Statistics concerning numbers of loci and numbers of sites as well as polymorphic sites are shown in Table 1.
Testing for variation in diversity and the effective population size
We investigated whether there was significant variation in the level of diversity across the genome, using two tests. If we assume there is free recombination within and between loci (or no recombination within and between loci), then variation in diversity can be tested using a simple (2 × k) χ2-test of independence across the k loci within each species, where for each locus we have the number of sites with a polymorphism and the number of sites without a polymorphism. Note that this test is valid only when the same numbers of chromosomes have been sampled across all loci. However, some of the variation in diversity between loci might be due to variation in the genealogy if there is limited or no recombination between loci. We therefore applied a variant of the classic HKA test, but we removed the divergence information from the test. The test statistic X2 is set up as
| (1) |
where and are the expected value and variance of the number of segregating polymorphisms, P, in gene i,
| (2) |
| (3) |
with n being the number of alleles, M the number of loci, θ = 4Neμ, and Li the number of sites in gene i. Estimates of θ were obtained by minimizing the value of X2. X2 is expected to be χ2-distributed with (L − 1) d.f.
Any variation that we detect in diversity might be due to variation in the mutation rate or variation in the local effective population size. We therefore performed two further analyses to investigate whether there was variation in diversity that could not be explained by variation in the mutation rate, as measured by synonymous divergence between species. The first test was a second approximate (2 × k) χ2-test of independence, performed as follows. For each locus we have the number of sites used to estimate the level of silent site divergence (Ld), the estimated number of substitutions (D), the number of sites used to estimate silent site diversity (Lp), and the number of sites with a polymorphism (P). Since Ld and Lp can be different, we reduced the divergence or polymorphism data set, whichever was larger, to the size of the other, resampling without replacement the numbers of substitutions or polymorphisms as appropriate; for example, if Ld was half Lp, we sampled Lp sites from the divergence data to generate a subsample of the substitutions (D′) over sites. We can then perform a (2 × k) χ2-test where the cells for each gene are the number of sites with a substitution (D′) and the number of sites with a polymorphism (P′). Note that the data set will be reduced using this method, resulting in a loss of power. Furthermore, this test is only approximate because we assume that the number of substitutions is binomially distributed, whereas in fact it has a more complex distribution because of the correction for multiple hits. Some of the expected values can be very small in both χ2-tests: we therefore checked the P-values from the χ2-tests by generating the null distribution for the test. This was performed by randomly assigning polymorphisms and substitutions across the contingency table preserving the marginal totals. We then recalculated the statistic and performed this 1000 times. The P-value was the proportion of such randomly generated values that exceeded the observed value. Generally we found that the P-value from randomization and the P-value assuming our test statistics were χ2-distributed were similar (Supporting Information, Table S1). We therefore present the results from the standard χ2-test.
This test assumes free recombination between sites within loci and loci (or no recombination between sites and loci). A more conservative test is the classic HKA test that tests for heterogeneity in the ratio of diversity divided by divergence between loci assuming no recombination within loci, but free recombination between loci. We performed the multiple-locus HKA test using software provided by J. Hey (http://genfaculty.rutgers.edu/hey/software#HKA). To perform this test we had to exclude loci with zero divergence; for most species this constituted a small fraction of the total number of loci. However, we had to exclude Sorghum bicolor from the analysis because too many loci showed zero divergence.
Recombination and density of selected sites
We obtained estimates of recombination rate variation along chromosomes for A. thaliana (Singer et al. 2006), D. melanogaster (Hey and Kliman 2002), H. sapiens (Kong et al. 2002), and Mus musculus (Dumont et al. 2011). Gene density was estimated as the proportion of coding sites in window sizes of 50 kb, 500 kb, and 5 Mb. Since results are qualitatively similar, we discuss only results for the window size of 500 kb. Conservation scores (Siepel et al. 2005) were obtained from the UCSC genome browser (http://genome.ucsc.edu/) for D. melanogaster across 15 species, H. sapiens across 17 species, and M. musculus across 30 species.
Bayesian analysis
To estimate the distribution of Ne we used a hierachical Bayesian analysis in which we estimate the parameters of the distribution of Ne (Figure S1). If we assume that the population size is stationary, the expected number of polymorphisms segregating in a sample of n sequences, P, and the number of differences between the outgroup and a single sequence from the ingroup, Ds, are
| (4) |
| (5) |
where Lp and Ld are the numbers of sites that can have a polymorphism or a substitution, respectively, μ is the nucleotide mutation rate per generation, and t is the time of divergence. We are interested in the distribution of Ne. To estimate this distribution we assume that Ne and μ follow a log-normal or a gamma distribution. Assuming free recombination and using Equations 4 and 5 above, we can write the likelihood of observing polymorphisms and substitutions,
| (6) |
where X(S, S(x)) is the Poisson distribution and is the probability density of the distribution of Ne, and M(μ|σμ) is the probability density of the distribution of the mutation rate; these distributions are parameterized such that the mean is fixed at unity, leaving us to estimate the shape parameter. If there is no recombination within a locus, then we can rewrite Equation 4 as
| (7) |
where τ is the length of the genealogy scaled such that E[τ] = 1. We can rewrite Equation 6, and the likelihood then becomes
| (8) |
To calculate the probability density distribution M(τ | n) of genealogy lengths we randomly simulated 10,000 genealogies, scaling them such that the average total length was unity. In theory it is possible to accommodate ancestral polymorphism into the method; however, we found that the method rarely gave stable estimates of , particularly in the no recombination model. We therefore concentrated on data sets in which the influence of ancestral polymorphism was likely to be minimal—i.e., in which the average divergence was more than five times the average of θW (Table 1). If we assume that the ancestral Ne of a locus is correlated to the current Ne, we expect ancestral polymorphism to decrease the apparent variation in Ne.
To estimate the posterior distribution of the parameters and σμ we used a Monte Carlo Markov chain running the Metropolis–Hastings algorithm (Hastings 1970). Unfortunately because we have very few synonymous polymorphisms per gene, this method tends to underestimate the true value of . For most data sets this underestimation is small, but it can be large. We therefore estimated the extent of bias by simulating data under a range of parameter values, using the actual numbers of sites from the real data such that the expected numbers of polymorphisms and substitutions were equal to the mean values. For example, if we estimated to be 0.5 and σμ to be 0.1, we simulated data for σμ-values of 0.1, 0.2, and 0.3 and for -values between 0.4 and 1.0 in steps of 0.05. For each simulated data set we estimated and using linear regression we inferred the relationship between (estimated) and (true). Using this relationship we inferred the true value of from the value estimated from the real data (Figure S2 and Figure S3). To obtain a corrected SE we multiplied the observed standard error by the ratio of the corrected estimate of divided by the observed estimate of . This slightly underestimates the true SE since we have not taken into account the small amount of error associated with estimating the regression line. To test for heterogeneity in between species we assumed that the estimate of was normally distributed; under this assumption is χ2-distributed with k − 1 d.f. for k species. was calculated as a weighted average, where the weights were inversely proportional to the variance of the estimate (Eyre-Walker 1996).
Variation of efficiency of selection
We tested whether the strength of selection on nonsynonymous mutations was correlated to the effective population size, which can be seen as testing whether the fraction of deleterious mutations varies with Ne. This can be done by considering the correlation of Pn/Ps and θs or Pn/Ps and Ne (= θs/(4μ)), where Pn and Ps are the numbers of nonsynonymous and synonymous mutations, respectively, and Ne values are point estimates from the genetic diversity and mutation rates taken from the literature. However, Ps and θs are not independent. We overcome this problem by splitting Ps into two independent values by generating a random hypergeometric variable as follows:
| (9) |
| (10) |
(Piganeau and Eyre-Walker 2009; Stoletzki and Eyre-Walker 2011). One of the Ps values is used to estimate Pn/Ps (see below) and the other one is used to estimate θS. There are two further problems to consider with this method: first, Pn/Ps can be an overestimate or an underestimate of the true value of Pn/Ps and, second, the ratio Pn/Ps is undefined if Ps = 0. Both of these problems can be overcome by considering the correlation between ψ and θs:
| (11) |
(Piganeau and Eyre-Walker 2009). Hence, using our method to split Ps into independent values, we have two independent pairs of θS and ψ; we present results from only one pair. Some of the data sets contain relatively little polymorphism, which results in substantial variance of ψ. To overcome this problem we sum data across loci. For this we ranked loci according to their neutral diversity obtained from θs2 and binned them into groups of size n (e.g., 2, 4, 8, and 16). For each group average θs2 and corresponding Ne2 values were calculated. Furthermore, for each group, the sums of Pn and Ps1 were calculated to calculate ψ1. Note that ψ2 can be obtained in a similar manner; however, results were qualitatively comparable and we therefore show only results for ψ1 vs. θ2 and Ne2. Also we show only results for group size 4 because results for group sizes >2 were similar. The correlations were performed by calculating Spearman’s rank correlation and probabilities were combined using the unweighted Z method (Whitlock 2005). For summary of symbols see File S1.
Results
To investigate variation in the effective population size within genomes, we assembled protein-coding sequences from 10 species. The data sets are from 6 plant species, 3 animal species, and 1 fungus. The data sets range in size from 66 to 918 loci per species and from 8 to 40 sequences per gene (Table 1). In all analyses we assume that synonymous mutations are neutral.
Variation of diversity and Ne within a genome using χ2- and HKA tests
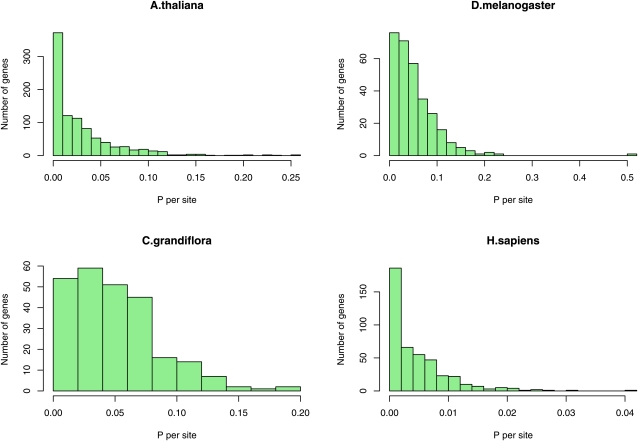
The level of genetic diversity appears to vary considerably within each genome (Figure 1); however, the number of polymorphisms per gene is generally quite low and hence this variation might be due to sampling error. To test whether the variation is significant we used two tests, which make different assumptions about the rate of recombination within loci: either free or no recombination. Both tests suggest that there is variation in the level of diversity in most species: all species are significant assuming free recombination and 6 of 10 are significant assuming no recombination (Table 2). This variation in diversity between loci could be due to variation in the effective population size or to variation in the mutation rate. To investigate whether variation in the mutation rate might be responsible, we estimated the number of synonymous substitutions for each locus (DS), between the species of interest and an outgroup species. In many species there is a significant positive correlation between the numbers of synonymous substitutions ds and polymorphisms ps per site (Table 3), suggesting that part of the variation in diversity is due to variation in the mutation rate. However, if we test whether there is significant variation between loci, taking into account the mutation rate, as estimated from the divergence between species, using either a χ2-test of independence or the more conservative HKA test, then we find significant evidence in the majority of species, whether or not we assume free or no recombination within loci: 9 of 10 loci for the free recombination test and 6 of 9 loci for the no recombination test (the HKA test could not be performed on S. bicolor due to the large number of genes in which the divergence was zero) (Table 2).
Figure 1 .
Distribution of the number of polymorphisms per site across genes for four species.
Table 2 . Results of the χ2-tests of independence and HKA tests.
| Diversity | Diversity and divergence | |||
|---|---|---|---|---|
| Species | P-value (χ2) | P-value (HKA) | P-value (χ2) | P-value (HKA) |
| D. melanogaster | <1 × 10−3 | 0.015 | <1 × 10−3 | <1 × 10−3 |
| H. sapiens | <1 × 10−3 | <1 × 10−3 | <1 × 10−3 | <1 × 10−3 |
| M. castaneus | <1 × 10−3 | 0.432* | 0.066* | 0.429* |
| A. thaliana | <1 × 10−3 | <1 × 10−3 | <1 × 10−3 | <1 × 10−3 |
| C. grandiflora | <1 × 10−3 | 0.462* | <1 × 10−3 | 0.565* |
| S. bicolor | <1 × 10−3 | <1 × 10−3 | 5.3 × 10−3 | NA |
| B. stricta | <1 × 10−3 | 0.434* | 6 × 10−3 | 0.01 |
| A. lyrata | <1 × 10−3 | 5.4 × 10−3 | <1 × 10−3 | 2.3 × 10−3 |
| C. rubella | <1 × 10−3 | <1 × 10−3 | <1 × 10−3 | <1 × 10−3 |
| S. paradoxus | 1.9 × 10−3 | 0.94* | <1 × 10−3 | 0.35* |
Results of the χ2-tests of independence and HKA tests for diversity and diversity/divergence data are shown. For details see Materials and Methods. P-values are given for each species. *P > 0.05 (not significant).
Table 3 . Results of correlates of Ps.
| psvs.ds | psvs.Ne | |||
|---|---|---|---|---|
| Species | ρ | P-value | ρ | P-value |
| D. melanogaster | 0.18 | 3.82 × 10−3 | 0.46 | 1.87 × 10−17 |
| H. sapiens | 0.29 | 1.62 × 10−6 | 0.38 | 3.02 × 10−16 |
| M. m. castaneus | 0.38 | 5.98 × 10−3 | 0.33 | 3.55 × 10−3 |
| A .thaliana | 0.16 | 1.13 × 10−4 | 0.44 | 5.94 × 10−45 |
| C. grandiflora | 0.35 | 3.00 × 10−8 | 0.52 | 2.53 × 10−19 |
| S. bicolor | 0.54 | 3.17 × 10−3 | 0.40 | 7.31 × 10−7 |
| B. stricta | 0.10* | 4.02 × 10−1 | 0.14* | 6.18 × 10−2 |
| A. lyrata | 0.22* | 1.03 × 10−1 | 0.62 | 1.21 × 10−8 |
| C. rubella | 0.12* | 6.34 × 10−1 | 0.65 | 2.35 × 10−7 |
| S. paradoxus | 0.04* | 7.91 × 10−1 | 0.42 | 1.01 × 10−5 |
Results of Spearman’s rank correlates of ps are shown. The nonindependence of ps and Ne is taken into account by splitting the data set into independent halves (see Materials and Methods). Correlation coefficients (ρ) and P-values are given for each species. *P > 0.05 (not significant).
Correlates of Ne
The variation in Ne across the genome is likely to be due to genetic hitchhiking and background selection. Both processes are expected to be stronger in regions of the genome with low rates of recombination and a high density of sites subject to natural selection. To investigate which or whether either of these factors is responsible for the variation in Ne, we investigated whether the variation in Ne was correlated to the rate of recombination and density of selected sites in four species for which these data were available: D. melanogaster, human, mouse, and A. thaliana. We measured the density of selected sites as either the number of nucleotides in annotated exons (genic density) or the number of nucleotides in conserved regions (conserved site density), as annotated in the UCSC conservation track, in windows of size 50 kb, 500 kb, and 5 Mb, where the window is centered on the gene from which the polymorphism data were taken (there is no conservation track for A. thaliana, so in this species we investigated just the density of genic sites). Results for the different window sizes were generally consistent, so we present the results from the 500-kb window size. We estimated Ne as the synonymous diversity divided by synonymous divergence.
In D. melanogaster we find, as others have done, that our estimate of Ne is positively correlated to recombination rate (Spearman’s correlation coefficient r = 0.45, P < 0.01). It is, however, also positively correlated to the density of conserved sites (r = 0.24, P < 0.01), which is unexpected, although not genic sites (r = 0.03, P = 0.65). The positive correlation with conserved site density might be due to the positive correlation that exists between the density of conserved sites and the rate of recombination (r = 0.56, P < 0.01), and indeed if we perform a multiple regression, we find that the correlation between Ne and the density of conserved sites disappears (P = 0.74), while the positive correlation between Ne and recombination rate remains (P < 0.01).
In humans we find, as others have done, that both diversity (r = 0.14, P = 0.02) and divergence (r = 0.18, P < 0.01) are positively correlated to the rate of recombination (Lercher and Hurst 2002; Hellmann et al. 2005), and there is, as a consequence, no correlation between estimates of Ne and the rate of recombination (r = 0.026, P = 0.69). Ne is significantly negatively correlated to the density of genic sites (r = −0.19, P < 0.01), but not conserved sites (r = −0.085, P = 0.17). Using multiple regression does not alter this picture: Ne is correlated only to the density of genic sites.
In mouse we see no significant correlations between estimates of Ne and the rate of recombination (r = 0.054, P = 0.72) and the density of genic (r = 0.089, P = 0.53) or conserved sites (r = 0.093, P = 0.51). This picture is unaffected by the use of multiple regression.
In A. thaliana we see a pattern like that in humans: both diversity (r = 0.10, P = 0.04) and to a lesser extent divergence (r = 0.064, P = 0.11) are positively correlated to recombination rate, and Ne is positively but not significantly correlated to recombination rate (r = 0.080, P = 0.11). Ne is significantly negatively correlated to genic density (r = −0.11, P = 0.02). Unfortunately there are no data on conserved sites in this species.
Quantifying variation of Ne
Since we find evidence for variation in Ne in many of our species, we attempted to quantify the amount of variation using a hierarchical Bayesian model. We assume underlying distributions for Ne and μ (e.g., log-normal distributions) and estimate the shape parameters and σμ and hence the variances of these distributions; the mean of each distribution is constrained to be equal to one (see Materials and Methods). We investigate two different models: in the first we assume free recombination and in the second we assume no recombination within loci, but free recombination between loci. These two models are likely to set the upper and lower bounds on the true level of variation in Ne. Under the free recombination model all the variation in diversity is attributed to variation in Ne, variation in the mutation rate, and sampling error. In the model with no recombination, variation in diversity may additionally be due to variation in the coalescent process. Hence, the free recombination model gives an upper estimate on the variation in Ne and the no recombination model gives a lower bound.
We applied our method to the polymorphism data from each of the 10 eukaryotic species to estimate the variation of Ne within each genome along with the variation in the mutation rate, σμ (Table 4). As expected, in all cases the estimate of is larger when free recombination is assumed, but the estimates from the two models are highly correlated (r = 0.95). The estimate of σμ is unaffected by the model of recombination assumed. We find evidence that the value of varies between species for both the free and the no recombination models (P = 2.5 × 10−9 and P = 4.2 × 10−8, respectively). We find that the level of variation of Ne is the lowest for M. musculus and highest for Capsella rubella for both recombination models. The estimates of and σμ were of similar magnitude for each taxon, suggesting that overall variation in the mutation rate and variation in the effective population size contribute a similar amount to the variation in diversity.
Table 4 . Estimates of the variation of Ne in 10 eukaryotic species.
| Free recombination | No recombination | |||
|---|---|---|---|---|
| Species | σμ (SD) | (SD) | σμ (SD) | (SD) |
| D. melanogaster | 0.370 (0.024) | 0.743 (0.048) | 0.372 (0.024) | 0.516 (0.072) |
| H. sapiens | 0.522 (0.021) | 0.682 (0.07) | 0.52 (0.02) | 0.578 (0.11) |
| M. m. castaneus | 0.369 (0.045) | 0.35 (0.119) | 0.372 (0.045) | 0.247 (0.15) |
| A. thaliana | 0.419 (0.015) | 0.83 (0.04) | 0.423 (0.015) | 0.809 (0.065) |
| C. grandiflora | 0.355 (0.021) | 0.475 (0.043) | 0.351 (0.021) | 0.165 (0.067) |
| S. bicolor | 0.689 (0.092) | 0.903 (0.263) | 0.710 (0.095) | 0.675 (0.292) |
| B. stricta | 0.441 (0.039) | 0.503 (0.174) | 0.443 (0.0379) | 0.411 (0.178) |
| A. lyrata | 0.276 (0.053) | 0.729 (0.119) | 0.278 (0.054) | 0.651 (0.139) |
| C. rubella | 0.263 (0.042) | 1.191 (0.21) | 0.258 (0.043) | 1.126 (0.243) |
| S. paradoxus | 0.23 (0.023) | 0.566 (0.208) | 0.23 (0.0218) | 0.387 (0.131) |
Estimates of the variation of Ne in 10 eukaryotic species are shown. Results are for an underlying log-normal distribution for Ne and μ, assuming either free recombination or no recombination (see Materials and Methods). For each data set the mean shape parameters and σμ and in parentheses their standard deviations (SD) obtained from the posterior distribution are given.
The level of variation in Ne we estimate using our method is quite modest. For example, C. rubella has the highest estimate of , but under this distribution the genes in the 90th percentile have an Ne that is only 7.2-fold greater than that of those in the 10th percentile; i.e., 80% of genes have an effective population size within 7.2-fold of each other. Four species have estimates of < 0.6, meaning that the difference between the 90th and the 10th percentile is <4-fold.
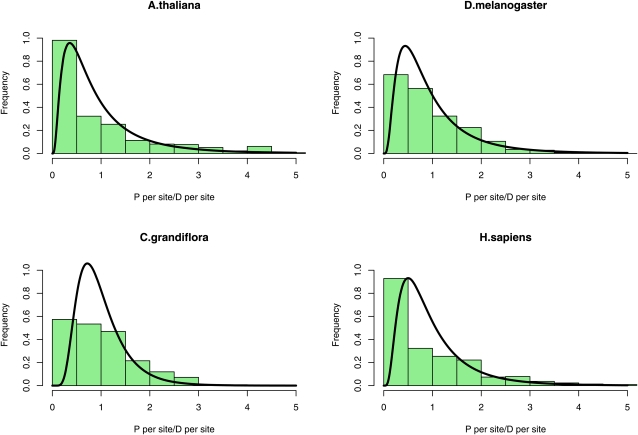
The estimated distribution appears to fit the data reasonably well (Figure 2). We would not expect the fit to be perfect, particularly at the lower end of the distribution, since this is where sampling error is a major issue; e.g., many genes have no polymorphism because of sampling error, not because they have an effective population size of zero. It is possible that assuming a log-normal distribution places some unwanted constraints on the estimation procedure; in particular, the probability density tends to zero for low Ne. We therefore also fitted a gamma distribution to the data (Table S3); with this distribution the probability density does not necessarily decline to zero near the origin. However, the estimated distributions are very similar to those obtained assuming a log-normal distribution (Figure S4 and Figure S5). The species that show low variation in Ne are also those that tend to show little evidence of variation in Ne, as judged by the χ2- and HKA tests. This implies that failure to detect variation in Ne is largely because there is limited variation in Ne rather issues with statistical power.
Figure 2 .
Distribution of the per site polymorphism/divergence ratio across genes for four species and corresponding distributions of Ne (solid line) estimated by hierarchical Bayesian analysis assuming a log-normal distribution.
Variation in the efficiency of selection
Although we estimate the variation in the effective population size to be modest, it is of interest to investigate whether this translates into significant differences in the efficiency of natural selection across the genome. To investigate this we tested whether there was a correlation between ψ = Pn/(Ps + 1) and either θs or Ne for each locus in a manner that controls for the obvious nonindependence of the two variables (see Materials and Methods). We remove the nonindependence by splitting Ps into two independent parts and we use ψ because it reduces the bias inherent in the estimation of Pn/Ps; furthermore it allows Pn/Ps to be calculated for all genes (Piganeau and Eyre-Walker 2009). This test is not very powerful since ψ has a large variance; furthermore, it is statistically biased in a manner that tends to generate a positive correlation between ψ and θs or Ne. We therefore follow the approach suggested by Piganeau and Eyre-Walker (2009) and grouped genes according to their θ or Ne value. The results are qualitatively similar for groupings of 4, 8, and 16 genes, so we present the results for groups of 4. There is a significant negative correlation between both θs and ψ and Ne and ψ in A. lyrata and C. grandiflora, and a marginally significant correlation between ψ and θs in D. melanogaster, although only the correlations in Cryptostegia grandiflora are significant after correction for multiple tests; otherwise the correlations are generally weak and nonsignificant. However, overall we find significant evidence for a negative correlation between ψ and θs or Ne if we combine probabilities: between ψ and θs P = 0.043 and between ψ and Ne P = 0.021.
The relationship between ψ and Ne can potentially yield information about the distribution of fitness effects (DFE) (Loewe and Charlesworth 2006; Loewe et al. 2006; Woolfit 2006; Elyashiv et al. 2010). If we assume that the DFE for nonsynonymous mutations is a gamma distribution and that synonymous mutations are neutral, then Pn/Ps is expected to be proportional to , where β is the shape parameter of the gamma distribution (Welch et al. 2008). Hence we can estimate β by considering the slope of the regression line between log(ψ) and log(Ne). Since the log of zero is undefined, we grouped genes in groups of size n such that no group had a zero estimate of ψ or Ne. We attempted to estimate β in the species that individually showed a significant correlation between ψ and Ne. However, we could not perform the analysis of A. lyrata because the diversity is so low that it was impossible to define groups that did not have zero values for both ψ and Ne. The estimates of β using this method are 0.41 (SE = 0.15) in C. grandiflora and 0.23 (0.15) in D. melanogaster; these are similar to those obtained using an independent method that uses the site frequency spectrum (Keightley and Eyre-Walker 2007): 0.27 (0.08) for C. grandiflora and 0.29 (0.07) for D. melanogaster (Table S2). This suggests that the gamma distribution is a reasonable approximation to the DFE, at least for mutations of weak effect.
Discussion
The effective population size (Ne) is one of the most important parameters in population genetics and evolutionary biology. It has been shown that Ne varies across the genome of D. melanogaster and some plant species, and it is thought that it might vary across the human genome (Hellmann et al. 2005). Here we have shown that it varies in most species that we have considered. However, the variation in Ne is not consistently correlated to either the rate of recombination or the density of selected sites. This might in part be because the variation in Ne is quite limited: most genes in a genome have an Ne that is within a few fold of that of most other genes. Nevertheless the variation is sufficient to cause differences in the effectiveness of natural selection on segregating nonsynonymous polymorphisms.
There are a number of factors that might have led us to over- or underestimate the variation in Ne. First, we have assumed that there is either free recombination or no recombination within loci to estimate the variation in the effective population size. This is unsatisfactory since we know that recombination is one of the factors that generates variation in the effective population size, at least in species like Drosophila, in which there is a correlation between diversity and the rate of recombination. Unfortunately it is not easy to get around this problem. However, as we have noted earlier, the estimate assuming free recombination should give an upper estimate on the amount of variation, because under this method all variation in the diversity is assumed to arise from sampling error and variation in the mutation rate and Ne. In reality, some of the variation between genes is a consequence of variation in the length of the genealogy in genes with little or no recombination.
Second, we have used the divergence between species as an estimate of the mutation rate, but if the mutation rate at a locus changes through time, for which there is evidence (Aguileta et al. 2006; Hodgkinson and Eyre-Walker 2011), then we will tend to overestimate the variation in Ne: this is most easily seen by assuming there is variation in the mutation rate, but no variation in Ne; if the mutation rate has changed through time, then the divergence will not be a perfect measure of the recent mutation rate and there will appear to be variation in Ne.
Third, we have assumed that synonymous mutations are neutral, but there is evidence of selection in humans (Iida and Akashi 2000) and other species (Duret 2002; Pond and Muse 2005); although it is clear that selection has acted upon synonymous mutations in the past in D. melanogaster, the evidence of selection currently acting is contradictory (Akashi 1996; McVean and Vieira 2001; Zeng and Charlesworth 2010) and biased gene conversion may be acting (Galtier et al. 2006; Zeng and Charlesworth 2010). Most of the other species we have analyzed have not been investigated in any detail. We need to consider two models. In the first model, let us assume that there is no variation in Ne but that there is variation in the strength of selection on synonymous codons. Such a model would generate apparent variation in Ne with the genes subject to the strongest selection apparently having the highest Ne, because negative selection affects divergence to a greater extent than polymorphism (Kimura 1983). However, this would lead to the regions of the genome with the lowest diversity apparently having the highest effective population size. This is clearly not the case. If we split Ps into two independent samples, using a hypergeometric distribution, then we find a positive correlation between our estimate of Ne and Ps (Table 3). In the second model, let us imagine that there is variation in Ne and variation in the strength of selection on codon usage bias, but that they are uncorrelated to each other. In this case selection on codon usage bias will tend to generate an overestimate of the variation in Ne: as Ne increases, selection becomes more effective, but this reduces the divergence more than the level of polymorphism, yielding a higher apparent effective population size. So genes in regions of high Ne will tend to have an exaggerated Ne. There is also another effect that needs to be considered. We have estimated the level of synonymous divergence using the method of Goldman and Yang (1994; Yang and Nielsen 1998), which assumes that codon bias is due to mutation bias; however, this method will tend to overestimate the synonymous substitution rate if codon bias is due to selection, because it will incorrectly infer that genes with high bias have a small number of synonymous sites and hence a relatively large number of substitutions (Bierne and Eyre-Walker 2003; Yang 2006). As a consequence, the divergence in high-biased genes will be overestimated, but at the same time the mutation rate will tend to be underestimated because of the action of selection. These two factors may cancel each other out.
Fourth, we have applied our method only to protein-coding sequences, so we are estimating the variation in the effective population size that applies to the proteome. There might be further variation in Ne in regions that are relatively devoid of protein-coding sequences, such as heterochromatin. Whether this is important depends on whether there are functional sequences within these regions. We have also considered genes only on the autosomes and occasionally the homogametic sex chromosome (14 loci in H. sapiens). We have not considered genes on the heterogametic sex chromosome, which often appear to have much lower effective population sizes. However, the heterogametic sex chromosome usually has very few genes (Graves 2006).
Fifth, in estimating the variation in Ne we have assumed that there is either free recombination or no recombination and the population size has been stationary. Variation in population size can generate variation in diversity between loci, which may for example be mistaken for the signature of genetic hitchhiking (Tajima 1989; Pluzhnikov et al. 2002). In principle we could take this into account by estimating a demographic model from the polymorphism data while simultaneously estimating the variation in Ne. This is difficult and is beyond the scope of the current work.
Finally, we have not taken into account ancestral polymorphism within our method. Ignoring ancestral polymorphism will lead us to underestimate the variation in Ne because loci with large Ne will tend to have higher divergences than loci with small Ne and this will appear as though these loci have higher mutation rates; variation in Ne will therefore be underestimated because the mutation rate has been overestimated. In principle it is possible to include ancestral polymorphism within the method, but we observe a lack of convergence, probably because the number of polymorphisms for each gene was so low. However, we have chosen data sets in which divergence is generally considerably larger than diversity; for example, we chose macaque as the outgroup to humans because variation in Ne does appear to generate variation in the divergence between human and chimpanzee (McVicker et al. 2009).
Despite finding variation in Ne in many of the species we tested, we find no consistent evidence that Ne is correlated to either the rate of recombination or the density of selected sites, the two factors that we would have expected variation in Ne to depend upon. This is probably in part due to the fact that we are using synonymous diversity; as such, our estimates of diversity are subject to considerable error. The lack of a strong correlation between recombination rate and Ne may also be due to the fact that the genetic maps in A. thaliana and mouse are relatively crude. Furthermore, for our mouse species we are using an F2 genetic linkage map constructed from intercrosses between M. m. domesticus and M. m. castaneus to infer recombination rates for M. m. castaneus. In humans it has previously been shown that diversity over divergence is correlated positively to recombination rate (Hellmann et al. 2005) and that dn/ds is correlated to gene density (Bullaughey et al. 2008). In contrast to Hellmann et al. (2005), we do not find a significant correlation between Ne and recombination rate, but they used long noncoding sequences to investigate diversity over divergence; their estimates were therefore subject to much less error than ours. It is surprising that there is a correlation between genic density but not conserved site density in humans. This might be due to the fact that there is approximately twice as much variation in genic density as in conserved site density (coefficient of variation: 0.79 vs. 0.30). It might also be due to differences in the DFE between the two types of sites: background selection is most effective when the strength of selection acting upon deleterious mutations is similar in magnitude to the rate of recombination (Nordborg et al. 1996).
In contrast, genetic hitchhiking depends upon the rate of advantageous mutation and sequences undergoing considerable adaptive evolution may not appear as conserved. The correlation between Ne and the density of genic sites may therefore suggest that hitchhiking is more important in generating variation in Ne than background selection. The lack of a correlation between Ne and the density of selected sites in Drosophila, once correlations to the rate of recombination have been taken into account, may reflect the fact that the variation in Ne is generated by genetic hitchhiking and a lot of adaptive evolution goes on outside coding sequences (Andolfatto 2005).
Across species we find evidence that variation in Ne leads to variation in the effectiveness of natural selection on nonsynonymous mutations across the genome (Table 5). However, this is individually significant for just two genomes: C. grandiflora and A. lyrata. A lack of a correlation in other genomes may be due to the fact that we have little power to detect the correlation since (i) some of the data sets are quite small, (ii) there is limited variation in Ne, and (iii) in most of these species the DFE is very leptokurtic. The kurtosis of the DFE is such that changes in effective population size do not greatly change the proportion of mutations that are effectively neutral. It can be shown that under a gamma DFE the proportion of effectively neutral mutations is proportional to (Ohta 1977; Kimura 1979, 1983; Welch et al. 2008). Since β-values are typically between 0.1 and 0.3 in most species (Table S2), changes in Ne tend to cause small changes in the proportion of effectively neutral mutations; for example, a 10-fold increase in effective population size will reduce the proportion of effectively neutral mutations by only 37% if β = 0.2. We find no evidence of a significant negative correlation between ψ and either θS or Ne in humans, in agreement with the work of Bullaughey et al. (2008). They found no evidence that the ratio of the nonsynonymous (dN) to the synonymous (dS) substitution rate between human, chimpanzee, and macaque was correlated to the rate of recombination.
Table 5 . The correlation of Pn/(Ps + 1) = ψ and θs and Ne, respectively, in 10 eukaryotic species.
| ψ vs. θs (groups of 4) | ψ vs.Ne (groups of 4) | |||||
|---|---|---|---|---|---|---|
| Species | n | ρ | P-value | n | ρ | P-value |
| D. melanogaster | 77 | −0.172 | 0.067 | 77 | −0.1 | 0.194 |
| H. sapiens | 110 | −0.068 | 0.239 | 110 | 0.016 | 0.564 |
| M. m. castaneus | 18 | −0.253 | 0.155 | 18 | −0.261 | 0.147 |
| A. thaliana | 231 | 0.051 | 0.781 | 231 | 0.055 | 0.799 |
| C. grandiflora | 64 | −0.357 | 0.002 | 64 | −0.483 | 2.673 × 10−5 |
| S. bicolor | 35 | 0.093 | 0.702 | 35 | 0.001 | 0.504 |
| B. stricta | 33 | 0.164 | 0.818 | 33 | −0.168 | 0.175 |
| A. lyrata | 18 | −0.477 | 0.022 | 18 | −0.507 | 0.016 |
| C. rubella | 13 | 0.451 | 0.939 | 13 | 0.491 | 0.955 |
| S. paradoxus | 25 | −0.219 | 0.146 | 25 | −0.019 | 0.462 |
| Combined (Z method) | 0.043 | 0.021 | ||||
The nonindependence of ψ and θs is taken into account by splitting the data set into independent halves (see Materials and Methods). Correlation coefficients (ρ) and P-values (one-tailed) are given for each species.
We find evidence that the amount of variation in Ne varies between species; however, there are no obvious correlates of this variation. Both plants and animals have species with high and low levels of variation. Surprisingly we find no obvious effect of self-fertilization as suggested by previous studies (Cutter and Payseur 2003; Roselius et al. 2005). A. thaliana, C. rubella, and Boechera stricta are all self-fertile with selfing rates of ∼0.95, 1, and 0.94, respectively (Charlesworth and Vekemans 2005; Song et al. 2006; Foxe et al. 2009), whereas the closely related species A. lyrata and C. grandiflora are obligate outcrossing species. However, the variation in Ne seems to be relatively low for C. grandiflora and B. stricta and similar for the two Arabidopsis species. It also should be noted that the confidence intervals on the estimate of Ne in C. rubella are very large and a substantial amount of variation is still shared between C. grandiflora and C. rubella so these estimates are not independent. Moreover, the lack of an effect for self-compatibility in our estimates of Ne for Arabidopsis may be not surprising as self-compatibility might evolved relatively recently in Arabidopsis (Bechsgaard et al. 2006; Tang et al. 2007). Furthermore, both Arabidopsis species have high sequence diversity in pericentromeric regions (Borevitz et al. 2007; Kawabe et al. 2008) that is not caused by varying mutation rates. Therefore this could be a major determinant of variation in Ne in those species and interfere with the effects of the breeding system.
Although the variation we observe in the effective population size appears to be modest, it does appear to influence both the level of neutral genetic diversity and the effectiveness of selection. This potentially has important implications. If slightly deleterious mutations contribute substantially to phenotypic traits, then variation in the effective population size may affect where the genetic variation underlying fitness and other traits is distributed. For example, Rockman et al. (2010) have recently shown that expression QTL (eQTL) tend to be present in regions of the Caenorhabditis elegans genome with the highest rates of recombination and lowest density of genes, where Ne is expected to be largest. However, population genetic theory also suggests that such weakly selected mutations are unlikely to contribute much to the overall genetic variance in fitness unless the proportion of mutations under such weak selection is large (Eyre-Walker 2010). Variation in the effective population might also affect the rate of adaptive evolution, as appears to be the case in Drosophila (Betancourt and Presgraves 2002). Advantageous mutations can potentially come from three sources. They can be generated de novo, in which case we expect regions of the genome with large Ne to adapt faster because the number of chromosomes an advantageous mutation can occur in is larger, and selection will be more effective on a greater proportion of the advantageous mutations. Advantageous mutations can also arise from standing genetic variation (Pritchard and Rienzo 2010; Pritchard et al. 2010). If these mutations were previously strongly deleterious, the genetic variation is not expected to depend upon Ne, unless the mutations are highly recessive. If, however, the advantageous mutations were previously neutral or weakly selected, regions of the genome with high Ne are expected to have more genetic variation and hence adapt more rapidly.
Acknowledgments
We are grateful to several anonymous referees for comments. T.I.G. was financially supported by the John Maynard Smith studentship.
Literature Cited
- Aguileta G., Bielawski J. P., Yang Z., 2006. Evolutionary rate variation among vertebrate beta globin genes: implications for dating gene family duplication events. Gene 380: 21–29 [DOI] [PubMed] [Google Scholar]
- Akashi H., 1996. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics 144: 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andolfatto P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Arguello J. R., Zhang Y., Kado T., Fan C., Zhao R., et al. , 2010. Recombination yet inefficient selection along the Drosophila melanogaster subgroup’s fourth chromosome. Mol. Biol. Evol. 27: 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Charlesworth B., 2002. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416: 323–326 [DOI] [PubMed] [Google Scholar]
- Bechsgaard J. S., Castric V., Charlesworth D., Vekemans X., Schierup M. H., 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23: 1741–1750 [DOI] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520 [DOI] [PubMed] [Google Scholar]
- Berlin S., Ellegren H., 2006. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J. Mol. Evol. 62: 66–72 [DOI] [PubMed] [Google Scholar]
- Betancourt A. J., Presgraves D. C., 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99: 13616–13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Welch J. J., Charlesworth B., 2009. Reduced effectiveness of selection caused by a lack of recombination. Curr. Biol. 19: 655–660 [DOI] [PubMed] [Google Scholar]
- Bierne N., Eyre-Walker A., 2003. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics 165: 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J. O., Hazen S. P., Michael T. P., Morris G. P., Baxter I. R., et al. , 2007. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 12057–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullaughey K., Przeworski M., Coop G., 2008. No effect of recombination on the efficacy of natural selection in primates. Genome Res. 18: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2009. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Vekemans X., 2005. How and when did Arabidopsis thaliana become highly self-fertilising. BioEssays 27: 472–476 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Payseur B. A., 2003. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol. Biol. Evol. 20: 665–673 [DOI] [PubMed] [Google Scholar]
- Dumont B. L., White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome Res. 21: 114–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., 2002. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 12: 640–649 [DOI] [PubMed] [Google Scholar]
- Elyashiv E., Bullaughey K., Sattath S., Rinott Y., Przeworski M., et al. , 2010. Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species. Genome Res. 20: 1558–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., 1996. Synonymous codon bias is related to gene length in Escherichia coli: Selection for translational accuracy? Mol. Biol. Evol. 13: 864–872 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., 2010. Evolution in health and medicine Sackler colloquium: Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci. USA 107(Suppl. 1): 1752–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov D. A., Laporte V., Vitte C., Charlesworth D., 2001. DNA diversity in sex-linked and autosomal genes of the plant species Silene latifolia and Silene dioica. Mol. Biol. Evol. 18: 1442–1454 [DOI] [PubMed] [Google Scholar]
- Foxe J. P., Dar V.-u.-N., Zheng H., Nordborg M., Gaut B. S., et al. , 2008. Selection on amino acid substitutions in Arabidopsis. Mol. Biol. Evol. 25: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxe J. P., Slotte T., Stahl E. A., Neuffer B., Hurka H., et al. , 2009. Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA 106: 5241–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Bazin E., Bierne N., 2006. GC-biased segregation of noncoding polymorphisms in Drosophila. Genetics 172: 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Hammer M. F., 2006. Reconstructing human origins in the genomic era. Nat. Rev. Genet. 7: 669–680 [DOI] [PubMed] [Google Scholar]
- Goldman N., Yang Z., 1994. A codon-based model of nucleotide substitution for protein-coding dna sequences. Mol. Biol. Evol. 11: 725–736 [DOI] [PubMed] [Google Scholar]
- Gossmann T. I., Song B.-H., Windsor A. J., Mitchell-Olds T., Dixon C. J., et al. , 2010. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27: 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. A. M., 2006. Sex chromosome specialization and degeneration in mammals. Cell 124: 901–914 [DOI] [PubMed] [Google Scholar]
- Guo Y.-L., Bechsgaard J. S., Slotte T., Neuffer B., Lascoux M., et al. , 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D. L., Oliver F., Eyre-Walker A., Harr B., Keightley P. D., 2010. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 6: e1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., Casa A. M., Sun H., Murray S. C., Paterson A. H., et al. , 2006. Challenges of detecting directional selection after a bottleneck: lessons from Sorghum bicolor. Genetics 173: 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings W. K., 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57: 97–109 [Google Scholar]
- Hellborg L., Ellegren H., 2004. Low levels of nucleotide diversity in mammalian Y chromosomes. Mol. Biol. Evol. 21: 158–163 [DOI] [PubMed] [Google Scholar]
- Hellmann I., Ebersberger I., Ptak S. E., Pääbo S., Przeworski M., 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I., Prüfer K., Ji H., Zody M. C., Pääbo S., et al. , 2005. Why do human diversity levels vary at a megabase scale? Genome Res. 15: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., Kliman R. M., 2002. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics 160: 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A., Eyre-Walker A., 2011. Variation in the mutation rate across the mammalian genome. Nat. Rev. Genet. 12: 756–766 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguadé M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K., Akashi H., 2000. A test of translational selection at ‘silent’ sites in the human genome: base composition comparisons in alternatively spliced genes. Gene 261: 93–105 [DOI] [PubMed] [Google Scholar]
- Ingvarsson P. K., 2004. Population subdivision and the Hudson-Kreitman-Aguade test: testing for deviations from the neutral model in organelle genomes. Genet. Res. 83: 31–39 [DOI] [PubMed] [Google Scholar]
- Innan H., 2006. Modified Hudson-Kreitman-Aguade test and two-dimensional evaluation of neutrality tests. Genetics 173: 1725–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A., Forrest A., Wright S. I., Charlesworth D., 2008. High DNA sequence diversity in pericentromeric genes of the plant Arabidopsis lyrata. Genetics 179: 985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1979. Model of effectively neutral mutations in which selective constraint is incorporated. Proc. Natl. Acad. Sci. USA 76: 3440–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge/London/New York [Google Scholar]
- Kliman R. M., Hey J., 2003. Hill-Robertson interference in Drosophila melanogaster: reply to Marais, Mouchiroud and Duret. Genet. Res. 81: 89–90 [DOI] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D. F., Sainz J., Jonsdottir G. M., Gudjonsson S. A., et al. , 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247 [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Sackton T. B., Greenberg A. J., Wong A., Singh N. D., et al. , 2008. Evolution of protein-coding genes in Drosophila. Trends Genet. 24: 114–123 [DOI] [PubMed] [Google Scholar]
- Lercher M. J., Hurst L. D., 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337–340 [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2006. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., Bartolomé C., Nöel V., 2006. Estimating selection on nonsynonymous mutations. Genetics 172: 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C. A., Kliman R. M., Markert J. A., Hey J., 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19: 472–488 [DOI] [PubMed] [Google Scholar]
- Marais G., Mouchiroud D., Duret L., 2003. Neutral effect of recombination on base composition in Drosophila. Genet. Res. 81: 79–87 [DOI] [PubMed] [Google Scholar]
- McVean G. A., Vieira J., 2001. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics 157: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G., Gordon D., Davis C., Green P., 2009. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5: e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell H., Fridolfsson A. K., Ellegren H., 2001. Contrasting levels of nucleotide diversity on the avian Z and W sex chromosomes. Mol. Biol. Evol. 18: 2010–2016 [DOI] [PubMed] [Google Scholar]
- Moriyama E. N., Powell J. R., 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277 [DOI] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996. The effect of recombination on background selection. Genet. Res. 67: 159–174 [DOI] [PubMed] [Google Scholar]
- Nordborg M., Hu T. T., Ishino Y., Jhaveri J., Toomajian C., et al. , 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., 1977. Extension to the nearly neutral random drift hypothesis, pp. 148–167 Evolution and Polymorphism, edited by Kimura M. National Institute of Genetics, Mishima, Japan [Google Scholar]
- Piganeau G., Eyre-Walker A., 2009. Evidence for variation in the effective population size of animal mitochondrial DNA. PLoS ONE 4: e4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov A., Rienzo A. D., Hudson R. R., 2002. Inferences about human demography based on multilocus analyses of noncoding sequences. Genetics 161: 1209–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S. K., Muse S. V., 2005. Site-to-site variation of synonymous substitution rates. Mol. Biol. Evol. 22: 2375–2385 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 15: 1651–1656 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Rienzo A. D., 2010. Adaptation—not by sweeps alone. Nat. Rev. Genet. 11: 665–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Skrovanek S. S., Kruglyak L., 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330: 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselius K., Stephan W., Städler T., 2005. The relationship of nucleotide polymorphism, recombination rate and selection in wild tomato species. Genetics 171: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J., Wright S. I., Foxe J. P., Kawabe A., DeRose-Wilson L., et al. , 2008. Patterns of polymorphism and demographic history in natural populations of Arabidopsis lyrata. PLoS ONE 3: e2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K. J., Ramos-Onsins S., Ringys-Beckstein H., Weisshaar B., Mitchell-Olds T., 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169: 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C., Hubisz M. J., Lu J., et al. , 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., et al. , 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T., Fan Y., Chang H.-S., Zhu T., Hazen S. P., et al. , 2006. A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet. 2: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Foxe J. P., Hazzouri K. M., Wright S. I., 2010. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 27: 1813–1821 [DOI] [PubMed] [Google Scholar]
- Smith J. M., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- Song B.-H., Clauss M. J., Pepper A., Mitchell-Olds T., 2006. Geographic patterns of microsatellite variation in Boechera stricta, a close relative of Arabidopsis. Mol. Ecol. 15: 357–369 [DOI] [PubMed] [Google Scholar]
- Song B.-H., Windsor A. J., Schmid K. J., Ramos-Onsins S., Schranz M. E., et al. , 2009. Multilocus patterns of nucleotide diversity, population structure and linkage disequilibrium in Boechera stricta, a wild relative of Arabidopsis. Genetics 181: 1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Li H., 2007. The recent demographic and adaptive history of Drosophila melanogaster. Heredity 98: 65–68 [DOI] [PubMed] [Google Scholar]
- Stoletzki N., Eyre-Walker A., 2011. Estimation of the neutrality index. Mol. Biol. Evol. 28: 63–70 [DOI] [PubMed] [Google Scholar]
- Sundström H., Webster M. T., Ellegren H., 2004. Reduced variation on the chicken Z chromosome. Genetics 167: 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y.-L., et al. , 2007. The evolution of selfing in Arabidopsis thaliana. Science 317: 1070–1072 [DOI] [PubMed] [Google Scholar]
- Tenaillon M. I., U’Ren J., Tenaillon O., Gaut B. S., 2004. Selection vs. demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 21: 1214–1225 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai I. J., Bensasson D., Burt A., Koufopanou V., 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl. Acad. Sci. USA 105: 4957–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. J., Eyre-Walker A., Waxman D., 2008. Divergence and polymorphism under the nearly neutral theory of molecular evolution. J. Mol. Evol. 67: 418–426 [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 2005. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J. Evol. Biol. 18: 1368–1373 [DOI] [PubMed] [Google Scholar]
- Woolfit M. R. Q., 2006. Effective population size and its effects on molecular evolution. Ph.D. Thesis, University of Sussex, East Sussex, UK [Google Scholar]
- Wright S. I., Charlesworth B., 2004. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics 168: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff G. J., Li J., Wu C.-I., 2002. Molecular evolution of functional genes on the mammalian Y chromosome. Mol. Biol. Evol. 19: 1633–1636 [DOI] [PubMed] [Google Scholar]
- Yang Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yang Z., 2006. Computational Molecular Evolution. Oxford University Press, New York [Google Scholar]
- Yang Z., Nielsen R., 1998. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 46: 409–418 [DOI] [PubMed] [Google Scholar]
- Zeng K., Charlesworth B., 2010. Studying patterns of recent evolution at synonymous sites and intronic sites in Drosophila melanogaster. J. Mol. Evol. 70: 116–128 [DOI] [PubMed] [Google Scholar]
- Zhang J., Webb D. M., Podlaha O., 2002. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics 162: 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]