Abstract
Actin dynamics is a tightly regulated process involved in various cellular events including transport carrier biogenesis such as clathrin-, AP1-coated carriers connecting the trans-Golgi network (TGN) and the endocytic pathway. However, the mechanisms coordinating coat assembly, membrane and actin remodeling during post-TGN transport remain poorly understood. Here, we show that the ARF1 GTPase synchronizes the TGN association of clathrin-AP-1 coats and protein complexes comprising CYFIP (Sra, PIR121), a clathrin heavy chain binding protein associated with mental retardation. The RAC1 GTPase and its exchange factor β-PIX activate these complexes allowing N-WASP- and ARP2/3-dependent actin polymerization towards membranes thus promoting tubule formation. These phenomena can be recapitulated using synthetic membranes. This protein-network-based mechanism facilitates the sequential coordination between ARF1-dependent membrane priming via recruitment of coats and CYFIP (Sra, PIR121)-containing complexes, and RAC1-dependent actin polymerization and provides complementary but independent levels of regulation during early stages of clathrin-, AP1-coated carrier biogenesis.
Keywords: Clathrin, AP-1 assembly proteins, CYFIP, Wave/Scar, actin, X-linked mental retardation
In mammalian cells, clathrin and the AP-1 adaptor mediate the trafficking of specific cargos, including the mannose 6-phosphate receptors (MPRs) and their bound lysosomal enzymes, from the TGN to the endosomal system1–3. The small adenosine diphosphate ribosylation factor 1 (ARF1) GTPase plays a central role in clathrin-, AP-1 coat recruitment onto membranes. GTP-ARF1 activates phosphatidylinositol (PI)-4 kinases4,5 and, together with PI-4P and sorting signals present in cytoplasmic domains of selected cargos, binds AP-16–8, leading to coat recruitment to membranes followed by membrane remodeling and transport carrier formation. Actin dynamics also regulates post-Golgi transport9,10 while maintaining Golgi integrity in non-dividing cells11 and controlling Golgi dynamics during cell division12. The actin cytoskeleton and its regulators huntingtin-interacting protein 1-related protein (HIP1R)13 and cortactin14 are involved in both clathrin-, AP-1- and endocytic, clathrin-, AP-2-coated vesicle biogenesis15–17. Whether actin dynamics at these different locations or during the cell cycle requires the same protein networks and the same regulatory mechanisms is unclear.
A key regulator of actin nucleation is the ARP2/3 complex18. Its activity at various cellular locations depends on Rho GTPase-regulated factors19 comprising WASP, N-WASP (Wiskott-Aldrich syndrome proteins), WAVE1-WAVE3 (WASP family Verprolin-homologous proteins) and WHAMM (WASP homolog associated with actin, membranes, and microtubules)20. These related WAVE and WASP proteins associate with a complex formed of ABI1 (Abelson-interacting protein), NAP1 (Nck-associated protein or p125NAP1), HSPC 300 (BRICK), and CYFIP1 and CYFIP2 (cytoplasmic Fragile X-mental retardation interacting proteins 1 and 2; or Sra1 and PIR121 respectively)19 that interact with the fragile X mental retardation protein21. This protein complex regulates RAC1-dependent WAVE2 function at cellular leading edges during cell motility22–25. ABI1 also binds N-WASP and plays a role in endocytosis following CDC42-mediated activation26,27. Thus, the activity of this CYFIP-containing complex may require the combinatorial use of different proteins to promote actin polymerization at various locations.
Although it is clear that membrane and actin remodeling are intimately linked, the mechanisms coordinating these processes remain poorly understood. We have previously identified, among the 40 proteins associated in vitro with clathrin-, AP-1 coated membranes, the ARP2/3 complex, WAVE proteins and a protein complex containing CYFIP1, CYFIP2, ABI1, ABI2, and NAP18. Here, we demonstrate the functional significance of ARF1, clathrin, CYFIP1, CYFIP2, N-WASP, WASP, RAC1 and its exchange factor β-PIX (ARHGEF7) as key components of the different cellular machines shaping clathrin-, AP-1-coated carriers and regulating their biogenesis.
RESULTS
Subcellular distribution of CYFIP1 and CYFIP2
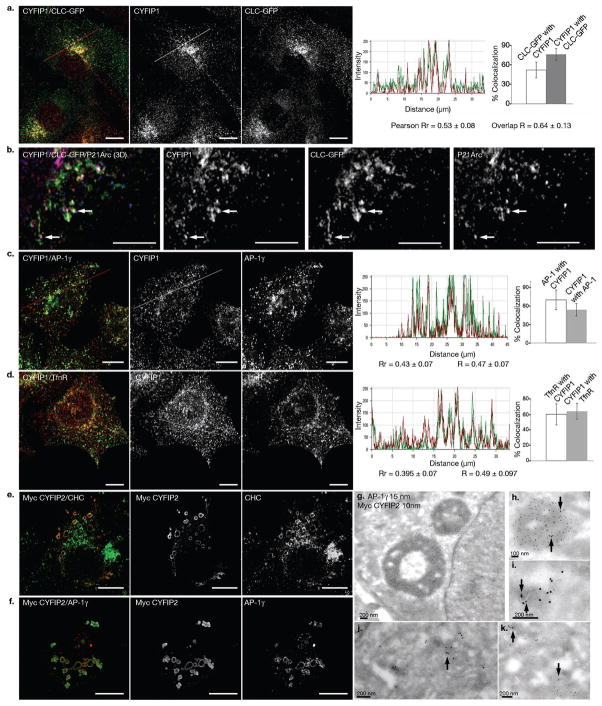
We first analyzed the cellular distribution of CYFIP1 and CYFIP2, which are believed to form heterodimers28. Statistical image analysis indicated that half of the endogenous CYFIP1 was detected in the perinuclear region on clathrin- and AP-1-positive structures (Fig. 1a-c) containing part of the transferrin receptor (TfnR) (Fig. 1d). These structures also contained a transmembrane protein made of GFP fused to the transmembrane and cytoplasmic domains of the cation-independent MPR (here referred to as GFP-MPR tail), which mostly localizes to the TGN of HeLa cells29 (Fig. 2a). A significant amount of P21-Arc, a subunit of the ARP2/3 complex, was detected on these clathrin- and CYFIP-coated structures (Fig. 1b). In the absence of anti-CYFIP2 antibodies, a myc-tagged CYFIP2 was expressed in HeLa cells. Myc-CYFIP2 was detected on enlarged, perinuclear, clathrin-, AP-1-coated compartments (Fig. 1e, f), also containing CYFIP1 and Rab11 (Supplementary Information, Fig. S1b, c). They also contained a fraction of the endogenous cation-independent MPR (Supplementary Information, Fig. S1a) and the TfnR (Supplementary Information, Fig. S1d), although most of these receptors were detected in small vesicular structures surrounding the CYFIP2-positive compartments, suggesting that their trafficking is affected by CYFIP2 over-expression. However, these enlarged structures remained devoid of early EEA1 and late LAMP-2 endosomal markers (Supplementary Information, Fig. S1e, f). The GM-130 positive cis-Golgi remained unaffected (data not shown). Similar CYFIP2- and AP-1-positive enlarged structures were detected in CYFIP2 expressing cells using electron microscopy (Fig. 1g-k). Expression of GFP-CYFIP1 also induced formation of similar, enlarged structures (data not shown). Thus, endogenous CYFIP1 and ectopically expressed CYFIP2 associate with the TGN and to a lower extent with recycling endosomes where clathrin-, AP-1 coats are found30.
Figure 1. Localization of CYFIP1 and CYFIP2.
(a, b) BSC-1 cells stably expressing clathrin light chain (CLC)-EGFP were labeled with antibodies against CYFIP1 (red) and (b) P21-Arc (blue). (c, d) HeLa cells were labeled with antibodies against CYFIP1 (red) and (c) AP-1γ (green) or (d) transferrin receptor (green). (e, f) HeLa cells transiently expressing myc-tagged CYFIP2 were labeled with antibodies against myc, (e) clathrin heavy chain (CHC, green) or (f) AP-1γ (red). Co-localization was analyzed and quantified with Volocity 5.2 software. R represents the overlap coefficients, Rr the Pearson correlation coefficients. Data is presented as mean ± s.d. (in each case, 20 – 25 cells from n= 3 different experiments). Scale bars, (a, c-f) 10 μm, (b) 5 μm. (g-k) HeLa cells transiently expressing myc-tagged CYFIP2 were processed for electron microscopy. Thawed cryosections were co-labeled with anti-AP-1 (15 nm gold particles) and anti-myc antibodies (10 nm gold particles). (g, h) Arrows indicate AP-1 present on myc-tagged CYFIP2 enlarged intracellular structures, or (i-k) AP-1 and CYFIP2 co-localization on intracellular membranes.
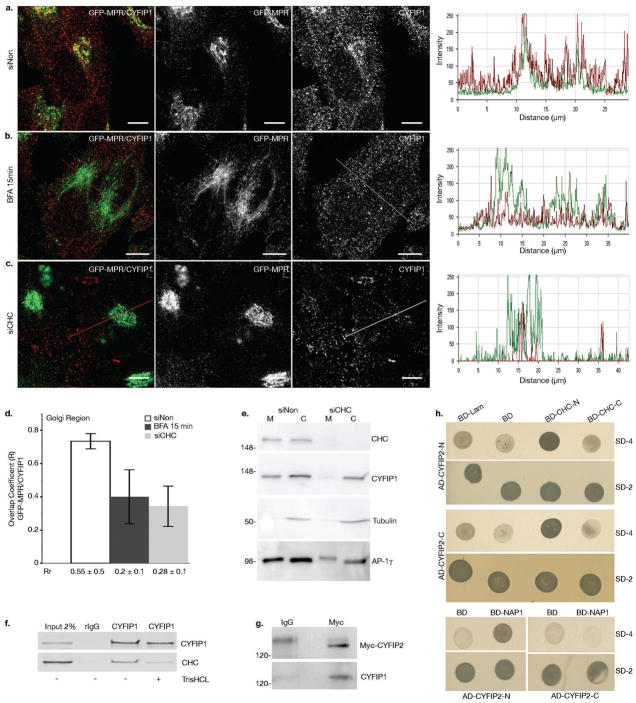
Figure 2. CYFIP1 and CYFIP2 interact with CHC and CYFIP1 recruitment to the TGN is regulated by ARF1.
(a-d) HeLa cells stably expressing GFP-MPR were treated with (a) control siRNA or (c) siRNAs to deplete CHC or (b) incubated with 5 μg ml−1 BFA for 15 min. Cells were then labeled with anti-CYFIP1 (red), and (d) the overlap (R) and Pearson correlation (Rr) coefficients between GFP-MPR and CYFIP1 in the TGN region were quantified for each condition (20 cells from n = 3 independent experiments were analyzed per condition; data represent the mean ± s.d.). Scale bars, 10 μm. (e) The membrane (M) and cytosolic (C) fractions of HeLa cells incubated either with siNon or with siCHC were analyzed by western blot (n = 3 independent experiments). (f) COS-7 cell lysates were incubated with anti-CYFIP1 or with control pre-immune rabbit IgG. Beads were washed with buffer with or without 0.5 M TrisHCl (pH 7.4), which induces clathrin cage depolymerization. The presence of CHC and CYFIP1 in the immunoprecipitates was determined by western blotting using the corresponding antibodies. CHC was co-immunoprecipitated with CYFIP1 only in the absence of TrisHCl (n = 3 independent experiments). (g) Lysates of HeLa cells transiently expressing myc-tagged CYFIP2 were incubated with anti-myc or control mouse IgGs and the immunoprecipitates were analyzed by western blotting. Full scans of all gels are shown in Supplementary Information, Fig. S9. (h) The N-terminal (AD-CYFIP2-N, AA 2–623) and the C-terminal domains C (AD-CYFIP2-C, AA 674–1299) of CYFIP2 were expressed as fusions with GAL4AD (pGADT7). The N-terminal (BD-CLC-N, AA 1–690) and the C-terminal (BD-CLC-C, AA 821–1679) halves of clathrin heavy chain, as well as (i) full-length Nap1 were fused to the DNA-BD (pGBKT7), and co-expressed with the GAL4AD containing plasmids. Interactions were detected by growth on agar plates lacking leucine and tryptophane (SD-2), or adenine, histidine, leucine and tryptophane (SD-4). Plasmids expressing either fusion of lamin C to the DNA-BD (BD-Lam) or DNA-BD alone (BD) were used as negative controls. N = 3 independent experiments.
CYFIP interacts with clathrin and ARF1 regulates its recruitment
The recruitments of clathrin-, AP-1 coats and the CYFIP-containing complexes on synthetic membranes are coordinated, presumably via interactions between components of the two machineries8. To address this issue, we treated cells with Brefeldin A (BFA), which prevents the ARF1 activation required for clathrin-, AP-1 recruitment. Statistical image analysis indicated that this treatment also resulted in a loss of CYFIP1 from the TGN containing most of the GFP-MPR tail (Fig. 2a, b, d). This prompted us to perform co-immunoprecipitation experiments from cell lysates. Anti-CYFIP1 co-immunoprecipitated a significant fraction of clathrin heavy chain (CHC), and anti-myc co-immunoprecipitated a fraction of CYFIP1 from lysates of myc-CYFIP2 expressing cells (Fig. 2f, g4). The N- and C-terminal domains of CYFIP2, both containing putative, evolutionary conserved CHC binding motifs also present in CYFIP1 (Supplementary Information, Fig. S2), were found to interact with the N-terminal domain of CHC in a yeast two-hybrid system (Fig. 2h). The known interaction between CYFIP2 and NAP1 was used as a control. Consistent with these findings, the siRNA-mediated CHC knockdown resulted in a loss of CYFIP1 from the GFP-MPR tail-containing TGN as observed by fluorescence microscopy and statistical image analysis (Fig. 2c, d), and from membranes as detected by cell fractionation (Fig. 2e). Thus, clathrin-, AP-1 coats and CYFIP-containing complexes were physically linked via CHC-CYFIP interactions thereby explaining why ARF1 regulated their concomitant recruitment onto TGN membranes.
CYFIP activity requires N-WASP and RAC1
The CYFIP-containing complexes appear to be versatile, being able to associate with various downstream components in order to activate actin polymerization19. Therefore, it was critical to determine which components regulate actin polymerization together with these modules at the TGN. We hypothesized that the formation of enlarged structures triggered by CYFIP2 expression resulted from higher actin polymerization around membranes. Indeed, these structures were enriched in P21-Arc and surrounded by F-actin (Supplementary Information, Fig. S3a–c). We reasoned that the knockdown of any component functioning together with CYFIP2 in actin polymerization would prevent the formation of these enlarged structures. Actin polymerization at the TGN may involve two GTPases previously shown to associate with clathrin-, AP-1 membranes8: first, CDC42 that activates ARP2/3 and is itself recruited in an ARF1-dependent manner by a yet unknown mechanism31,32 and, second, RAC1 that binds to CYFIP21,33. The knockdown of RAC1, but not that of CDC42, reduced the number of CYFIP2-enriched structures, as did Latrunculin B, which prevents actin polymerization (Supplementary Information, Fig. S3e, f, h, j). Moreover, RAC1 was detected on these enlarged structures (Supplementary Information, Fig. S3d). Our proteomic studies using brain cytosol also detected WAVE1 and WAVE3, proteins predominantly expressed in brain19. In HeLa cells, their counterpart WAVE2 may activate ARP2/3. However, only the knockdown of N-WASP or WASP, but not that of WAVE2 reduced the number of CYFIP2-positive large structures (Supplementary Information, Fig. S3i, j). Together, these results indicate that CYFIP2 and RAC1 regulate actin nucleation at the TGN with N-WASP or WASP.
RAC1 and its exchange factor β-PIX regulate ARP2/3 recruitment onto the TGN
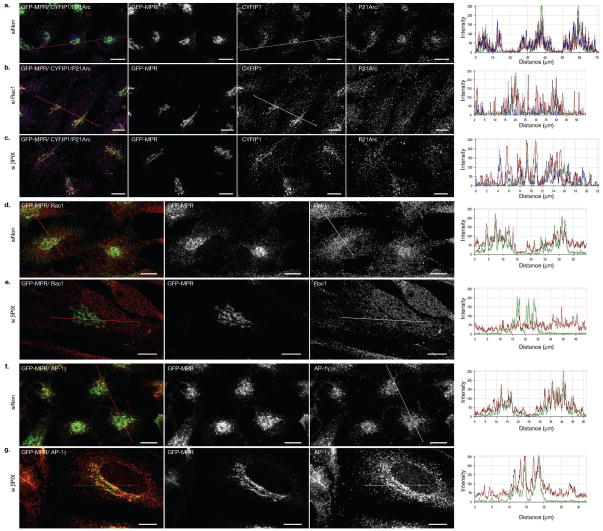
RAC1, when activated by a specific exchange factor, could regulate the membrane association of CYFIP-containing complexes, their activity in ARP2/3-dependent actin polymerization or both. We previously identified β-PIX as a unique Rho-GTPase exchange factor associated with clathrin-, AP-1 coats8. Statistical image analyses indicated that the knockdown of RAC1 (70% inhibition) reduced the amount of P21-Arc, but not that of CYFIP1, on the GFP-MPR tail-positive TGN (Fig. 3a, Supplementary Information, Fig. S3g and Fig. S4a, b). Moreover, the knockdown of β-PIX (90% inhibition) resulted in a loss of RAC1 and P21-Arc from the GFP-MPR tail-rich perinuclear compartments without affecting the distributions of CYFIP1 and AP-1 (Fig. 3b-c, Supplementary Information, Fig. S4c, d). The siRNA-mediated knockdown of CYFIP1 or CYFIP2 (Fig. 4a, b, d; Supplementary Information, Fig. S4e–g) also resulted in a significant loss of P21-Arc from the TGN. No changes were detected in the cell periphery. It is worth noting that the knockdown of CYFIP2 did not result in a complete loss of CYFIP1 and did not drastically affect the stability of NAP1, another component of the CYFIP-containing protein complex (Supplementary Information, Fig. S4f, g), an observation which differs from others27. Thus, RAC1 and β-PIX do not regulate the membrane association of CYFIP-containing complexes but rather their ability to recruit ARP2/3 complexes as nucleators of actin polymerization.
Figure 3. RAC1 and β-PIX control the recruitment of P21-Arc but not CYFIP1 to the TGN.
GFP-MPR expressing HeLa cells incubated with the indicated siRNAs were labeled with (a) anti-CYFIP1 (red) and anti-P21-Arc (blue), (b) anti-RAC1 (red) or anti-AP-1γ (red) and examined by confocal microscopy. 20–25 cells from n = 3 independent experiments were analyzed in each case. Scale bars, 10μm.
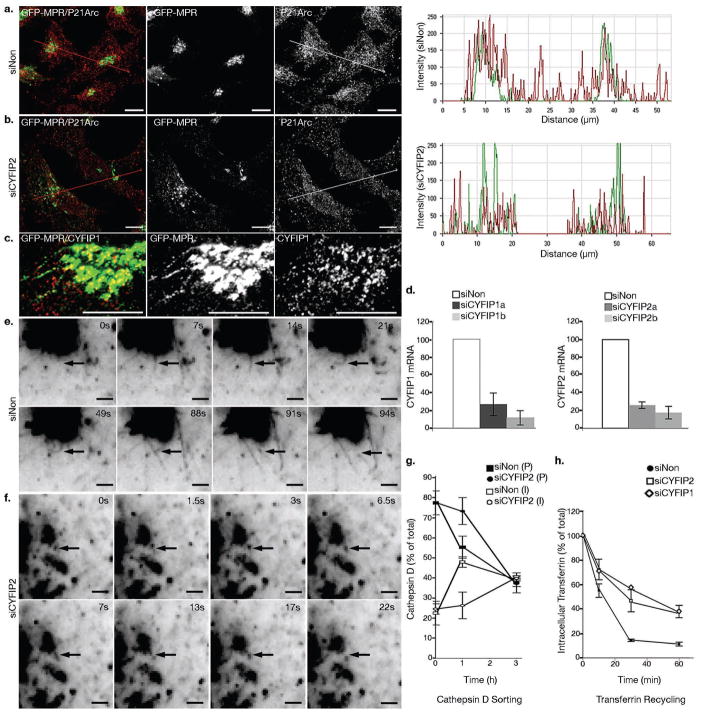
Figure 4. CYFIP2 depletion disrupts organelle integrity and decreases transport carrier biogenesis.
GFP-MPR expressing HeLa cells were treated with control siRNAs or with siRNAs targeting CYFIP2 or CYFIP1. (a) Cells were labeled with anti-P21-Arc (red) and co-localization between GFP-MPR and P21-Arc in the TGN region was analyzed. 20 cells from n = 3 independent experiments were analyzed for each condition. (c) Confocal fluorescence analysis indicated that CYFIP1 localized along GFP-MPR tubules (n = 20 cells). Scale bars, 10 μm. (d) The levels of CYFIP1 and CYFIP2 mRNAs following the indicated knock-downs were measured by Q RT-PCR; GAPDH was used as a control (n = 4 independent experiments; data represent the mean ± s.d.). (e, f) Cells were examined by time-lapse videomicroscopy. The number of GFP-MPR positive tubules exiting from the GFP-MPR rich compartment was analyzed as shown in Table I. Scale bars, 2 μm. (g) HeLa cells were labeled with 35S Methionine for 30 min, then pulse-chased for the indicated times. Cathepsin D was immunoprecipitated from the lysates and analyzed by SDS-PAGE. The relative levels of the precursor (P) and intermediate (I) forms were quantified relative to the total Cathepsin D levels (n = 3 independent experiments; data represent the mean ± s.d.). (h) For measuring Tfn recycling, cells were starved for 1 h, then incubated with fluorescent-labeled Tfn for 1h, and chased at 37°C in the presence of unlabeled Tfn for the indicated times. The amount of intracellular labeled Tfn was measured by fluorescence microscopy and quantified using ImageJ (n = 3 independent experiments for siNon and siCYFIP2; data represent the mean ± s.d.).
CYFIP1/2, RAC1, β-PIX and N-WASP promote clathrin/AP-1-coated carrier biogenesis in vivo
MPR trafficking depends on pleomorphic, clathrin-, AP-1-coated tubular carriers that form at the TGN in an actin- and microtubule-dependent manner29. Endogenous CYFIP1 was detected at proximal sites and along most of the GFP-MPR tail-labeled tubules (77.6 ± 7.8%; n = 20 cells) still connected with the TGN (Fig. 4c). Therefore, we analyzed the formation of these tubules in cells depleted of different regulators of actin polymerization. Consistent with our previous studies29, an average of 4.3 ± 1 GFP-MPR tail labeled tubules with an average length of 5.9 ± 2.8 μm were formed within 2 min in control cells (Fig. 4e, Table I and Supplementary Information, Movie 1). Both the number and the length of these tubules were significantly reduced in cells depleted of CYFIP1, CYFIP2, N-WASP, RAC1 or β-PIX by siRNA treatment (Fig. 4f, Table I and Supplementary Information, Movies 2, 4–9). In contrast, the knockdown of CDC42 had a milder effect on the dynamics of tubules that contained GFP-MPR tails (Table I) although it affected the integrity of the TGN containing GFP-MPR tails (Supplementary Movie 8) or endogenous MPR, as did the knockdowns of CYFIP1 or CYFIP2, N-WASP, RAC1 (Supplementary Information, Fig. S5, S6a), or ABI1 (not shown). CYFIP1 or CYFIP2 knockdowns also resulted in the redistribution of the GFP-MPR tail to TfnR-rich compartments, presumably recycling endosomes (Supplementary Information, Fig. S4h and Fig. S6b). However, the GM-130-positive cis-Golgi (Supplementary Information, Fig. S6d), as well as the integrity of microtubules (not shown), remained unaffected. In contrast, the knockdown of WAVE2 resulted in a compaction of the TGN (Supplementary Information, Fig. S5), thereby suggesting that a balance between the activity of N-WASP and WAVE2 may regulate compartment integrity. We conclude that CYFIP1 and CYFIP2, RAC1, β-PIX and N-WASP are essential for maintaining the integrity of the TGN and for promoting the formation of GFP-MPR-containing tubular carriers from this organelle.
Table I. Biogenesis of GFP-MPR tail-containing tubular transport carriers in siRNA-treated HeLa cells.
GFP-MPR tail expressing cells were incubated with the indicated siRNAs for 72h. The exit of GFP-MPR tubules from the TGN region was monitored using time-lapse fluorescence microscopy. The interval between the acquisition of 2 consecutive images t = 500 ms. N = 5 independent experiments for siCYFIP2 and siCYFIP1 and n = 3 independent experiments for the other conditions; data represent the mean ± s.d.;
| No Tubules growing form the TGN/cell/2min | Tubule length (μm) | No Cells | |
|---|---|---|---|
| siNon | 4.3 ± 1.02 | 5.9 ± 2.8 | 40 |
| siCYFIP2 | 1.72 ± 0.99* | 2 ± 0.55* | 50 |
| siCYFIP1 | 1.53 ± 0.99* | 42 | |
| siN-WASP | 2.07 ± 0.77* | 1.74 ± 0.43* | 25 |
| siCDC42 | 3.05 ± 1.14 | 3.44 ± 1.61* | 47 |
| siRAC1 | 1.98± 0.81* | 2.35 ± 0.81* | 100 |
| siβ-PIX | 1.83± 1.2* | 1.59± 0.42* | 36 |
p < 0.0001, Anova single factor analysis, α = 0.05.
CYFIP regulates cargo transport
To address the functional importance of the CYFIP-containing module in biosynthetic transport, we performed classical pulse-chase experiments to monitor the MPR-dependent transport of the newly synthesized cathepsin D to lysosomes. This lysosomal enzyme is synthesized as a 55 kDa precursor that is then processed to a 45 kDa intermediate form upon delivery to acidified endosomal compartments. This processing was significantly reduced after CYFIP2 knockdown (Fig. 4g and Supplementary Information, Fig. S7a), to an extent similar to what has been observed after clathrin light chain knockdown34.
We also monitored the recycling of the cell surface GFP-MPR tail back to the TGN in GFP-MPR-tail expressing HeLa cells. Whereas in control cells the internalized anti-GFP antibodies rapidly localized to the perinuclear region together with the bulk of GFP-MPR tail, in CYFIP2 depleted cells they remained in peripheral structures (Supplementary Information, Fig. S7b, c). This would be consistent with the observation that in CYFIP1- or CYFIP2-depleted cells GFP-MPR tails are more abundant in TfrR-positive endocytic compartments (Supplementary Information, Fig. S6f). CYFIP1 or CYFIP2 knockdown also reduced the recycling rate of endocytosed transferrin, which accumulated in peripheral structures (Fig. 4h and Supplementary Information, Fig. S7d). However, the endocytosis of transferrin remained unaffected (Supplementary Information, Fig. S7e). Whereas it is possible that CYFIP1 and CYFIP2 are involved in transport from these peripheral structures, these results may also be due to indirect effects (e.g. TGN fragmentation).
Clathrin/AP-1 coated carrier biogenesis on model membranes
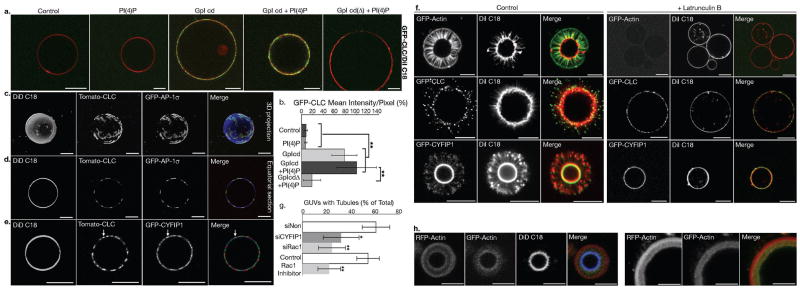
Clathrin-, AP-1 coats and CYFIP-containing actin nucleation modules bind onto model membranes minimally composed of sorting signals present in the cytoplasmic domains of specific cargos, ARF1 and PI-4P8. To image these model membranes and their bound components, we incubated functionalized giant unilamellar vesicles (GUVs) with pig brain cytosol spiked with cytosol of cells stably expressing various fluorescent-tagged proteins in the presence of GTP-γ-S, and then imaged these GUVs by fluorescence microscopy. Consistent with our previous studies, efficient clathrin binding depended on the presence of intact sorting motifs in cargo cytoplasmic tails (Fig. 5a, b). In the absence of ATP, clathrin and AP-1 co-assembled on these GUVs (Fig. 5c, d), where CYFIP1 was also present (Fig. 5e). In the presence of ATP, tubular membrane extensions labeled with the lipophilic dye DiI were detected (Fig. 5f and Supplementary Information, Fig. S8a and Movie 3). Clathrin was present at the tips of these tubules, as in HeLa cells29. CYFIP1 was also detected at the tips and along the tubules (Fig. 5f), as observed in HeLa cells (Fig. 4d). In the presence of Latrunculin B, as in the absence of ATP, these tubular membrane extensions were not formed, clathrin and CYFIP1 remaining on GUV surfaces. Similarly, the knockdown of CYFIP1 (50–60% inhibition) or RAC1 (70% inhibition) in GFP-actin expressing cells reduced tubule formation by more than 50%, as did NSC23766, a RAC1 inhibitor (Fig. 5g and Supplementary Information, Fig. S8b). A dense F-actin meshwork surrounded the tubules forming from GUVs (Fig. 5f). In pulse-chase experiments in which RFP-actin was added first, followed by GFP-actin, GFP-actin was detectable near the membrane, thus clearly indicating that actin polymerized towards the GUV membranes (Fig. 5h). We conclude that ATP-driven actin polymerization was sufficient to generate tubules from GUVs provided that clathrin coats and CYFIP-containing complexes were present on membranes and that RAC1 activated actin polymerization.
Figure 5. Reconstitution of clathrin-, AP-1-coated carrier biogenesis on model membranes.
(a) DiI C18 labeled giant unilamellar vesicles (GUVs), alone, or containing only PI-4P, only GpI cytoplasmic domains, or both GpI tails and PI-4P, or PI-4P and the GpI cytoplasmic domain devoid of sorting signals (GpIcdΔ), were incubated in the presence of GTP-γ-S with porcine brain cytosol spiked with cytosol of CLC-EGFP expressing cells. They were then imaged by confocal microscopy and (b) CLC-EGFP intensities were determined (n = 3 independent experiments; data represent the mean ± s.d.; pPI4P/no PI4P = 0.386, ≥ 7 GUVs per condition; pGpI/no GpI (no PI4P) = 9.88 × 10−9, 10GUVs per condition; pGpI/no GpI (with PI4P) = 2.18×10−7, 7 GUVs per condition; pGpI + PI4P/GpI (no PI4P) = 0.088, ≥ 7 GUVs per condition; pGpI/GpIcdΔ (with PI4P) = 2.5×10−23; ≥ 43 GUVs per condition; Anova single factor analysis). (c–e) DiD C18- labeled GUVs with GpI cytplasmic domains and PI-4P were incubated in the presence of GTP-γ-S and porcine brain cytosol spiked with a mixture of cytosols from cells expressing dTomato-CLC and EGFP-AP-1σ or GFP-CYFIP1. The samples were imaged by confocal microscopy. (f) GUVs with GpI tails and PI-4P were incubated, as in C, with an ATP regenerating system, in the absence (left panels) or the presence of 50 μM Latrunculin B (25 min) (right panels). (g) DiI C18-labeled GUVs with GpI tails and PI-4P were incubated with cytosols of EGFP-actin expressing HEK cells treated with the indicated siRNAs or with 100 nM RAC1 inhibitor (NSC23766). Actin polymerization and tubule formation were analyzed by confocal microscopy, and the number of GUVs displaying EGFP-actin tubes is shown as a percentage of the total DiI C18-positive GUVs (n = 3 independent experiments for siRNA-treated cells and n = 5 independent experiments for Rac1 inhibitor; data represent the mean ± s.d.; > 250 GUVs were analysed per condition; psiCYFIP1 = 0.02, psiRAC1 = 0.0017 compared with control siNon; pRac1 inhibitor = 0.002 compared to control cells; Anova single factor analysis). (h) DiD C18-labeled GUVs with GpI cytoplasmic domains and PI-4P were incubated at 37°C in the presence of GTP-γ-S and porcine brain cytosol spiked with cytosol from RFP-actin expressing HEK cells. After 15 min, cytosol from EGFP-actin expressing cells was added, and the GUVs were incubated for 10 additional min and analyzed by confocal microscopy. Scale bars: 10 μm.
DISCUSSION
Our study reveals that the biogenesis of clathrin-, AP-1 coated transport carriers results from the coordinated activities of coat components and actin nucleation machineries. It elucidates the functional links between ARF1, clathrin-, AP-1 coats, CYFIP-containing complexes, RAC1, its exchange factor β-PIX and N-WASP, which regulate ARP2/3-dependent actin polymerization, membrane remodeling and transport carrier biogenesis. Thus, our study provides a protein network-based mechanism that not only links different machineries at the TGN, but also allows the sequential coordination between ARF1 and RAC1 signaling. This mechanism provides complementary but independent levels of regulation during the early stages of post-Golgi transport.
Actin dynamics is involved in several aspects of post-Golgi transport; however, the associated protein networks are poorly characterized. Thus far, HIP1R, which binds to clathrin light chains has been shown to inhibit actin polymerization during post-Golgi transport of MPRs13,34 and endocytosis. Our study demonstrates the functional importance of an entire protein network in which the interaction between CHC and CYFIP plays a central role in coordinating coat assembly, actin polymerization and membrane remodeling during clathrin-, AP-1 coated carrier biogenesis. It remains possible that additional interactions exist between AP-1 subunits and other subunits of the CYFIP-containing complexes (ABI, NAP1 and HSPC 300). It is likely that these complexes bind to the edges of clathrin-, AP-1 coats, where CHC N-terminal domains protruding towards the membrane would be available for interactions with CYFIP. In contrast, HIP1R interacts with clathrin light chains located on the surface of clathrin coats. Therefore, the HIP1R covering the surface of clathrin coats may impair the accessibility of the CYFIP-containing complexes inside these coats, thus preventing actin polymerization at this location.
Actin dynamics at the Golgi complex is regulated by Rho GTPases whose activity is controlled by the ARF1 GTPase. However, the mechanisms connecting ARF1 and Rho signaling have thus far remained elusive. Membrane-bound ARF1-GTP recruits ARHGAP10, a CDC42-GAP, and therefore down-regulates ARP2/3 activity and F-actin dynamics through the control of Golgi-associated CDC42, thereby regulating Golgi integrity31. RAC1 is also recruited onto the Golgi in an ARF1-dependent manner35,36. Its over-expression leads to the formation of enlarged intracellular compartments surrounded by actin37, as observed here after CYFIP over-expression. Our study highlights a multi-step-process, in which ARF1 controls the recruitment of both clathrin-, AP1 coats and CYFIP-containing complexes, a process requiring CHC-CYFIP interactions. RAC1 is clearly dispensable for CYFIP recruitment onto TGN. On clathrin/AP-1 coated-model membranes we have also identified the ARF1-GAPs GIT1 or GIT2 and the Rho-GEF β-PIX8, known to form a complex38. This study illustrates a role for β-PIX in RAC1-dependent actin polymerization on TGN membranes. Similar to RAC1, it does not contribute to the recruitment of either clathrin/AP-1 coats or CYFIP-containing complexes. However after a β-PIX–mediated nucleotide exchange, activated RAC1 could in a later step bind to CYFIP21,33 thereby leading to N-WASP-, WASP- and ARP2/3-dependent actin polymerization, membrane remodeling and formation of clathrin-, AP-1 coated carriers. Thus, the control of ARF1-dependent RAC1 signaling at the TGN appears to be more complex than previously anticipated, implicating different, but interconnected levels of regulation based on different types of protein-protein interactions (Fig. 6). Several other studies have illustrated the role of Rho GTPases in post-Golgi transport. CDC42 regulates membrane traffic to the basolateral membrane of polarized cells39,40. The Golgi-associated CDC42 exchange factor FGD1 regulates post-Golgi transport of various cargos to the osteoblast surface and the formation of VSVG-containing tubules at the TGN41. Therefore, it would appear that RAC1 and CDC42 regulate actin dynamics and protein transport along different routes of post-Golgi traffic.
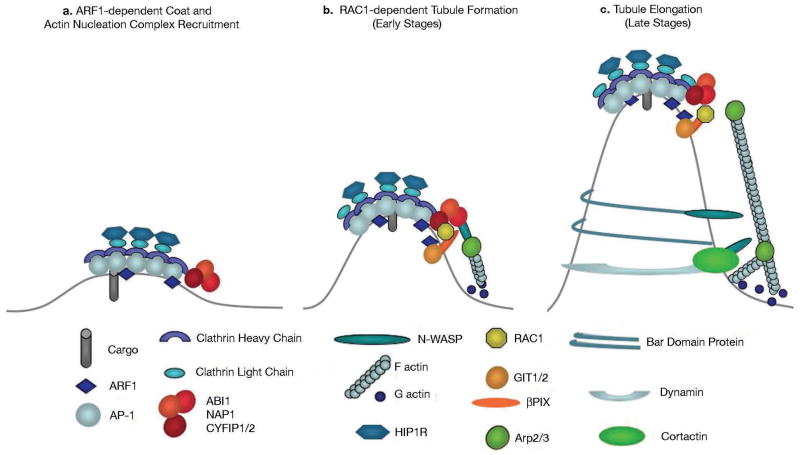
Figure 6. Model of clathrin-, AP-1-coated carrier biogenesis.
(a) ARF1 activation triggers the recruitment of AP-1 and clathrin onto TGN membranes leading to cargo sorting. The CYFIP, ABI, NAP1 complex is recruited at the edges of the clathrin-, AP-1 coated subdomains of the TGN. HIP1R binding to clathrin light chains could prevent actin polymerization on the surface of clathrin coats. (b) RAC1, activated by its GEF β-PIX, which forms a complex with the ARF1-GAP GIT1 and/or GIT2, binds to CYFIP and activates the actin nucleation complex leading to N-WASP-dependent activation of ARP2/3 and actin polymerization towards the TGN membrane during the initial stages of tubular carrier formation. (c) BAR domain proteins, dynamin2 and cortactin bind to tubular membranes. These molecules can bind N-WASP and thus sustain ARP2/3-dependent actin polymerization during tubule elongation and fission.
Actin polymerization is thought to occur during the late stages of transport carrier biogenesis. This notion arises from the molecules that connect membranes and the actin cytoskeleton identified thus far. Thus, the large GTPase dynamin2, which mediates carrier fission, binds the actin interacting protein cortactin during post-Golgi transport14 or endocytosis42. Several BAR domain containing proteins, which sense the curvature of tubular membranes, bind not only dynamin but also N-WASP43. Indeed, these proteins are detected by quantitative proteomics when tubular extensions are allowed to form from clathrin-, AP-1 coated synthetic membranes (our unpublished observation). Our study strongly suggests that actin polymerization also occurs during the early stages of clathrin-, AP-1 coated carrier biogenesis when TGN membranes have been primed with clathrin coats and their bound CYFIP-containing complexes. Actin polymerization, as shown here, occurs towards membranes and could thus provide the forces necessary to deform membranes in order to generate tubular transport carriers. It is likely that lipid modifications and/or partition also accompany membrane remodeling. BAR domain-containing proteins that bind to tubular membrane extensions could then stabilize this process and, together with N-WASP, further sustain actin polymerization. These molecular mechanisms could be similar to those regulating endocytosis, in which the recruitment of clathrin to the plasma membrane precedes those of N-WASP, ARP2/3 and actin, and actin polymerization towards membranes provides the forces needed for endocytic vesicle formation16,44–46.
While our study illustrates the functional importance of coordinating protein sorting, coat assembly, actin and membrane remodeling during post-Golgi transport, previous studies in Drosophila melanogaster have implicated CYFIP (Sra1, PIR) proteins in neuronal development47. Several proteins shown to connect ARF1-dependent clathrin-, AP-1 coat assembly and RAC1-dependent actin polymerization (8 and this study) are associated with human mental retardation. Mutations in genes encoding AP-1σ248,49 and p21 Activated Kinase 3 (PAK3)50, a kinase regulating β-PIX activity, cause the X-linked mental retardation syndrome. CYFIP1 and CYFIP2 link this pathway to the Fragile X mental retardation protein (FMRP) that is absent in the most common form of monogenic mental retardation21,51. This strongly indicates that clathrin-, AP-1-dependent sorting and actin polymerization may be perturbed in these cognitive disorders. In C. elegans, the AP-1μ1 subunit plays a role in polarized receptor trafficking to dendrites52,53, suggesting that dysfunctions of clathrin-, AP-1-dependent sorting supported by actin polymerization may impair normal trafficking of some neuronal receptors or cell adhesion molecules. Interestingly, in the absence of a functional FMRP, a CYFIP interactor, AMPA receptor (especially Glutamate Receptors 1 and 5) trafficking to dendrites is impaired54,55. Synaptic AMPA receptors are stored in recycling endosomes, from where they can be recruited to the surface of synapses upon stimulation56. Accordingly, AP-1μ and Rab11 regulate Glutamate Receptor 1 trafficking57,58. Thus, it is an exciting challenge to identify the specific cargos whose trafficking is altered in the absence of components that regulate clathrin-, AP-1-dependent protein transport and actin nucleation, in order to better understand the molecular bases of these neurological disorders.
METHODS
Reagents
Antibodies
rabbit polyclonal antibodies against human CYFIP1/PIR121/Sra1 (aa 872–886), Myc (aa 409–420) (Upstate, Lake Placid, NY), MPR (Waguri et al., 2003), N-WASP-WASP (C-terminus; ECM Biosciences, Versailles, KY); CDC42 (C-terminus, Santa Cruz, Santa-Cruz, CA), Cathepsin D (Biodesign, Asbach, Germany), NAP1 (aa 1–16; Sigma-Aldrich), Rab11a (C terminus, Zymed, San Francisco, CA), recombinant β-PIX SH3 domain (Millipore, Temecula, CA; mouse monoclonal antibodies against mouse AP-1γ, EEA1, LAMP-2, human P21-Arc (aa 10–118), rat GM130, rat clathrin heavy chain (BD Biosciences, Franklin Lakes, NJ), human transferrin receptor (clone H68.4, Zymed, San Francisco, CA); Myc (clone 9E10; MPI-CBG Antibody Facility, Dresden, Germany); green fluorescent protein (Roche Applied Science, Mannheim, Germany), RAC1 (Cell Biolabs, San Diego, CA); rat beta-tubulin (cl. 2.1, Sigma-Aldrich); sheep polyclonal anti-human rTGN46 (GeneTex, San Antonio, TX).
Secondary antibodies
Alexa Fluor 488, 546 or 647-conjugated against the corresponding primary antibodies (Molecular Probes, Invitrogen, Germany); horse-radish peroxidase (HRP)-conjugated goat anti-mouse IgG and goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, UK).
Latrunculin B, NSC23766 RAC1 inhibitor and D-Mannose-6-Phosphate (M6P) Disodium Salt were from Calbiochem (Germany), and Brefeldin A from Sigma.
Cell culture, transfection and RNA interference
HeLa cells or HeLa cells stably expressing an EGFP-tagged MPR29 were grown in DMEM containing 10% fetal calf serum. Transient expression of myc-CYFIP2 and GFP-CYFIP128 was performed using jetPEI (Biomol, Germany). BSC1 cells stably expressing EGFP- or dTomato-clathrin light chain59, or EGFP- AP-1σ1 and HEK 293 cells stably expressing GFP-CYFIP128, EGFP-β-actin or RFP-β-actin were generated as follows. Briefly, HEK 293 cells were grown in 24-well plates in DMEM with Glutamax (Gibco, Germany) with 10% FCS (Greiner Bio One, Germany), and incubated with 1 μg DNA and 3 μl jetPEI (Biomol); after 48 h, 0.8 mg ml−1 GIBCO™ Geneticin Selective Antibiotic (Gibco) was added. After 14 days, cells were transferred to 10 cm plates and grown in 10% FCS DMEM with 0.5 mg ml−1 Geneticin. Individual colonies were then picked and transferred to 96-well plates (Greiner Bio-One). In each case, 5–6 clones were then chosen using fluorescence microscopy and tested for the expression of the protein of interest by western blotting. For gene silencing, they were transferred into fresh medium and incubated for 72 h with 20 nM siRNAs (HeLa cells) or 50 nM siRNAs (EGFP-β-actin expressing HEK cells) and Interferin transfection reagent (Biomol). Western blotting or QRT-PCR was used to evaluate knockdown efficiencies.
siRNAs sequences (5′→3′):
AP-1γ (NM__001030007), ID 147048: GGAAGUUAUGUUCGUGAUGTT′ CAUCACGAACAUAACUUCCTG;
CYFIP2 (NM_001037333), IDa 134286: CCUUCCUCCAUCAUGUACCtt, GGUACAUGAUGGAGGAAGGtg; IDb* 134287: GGUACAUUGAGCAGGCUACtt, GUAGCCUGCUCAAUGUACCtt;
CYFIP1 (NM_014608, NM_001033028), Ida 21718: GGAAUUUCAAAGAGAUAAGtt, UGCAUUCUGAUCGUAACCGtt; IDb* 284749: CCAAUUUGUUUACAAGCUAtt, CUUAUCUCUUUGAAAUUCCtg;
WAVE2 (NM_006990), ID 138209: GGUAGGAUUAGAUCAUUAGtt, CUAAUGAUCUAAUCCUACCtt;
WASP (WAS, NM_000377), ID 138724: GCUGAUAUUGGUGCACCCAtt, UGGGUGCACCAAUAUCAGCtt;
N-WASP (WASL, NM_003941) ID s17132: CGACAGGGUAUCCAACUAAtt, UUAGUUGGAUACCCUGUCGta;
CDC42 (NM_001039802), ID s2765: UGGUGCUGUUGGUAAAACAtt, UGUUUUACCAACAGCACCAtc;
RAC1 (NM_198829), ID s11713: GGAACUAAACUUGAUCUUAtt, UAAGAUCAAGUUUAGUUCCca;
Clathrin heavy chain (CLTC; NM_004859), ID* 107566: GGGUGCCAGAUUAUCAAUUtt, AAUUGAUAAUCUGGCACCCtg. ID 107565 GGCUCAUACCAUGACUGAUtt, AUCAGUCAUGGUAUGAGCCtt;
ABI1 (NM_001012750), ID 137945: GGCAGAUAUCUCGACACAAtt, UUGUGUCGAGAUAUCUGCCtg;
Beta-PIX (ARHGEF7, NM_003899), ID 119397: GCAAAUGCUCGUACAGUCUtt, AGACUGUACGAGCAUUUGCtg.
Other two siRNAs targeting β-PIX (NM_003899) were from Invitrogen (GCAGACCAGUGAGAAGUUA; CCUUCAUGCGCCUGGAUAA*). * Indicates the siRNA used predominantly. These siRNAs and nontargeting controls Nontargeting (siNon) #2 and #6 were purchased from Ambion, Applied Biosciences (Foster City, CA).
Quantitative RT-PCR
Quantitative real-time polymerase chain reaction (Q RT-PCR) was performed using the MX400 Multiplex QPCR system (Stratagene, La Jolla, CA) and brilliant SYBR Green QPCR Master Mix.
Primers:
CYFIP2: 5′-GCAGGAAGGACTTTGTCTC-3′, 5′-CACTGGGTGATCCTGTTG-3′;
CYFIP1: 5′ – CAGGATGGAGAGCGTGTT-3′, 5′ – GGACTCTAGCATGGTTCTC-3′;
WAVE2: 5′-GGACGACTGGTCCGATTA-3′, 5′-GACTTGGAGGAAGCACTTG-3′;
WASP: 5′-CTGGACCAAGGAGCATTGT-3′, 5′-GTCATCTCCAGCGAAGGT-3′;
N-WASP: 5′-ACCTTCAAGAGCTCCCAC-3′, 5′-CAACACAGATGGAGGTGG-3′;
GAPDH: 5′-TCACCACCATGGAGAAGGC-3′, 5′-GCTAAGCAGTTGGTGGTGCA-3.
Immunoprecipitation
Cos-7 cells or HeLa cells transiently transfected with myc- CYFIP2 were harvested and lyzed in 100 mM NaCl, 2mM EDTA, 50 mM HEPES, pH 7.4, 5 mM MgCl2, 0.5% NP-40, 1 mM PMSF, and Complete Mini-protease inhibitor mixture (Roche, Germany). Lysates were centrifuged (15 min, 14,000 rpm, 4°C), precleared (1 h) with Protein A-Sepharose beads (GE Healthcare, Germany), and then incubated for 1 h or overnight at 4°C with the indicated antibodies (4μg ml−1). Protein A-Sepharose beads pre-blocked in 1 mg ml−1 BSA were then added to the lysates for 1 h at 4°C. Beads were recovered by centrifugation, washed either with Buffer 1 (150 mM NaCl, 50 mM HEPES, pH 7.4, 5 mM MgCl2, 0.5% NP-40, 0.25% TX-100, 1 mM PMSF, and protease inhibitor cocktail) or with Buffer 1 with 0.5 M TrisHCl, pH 7.4, and resuspended in Laemmli buffer; immunoprecipitates were analyzed by SDS-PAGE followed by western blotting. For immunoprecipitation of Cathepsin D, HeLa cells were labeled with 35S Methionine and processed as previously described60.
Subcellular Fractionation
HeLa cells grown in 10 cm plates were washed with PBS, collected in a lysis buffer (100 mM sodium phosphate, 50 mM NaCl, 1 mM MgCl2, 1 mM EGTA, pH 7.4, with protease inhibitor mixture and passed 15 times through a 27G needle on ice. The lysates were centrifuged 5 min at 1000 g, and the post-nuclear supernatants centrifuged for 1h at 100,000g (Optima Ultracentrifuge, TLA55 rotor; Beckman Coulter, Germany) at 4 °C. The membrane pellets and the cytosolic supernatants (equal protein amounts) were analyzed by western blot. Pig brain cytosol was prepared as previously described8. Cytosols of HEK cells expressing various GFP-tagged proteins were prepared as decribed above for HeLa cells.
Yeast Two Hybrid analysis
DNA sequences (from imaGenes) encoding CYFIP2 N-terminal (2–623 aa), CYFIP2 C- terminal (675–1299 aa), Clathrin Heavy Chain N-terminal (1–690 aa), Clathrin Heavy Chain Ct (821–1679 aa) and NAP1 were subcloned into pGADT7 and pGBKT7 plasmids (Clontech, Mountain View, CA) using restriction endonucleases (Fermentas, Germany) to create the GAL4 DNA-activation and DNA-binding domain fusions, respectively. The recombinant plasmids were analyzed by DNA sequencing. The Saccharomyces cerevisiae strain AH109 (Clontech) was used for transformations. Yeast transformants were selected on medium SDA-Leu-Trp (MP Biomedicals, LLC). To monitor protein-protein interactions, a selective medium SDA-Ade-Lys-Leu-Trp was used. As negative controls, yeast cells transformed either with the original yeast-two-hybrid vectors or with pGBKT7-Lam (Clontech) were used. As positive controls, interaction between SV40 large T-antigen and murine p53 as well as known protein interactions were used.
Cell imaging
Cells were washed with PBS, fixed with 3% PFA (15 min, room temperature), permeabilized (0.1% Triton X-100, 5 min, room temperature), blocked with 3% BSA (30 min, room temperature), incubated with the primary antibody (1 h, room temperature) and then with corresponding Alexa Fluor-labeled secondary antibodies (20 min, room temperature). Cells were then washed with PBS and mounted on microscope slides by using Mowiol (Calbiochem, UK). For measuring transferrin uptake, cells were starved for 1 h in DMEM with 0.2% BSA, incubated with 25 μg ml−1 Alexa Fluor 564-labeled transferrin (Molecular Probes) for the indicated times, placed on ice, washed with ice-cold PBS, fixed and analyzed by microscopy. For measuring transferrin recycling, cells were starved, incubated with Alexa Fluor 564-labeled transferrin for 1h at 37°C, washed with ice-cold PBS, then incubated with 2.5 mg ml−1 unlabeled holotransferrin (Calbiochem) in DMEM containing 0.2% BSA, for the indicated times. Cells were next fixed and analyzed by fluorescence microscopy. For anti-GFP uptake, GFP-MPR HeLa cells were incubated for 1h on ice with anti-GFP antibodies, then incubated at 37°C for 60 min, fixed, permeabilized and labeled with a secondary antibody to detect the internalized anti-GFP antibody. Samples were analyzed by confocal fluorescence microscopy, using a LSM 510 meta (Carl Zeiss Microimaging, Germany) and a Leica SP5 inverted microscope (Leica Microsystems, Germany) and a 63x, 1.4 plan apochromat objective (Carl Zeiss Microimaging). Images were analyzed using Adobe Photoshop 7 (Adobe Systems, Mountain View, CA), Volocity 5.2 (Improvision, UK) or ImageJ software (http://rsb.info.nih.gov/ij/); for statistical analyses we used Anova Single Factor (Excel).
High-speed time-lapse microscopy was done using an Axiovert 200 M with temperature, CO2 and humidity control and a 63x, 1.2 water-immersion objective (Carl Zeiss Microimaging). Images were collected every 500 ms during 2–4 min. Image analysis was done with Metamorph software (MDS, Canada).
Electron Microscopy
HeLa cells transiently expressing myc-tagged CYFIP2 were fixed with 2% paraformaldehyde (PFA) in 0.1 M phosphate buffer (0.1M Na2HPO4 + 0.1M NaH2PO4), pH 7.4, for 2.5 h. Sections of 70 nm were cut on Leica ultramicrotome (Leica, Austria). Thawed cryosections were double labeled using the indicated primary and secondary antibodies conjugated to 10- or 15-nm gold particles (Cell Microscopy Center, Utrecht Medical School). Sections were analyzed using a Philips CM120 (FEI) electron microscope, and images were acquired with a numeric camera (Keen View; Soft Imaging System).
In vitro reconstitution with model membranes
Liposomes coupled with varicella zoster virus glycoprotein I (GpI) cytoplasmic domains were prepared as previously described61. Giant unilamellar liposomes (GUVs) were grown by electroswelling62. Briefly, 20–50 μl of liposomes were dried onto ITO-slides (Praezisions Glas & Optik, Germany) and GUVs were formed in 330 mM Sucrose with alternating current (10 Hz, 1.8 V) for 2 h. Functionalized DiI C18 or DiD C18 (Molecular Probes) – labeled GUVs (12 μg) were then incubated as indicated with pig brain cytosol (13 mg ml−1) supplemented with 20 % cytosol (≈ 5mg ml−1 proteins) from cells stably expressing a fluorescently labeled protein of interest in the presence of GTP-γ-S and imaged by confocal microscopy. Where indicated, an ATP-regenerating system was added. Actin polymerization assay: cytosols of HEK cells stably expressing EGFP-actin and pre-incubated with the indicated siRNAs for 72 h or with 100 μM NSC23766 RAC1 inhibitor63 for 1h at 4°C. In each case 150 μl cytosol (4–5 mg ml−1 protein concentration) and 15 μl DiI C18-labeled GUVs were incubated for 20 min at 37°C and then imaged with an LSM 510 meta microscope with a 40x, 1.2 water immersion objective (Carl Zeiss Microimaging).
Supplementary Material
Acknowledgments
We thank the different lab members for their helpful discussions and critical comments. D. Thiel kindly provided her technical assistance. We are grateful to M. Zerial, T. Wassmer and C. Antos for their critical reading of the manuscript. This work was supported in part by grants from DFG (TRR 13/1-2010, HO 2584/1-1, HO 2584/2-1, HO 2584/6-1, HO 2584/8-1, HO 2584/9-1), BMBF (0313815B) and TU-Dresden (HWP-1207).
Footnotes
The authors declare that they have no competing financial interests.
Author contributions.
MA designed, performed and analyzed the in vivo experiments, CS designed, performed and analyzed the in vitro recruitment experiments, IP performed the Q-RT-PCR experiments and yeast two hybrid analyses, GR performed the electron microscopy, TB, AS and TK provided key reagents, BH designed, analyzed experiments, wrote the manuscript with MA and CS.
References
- 1.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–12. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 2.Edeling MA, Smith C, Owen D. Life of a clathrin coat: insights from clathrin and AP structures. Nat Rev Mol Cell Biol. 2006;7:32–44. doi: 10.1038/nrm1786. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 4.Godi A, et al. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–7. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 5.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 6.Lee I, Doray B, Govero J, Kornfeld S. Binding of cargo sorting signals to AP-1 enhances its association with ADP ribosylation factor 1-GTP. J Cell Biol. 2008;180:467–72. doi: 10.1083/jcb.200709037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Borgne R, Hoflack B. Mannose 6-phosphate receptors regulate the formation of clathrin-coated vesicles in the TGN. J Cell Biol. 1997;137:335–45. doi: 10.1083/jcb.137.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baust T, Czupalla C, Krause E, Bourel-Bonnet L, Hoflack B. Proteomic analysis of adaptor protein 1A coats selectively assembled on liposomes. Proc Natl Acad Sci U S A. 2006;103:3159–64. doi: 10.1073/pnas.0511062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelish HE, et al. Secramine inhibits Cdc42-dependent functions in cells and Cdc42 activation in vitro. Nat Chem Biol. 2006;2:39–46. doi: 10.1038/nchembio751. [DOI] [PubMed] [Google Scholar]
- 10.Salvarezza SB, et al. LIM kinase 1 and cofilin regulate actin filament population required for dynamin-dependent apical carrier fission from the trans-Golgi network. Mol Biol Cell. 2009;20:438–51. doi: 10.1091/mbc.E08-08-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egea G, Lazaro-Dieguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006;18:168–78. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Kondylis V, van Nispen tot Pannerden HE, Herpers B, Friggi-Grelin F, Rabouille C. The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev Cell. 2007;12:901–15. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Carreno S, Engqvist-Goldstein AE, Zhang CX, McDonald KL, Drubin DG. Actin dynamics coupled to clathrin-coated vesicle formation at the trans-Golgi network. J Cell Biol. 2004;165:781–8. doi: 10.1083/jcb.200403120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao H, et al. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol. 2005;7:483–92. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- 15.Engqvist-Goldstein AE, et al. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell. 2004;15:1666–79. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–14. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 17.McNiven MA, Thompson HM. Vesicle formation at the plasma membrane and trans-Golgi network: the same but different. Science. 2006;313:1591–4. doi: 10.1126/science.1118133. [DOI] [PubMed] [Google Scholar]
- 18.Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–9. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 19.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 20.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–61. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schenck A, et al. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–98. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- 22.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–3. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 23.Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–75. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Steffen A, et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. Embo J. 2004;23:749–59. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautreau A, et al. Purification and architecture of the ubiquitous Wave complex. Proc Natl Acad Sci U S A. 2004;101:4379–83. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogdan S, Stephan R, Lobke C, Mertens A, Klambt C. Abi activates WASP to promote sensory organ development. Nat Cell Biol. 2005;7:977–84. doi: 10.1038/ncb1305. [DOI] [PubMed] [Google Scholar]
- 27.Innocenti M, et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat Cell Biol. 2005;7:969–76. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- 28.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–9. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waguri S, et al. Visualization of TGN to endosome trafficking through fluorescently labeled MPR and AP-1 in living cells. Mol Biol Cell. 2003;14:142–55. doi: 10.1091/mbc.E02-06-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folsch H, Pypaert M, Maday S, Pelletier L, Mellman I. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol. 2003;163:351–62. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubois T, et al. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–64. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- 32.Fucini RV, Chen JL, Sharma C, Kessels MM, Stamnes M. Golgi vesicle proteins are linked to the assembly of an actin complex defined by mAbp1. Mol Biol Cell. 2002;13:621–31. doi: 10.1091/mbc.01-11-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi K, et al. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–5. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- 34.Poupon V, et al. Clathrin light chains function in mannose phosphate receptor trafficking via regulation of actin assembly. Proc Natl Acad Sci U S A. 2008;105:168–73. doi: 10.1073/pnas.0707269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–7. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 36.Faucherre A, et al. Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum Mol Genet. 2003;12:2449–56. doi: 10.1093/hmg/ddg250. [DOI] [PubMed] [Google Scholar]
- 37.Jou TS, et al. Selective alterations in biosynthetic and endocytic protein traffic in Madin-Darby canine kidney epithelial cells expressing mutants of the small GTPase Rac1. Mol Biol Cell. 2000;11:287–304. doi: 10.1091/mbc.11.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Premont RT, et al. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16:1001–11. doi: 10.1016/j.cellsig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Kroschewski R, Hall A, Mellman I. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat Cell Biol. 1999;1:8–13. doi: 10.1038/8977. [DOI] [PubMed] [Google Scholar]
- 40.Musch A, Cohen D, Kreitzer G, Rodriguez-Boulan E. cdc42 regulates the exit of apical and basolateral proteins from the trans-Golgi network. Embo J. 2001;20:2171–9. doi: 10.1093/emboj/20.9.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egorov MV, et al. Faciogenital dysplasia protein (FGD1) regulates export of cargo proteins from the golgi complex via Cdc42 activation. Mol Biol Cell. 2009;20:2413–27. doi: 10.1091/mbc.E08-11-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNiven MA, et al. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J Cell Biol. 2000;151:187–98. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson JC, Legg JA, Machesky LM. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–8. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–8. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 45.Merrifield CJ, Qualmann B, Kessels MM, Almers W. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur J Cell Biol. 2004;83:13–8. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- 46.Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Schenck A, et al. WAVE/SCAR, a multifunctional complex coordinating different aspects of neuronal connectivity. Dev Biol. 2004;274:260–70. doi: 10.1016/j.ydbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Borck G, et al. Clinical, cellular, and neuropathological consequences of AP1S2 mutations: further delineation of a recognizable X-linked mental retardation syndrome. Hum Mutat. 2008 doi: 10.1002/humu.20531. [DOI] [PubMed] [Google Scholar]
- 49.Tarpey PS, et al. Mutations in the gene encoding the Sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am J Hum Genet. 2006;79:1119–24. doi: 10.1086/510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen KM, et al. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- 51.Nowicki ST, et al. The Prader-Willi phenotype of fragile X syndrome. J Dev Behav Pediatr. 2007;28:133–8. doi: 10.1097/01.DBP.0000267563.18952.c9. [DOI] [PubMed] [Google Scholar]
- 52.Dwyer ND, Adler CE, Crump JG, L’Etoile ND, Bargmann CI. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron. 2001;31:277–87. doi: 10.1016/s0896-6273(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 53.Sakaguchi-Nakashima A, Meir JY, Jin Y, Matsumoto K, Hisamoto N. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Curr Biol. 2007;17:592–8. doi: 10.1016/j.cub.2007.01.074. [DOI] [PubMed] [Google Scholar]
- 54.Nakamoto M, et al. Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc Natl Acad Sci U S A. 2007;104:15537–42. doi: 10.1073/pnas.0707484104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, et al. FMRP acts as a key messenger for dopamine modulation in the forebrain. Neuron. 2008;59:634–47. doi: 10.1016/j.neuron.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 56.Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–5. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- 57.Correia SS, et al. Motor protein-dependent transport of AMPA receptors into spines during long-term potentiation. Nat Neurosci. 2008;11:457–66. doi: 10.1038/nn2063. [DOI] [PubMed] [Google Scholar]
- 58.Margeta MA, Wang GJ, Shen K. Clathrin adaptor AP-1 complex excludes multiple postsynaptic receptors from axons in C. elegans. Proc Natl Acad Sci U S A. 2009;106:1632–7. doi: 10.1073/pnas.0812078106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehrlich M, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 60.Ludwig T, et al. Differential sorting of lysosomal enzymes in mannose 6-phosphate receptor-deficient fibroblasts. Embo J. 1994;13:3430–7. doi: 10.1002/j.1460-2075.1994.tb06648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baust T, et al. Protein networks supporting AP-3 function in targeting lysosomal membrane proteins. Mol Biol Cell. 2008;19:1942–51. doi: 10.1091/mbc.E08-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bacia K, Schuette CG, Kahya N, Jahn R, Schwille P. SNAREs prefer liquid-disordered over “raft” (liquid-ordered) domains when reconstituted into giant unilamellar vesicles. J Biol Chem. 2004;279:37951–5. doi: 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- 63.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–23. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.