Abstract
Background and Aims
Although there is evidence that both allopolyploid and homoploid hybridization lead to rapid genomic changes, much less is known about hybrids from parents with different basic numbers without further chromosome doubling. Two natural hybrids, Narcissus × alentejanus (2n = 19) and N. × perezlarae (2n = 29), originated by one progenitor (N. cavanillesii, 2n = 28) and two others (N. serotinus, 2n = 10 and N. miniatus, 2n = 30, respectively) allow us to study how DNA content and composition varies in such hybrids.
Methods
Flow cytometry measurements with two staining techniques, PI and DAPI, were used to estimate 2C values and base composition (AT/GC ratio) in 390 samples from 54 wild populations of the two natural hybrids and their parental species. In addition, 20 synthetic F1 hybrid individuals were also studied for comparison.
Key Results
Natural hybrids presented 2C values intermediate between those found in their parental species, although intra-population variance was very high in both hybrids, particularly for PI. Genome size estimated from DAPI was higher in synthetic hybrids than in hybrids from natural populations. In addition, differences for PI 2C values were detected between synthetic reciprocal crosses, attributable to maternal effects, as well as between natural hybrids and those synthetic F1 hybrids in which N. cavanillesii acted as a mother.
Conclusions
Our results suggest that natural hybrid populations are composed of a mixture of markedly different hybrid genotypes produced either by structural chromosome changes, consistent with classic cytogenetic studies in Narcissus, or by transposon-mediated events.
Keywords: Amaryllidaceae, base composition, DAPI, flow cytometry, genome size, interspecific hybrids, Narcissus, polyploidy, propidium iodide
INTRODUCTION
Changes in genome size as expressed by the DNA content of the non-replicated somatic genome (2C value sensu Greilhuber, 2005) and genome base composition following hybridization are far from completely understood in angiosperms, due to complex or as yet unknown mechanisms underlying intraspecific variation. Despite more detailed knowledge of the different molecular processes involved in chromosome evolution, such as duplications and expansions of satellite DNA, fixation of accessory chromosomes, transposable element activation or allopolyploidy (Bennetzen et al., 2005; Cavalier-Smith, 2005), an insightful interpretation of genome size changes has not been satisfactorily achieved in most plant groups studied (Bennett and Leitch, 2005; Greilhuber, 2005). The evidence, now widely accepted, that reticulate evolution has played an important role in the evolution of angiosperms only adds complexity to the already complex problem of looking for patterns in DNA content change across evolution in general. Nevertheless, understanding the influence of hybridization in those genome size changes is highly relevant when realizing that hybridization is not restricted to sporadic events between species but that it is involved in the origin of whole, even important, lineages (Wendel, 2000).
Classical studies on synthetic allopolyploids have shown that after hybridization extensive and rapid genome changes can occur (Soltis and Soltis, 1993; Wendel, 2000). These include rearrangement of chromosomes and sequences (Song et al., 1995; Wendel et al., 1995; Leitch and Bennett, 1997), regulation of gene expression (Scheid et al., 1996; Galitski et al., 1999; Matzke et al., 1999; Comai, 2000), activation of transposable elements (Zhao et al., 1998; Soltis and Soltis, 1999) and, most frequently, amplification, reassortment or elimination of highly repetitive sequences or low-copy genes (Feldman et al., 1997; Hanson et al., 1998; Liu et al., 1998a, b; Salina et al., 2000). Although the mechanisms responsible for some of these genomic changes are not fully understood, it is assumed that they may facilitate the viability and establishment of newly generated polyploid species (Song et al., 1995; Wendel et al., 1995; Feldman et al., 1997; Liu et al., 1998a, b; Ozkan et al., 2001).
Genomic alterations also occur in homoploid hybrids (i.e. those in which there is no change in chromosome number), but they are even less understood than in allopolyploid species (Loureiro et al., 2010). For instance, studies conducted on three homoploid hybrid species of Helianthus originating from the same parental species have revealed significant rearrangements in just a few hybrid generations (Lai et al., 2005; Karrenberg et al., 2007). Although two of these three hybrid species (H. anomalus and H. deserticola) appear to have arisen several times in nature (Schwarzbach and Rieseberg, 2002), the different populations exhibit similar increases in genome size, with almost 50 % more nuclear DNA than the parental species (Baack et al., 2005). Nevertheless, it is not clear whether this increase in genome size arose as a consequence of hybridization or only occurs under particular ecological conditions, such as stressful environments (Baack et al., 2005).
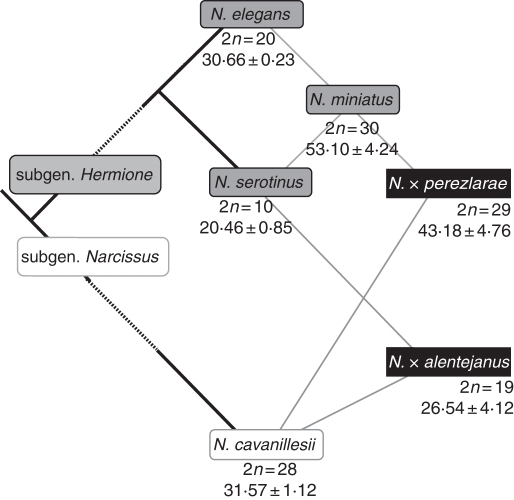
Hybridization between species differing in basic chromosome number and ploidy levels is not unusual in plants (Krahulcova et al., 1996; Bureš et al., 2004) although sometimes reports of these types of hybrids are misinterpreted as cases of ascending aneuploidy (Grant, 1981). However, studies focusing on genome size changes in such hybrids are scarce or non-existent. Narcissus L. (Amaryllidaceae) has been the subject of several cytogenetic and flow cytometry studies supporting the view that the genus has a highly dynamic cytological organization (Fernandes, 1951, 1967, 1968, 1975; Zonneveld, 2008). Here, we seek to understand the effects of natural interspecific hybridization in the genome size of two closely related hybrids: N. × alentejanus Fernández Casas and N. × perezlarae Font Quer (Amaryllidaceae). These natural hybrids provide a suitable system to study genome evolution because both the phylogenetic relationships between the progenitor species and their hybrid origin are well documented (Marques et al., 2010). The two morphologically similar hybrids share one parental species (N. cavanillesii A. Barra and G. López 2n = 4x = 28; subgenus Narcissus) but differ in the other. Narcissus serotinus L. (2n = 2x = 10; subgenus Hermione) is the progenitor of N. × alentejanus while N. miniatus Donnison-Morgan, Koopowitz and Zonneveld (2n = 6x = 30; subgenus Hermione) is the progenitor of N. × perezlarae (Marques et al., 2010). On the other hand, the latter two progenitors are closely related as N. miniatus is an allohexaploid that originated between N. elegans (Haworth) Spach (2n = 4x = 20; subgenus Hermione) and N. serotinus L. (Díaz-Lifante et al., 2009; Marques et al., 2010) (Fig. 1). The two hybrids exhibit odd chromosome numbers that are intermediate between those of their parental species (2n = 19, N. × alentejanus; 2n = 29, N. × perezlarae). In addition to being well documented on molecular grounds, the case studies allow a comparison of genome size in different natural hybrid populations. These hybrid populations arose independently and thus might exhibit different DNA content, as reported in some studies involving distinct ecological conditions (Bureš et al., 2004). Secondly, the inclusion of F1 synthetic hybrids in the study aims to complement the dynamic picture of DNA content changes in the early stages of hybridization events by comparison with those in established natural hybrids. We cannot, however, identify the generation of those natural hybrids because in previous studies we obtained partially contradictory evidence. On the one hand, chromosome numbers recorded in hybrids were intermediate between the parents (Marques et al., 2010). On the other, a thorough sampling of nuclear ribosomal cloned sequences in hybrids revealed that in a number of individuals there was only a single internally transcribed spacer (ITS) sequence type of one of the parents, usually the mother, which is not expected in F1s (Marques et al., 2010).
Fig. 1.
Scheme of the phylogenetic relationships among the studied species, based on DNA sequences from two mitochondrial regions (atpA and cob), two plastid regions (ndhF and matK) and nrITS; chromosome numbers are also reported (based on Marques et al., 2010). Mean PI 2C values gathered in this study are also indicated.
In this study, we used flow cytometry based on two different fluorochrome staining techniques to assess the quality and quantity of genomic changes in a group of naturally hybridizing species of Narcissus differing in basic chromosome number and ploidy level. While DNA-intercalating dyes such as PI (propidium iodide) bind to all bases homogeneously, DAPI (4′,6-diamidino-2-phenylindole) binds preferentially to AT-rich regions so that results are influenced by the differences in base composition of each species in comparison with the reference standard (Meister and Barow, 2007). Thus, comparing the results obtained with the two dyes allows some estimation of whether hybridization leads to bias in genome base composition (Šmarda and Bureš, 2006).
Specifically, this work tries to answer the following questions in the two hybrids Narcissus × alentejanus and N. × perezlarae: (1) Are the values of genome size of natural hybrids within the expected range given the genome size and ploidy level of the parental species? (2) Do synthetic hybrids resulting from reciprocal crosses exhibit differences in genome size? (3) Is genome size similar in synthetic F1 hybrids and hybrid plants from natural populations? (4) Does hybridization affect genome base composition in addition to genome size?
MATERIAL AND METHODS
Study system
A total of 390 samples from 59 wild populations were collected in the field: 14 populations of Narcissus cavanillesii, 15 populations of N. serotinus, 16 populations of N. miniatus, four populations of N. × alentejanus, four populations of N. × perezlarae and six populations of N. elegans (Supplementary Data Table S1, available online). Populations selected span the geographical range in which the two species hybridize in the southern part of the Iberian Peninsula although samples outside this area were also collected for comparison. Narcissus elegans, the other progenitor of the allopolyploid N. miniatus, was included in this study for comparison purposes. Nuclear DNA content was estimated from 4–8 individuals per population (Table S1). In addition to natural populations, we analysed 20 F1 synthetic hybrids obtained from controlled reciprocal pollinations between N. cavanillesii and N. serotinus (CS and SC) and between N. cavanillesii and N. miniatus (CM and MC). The first letter indicates the ovule-donor in the crossing experiment (C, N. cavanillesii; S, N. serotinus; M, N. miniatus). Experimental pollinations were performed in 2002 for analyses of crossability in both directions (Marques and Draper, 2004). All 410 individuals (390 natural plus 20 F1 synthetic hybrids) were grown in the greenhouse at the Real Jardín Botánico Madrid, Spain (40 °24′45″N, 3 °41′2″E; 667 m a.s.l). A previous study documented chromosome numbers in all the populations (Marques et al., 2010; Table S1).
Flow cytometry
Measurements were made on a Cell Lab Quanta Beckman Coulter flow cytometer (Beckman Coulter Inc., Brea, CA, USA) equipped with a 100-W Mercury arc lamp with excitation line optimized at 366 nm for UV measurements with DAPI, and a 2–22-mW, 488-nm laser diode using the 570-nm long-pass filter for PI measurements. Two types of fluorochromes were used for assays: intercalating PI for absolute nuclear DNA content estimations and AT-selective DAPI for estimation of the AT portion of nuclear DNA content. PI has been previously used to investigate systematic relationships in Narcissus (Zonneveld, 2008). For sample preparation, a piece of fresh young tissue collected from leaves was chopped using a razor blade together with an internal standard in a Petri dish containing 600 µL Galbraith's buffer (Galbraith et al., 1983) supplemented with 100 µg ml−1 RNase A (Boehringer, Meylan, France). Pisum sativum ‘Express Long’ (2C = 8·37 pg; GC = 40·5 %; AT = 59·5 %) was used as internal standard (seeds provided by the Institut des Sciences du Végétal, Gif-sur-Yvette, France). The suspension was filtered through a 33-μm SEFAR PA 1000 140/355-35W nylon filter (SEFAR, Barcelona, Spain) and subsequently supplemented with either 60 µg mL−1 PI (Sigma-Aldrich, Alcobendas, Madrid, Spain) or 2·0 µg mL−1 DAPI (Partec, Münster, Germany) depending of the staining technique. The two measurements were made on the same day using a portion of the leaf obtained from the same individual. Prior to the first measurement, the cytometer was checked for linearity using standard fluorescent beads (Coulter Electronics). Amplification settings were kept constant throughout the experiment. All measurements were taken twice on different days to avoid instrument drift. In each sample, 10 000 particles were counted. The 2C-value was calculated by multiplying the known DNA content of the standard by the quotient of the 2C peak positions of the target species and the standard in the histogram of fluorescence intensities, under the assumption that there is a linear correlation between the fluorescent signals from stained nuclei of the unknown specimen and the internal standard. The AT frequency was calculated using the equation:
where R-DAPIspecies is the ratio between mean fluorescence peaks for a sample and reference stained by DAPI, R-PIspecies is the ratio between mean fluorescence peaks for a sample and reference stained by PI and f(ATreference) is the AT frequency of the reference sample (Barow and Meister, 2002).
As generally accepted, genome size is the 2C-value divided by ploidy level (Bennett, 1998), so that the genome size corresponds to the 1C-value in diploids but can vary in polyploids due to the ‘Cx-paradox’ (Greilhuber, 2005). However, in our study we have used 2C-values instead of 1C, to allow comparisons between hybrids and between them and their progenitors. The cytogenetic complexity of hybrids between parents differing in basic chromosome number and ploidy level prevents a reliable calculation of 1C values in the hybrids.
Sampling design and statistical analyses
The mean and s.d. of each sample was calculated from the means of the two measurements performed. Only histograms with symmetrical peaks and with a coefficient of variation (CV) of the sample G1 peak below 2 % were considered. We also performed some simultaneous measurements of hybrid samples and their respective parental species. The normality of the distribution of genome size of all samples was tested using the Kolmogorov–Smirnov test. Differences of genome size (PI and DAPI) among species, populations and individuals were evaluated using analysis of variance (ANOVA). In those cases in which ANOVA revealed significant differences, the Tukey HSD post-hoc test was performed. In addition, a t-test was used to compare genome size values between the two natural hybrids and between the reciprocal synthetic F1 hybrids. To explore if the values of genome size obtained for N. × alentejanus and N. × perezlarae differed from those expected assuming an intermediate value between that of their parents, we used a chi-square test. To assess differences in base composition (DAPI) with respect to whole genome size (PI) we computed the DAPI/PI ratio for each individual, population and species.
RESULTS
Accuracy of measurements
CV of G0/G1 peaks for PI staining of the standard Pisum sativum ‘Express Long’ and for the measured samples were always below 3·01 % in 410 estimates (mean of standard CV = 1·99 ± 0·32 %, mean of samples CV = 2·10 ± 0·45 %). Histograms yielded by DAPI staining did not exceed 3·51 %, either for the standard or for the sample (mean of standard CV = 2·99 ± 0·33 %, mean of samples CV = 3·12 ± 0·15 %).
Genome size in natural hybrids and parental species
The mean 2C-value assessed with PI increased in the following order: Narcissus serotinus < N. × alentejanus < N. elegans < N. cavanillesii < N. × perezlarae < N. miniatus, which is congruent with the number of chromosomes reported for each taxon (Table 1). Nevertheless, 2C-values assessed with DAPI increased in a different order: N. cavanillesii < N. × alentejanus < N. serotinus < N. elegans < N. × perezlarae < N. miniatus. These values showed a significant interspecific variation (ANOVA test, PI: F5,51 = 28·876, P = 0·0001; DAPI: F5,51 = 39·444, P = 0·0001).
Table 1.
2C-values (pg) assessed with PI and DAPI from the studied species of Narcissus
| Species | 2n | N | Npop | PI |
DAPI |
%AT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Min. | Max. | Mean | s.d. | Min. | Max. | Mean | s.d. | |||||
| N. cavanillesii | 28 | 88 | 14 | 30·52 | 33·77 | 31·57 | 1·12 | 12·30 | 14·44 | 13·21 | 1·33 | 24·95 |
| N. serotinus | 10 | 94 | 15 | 19·52 | 21·30 | 20·46 | 0·85 | 13·67 | 19·78 | 17·64 | 2·89 | 51·37 |
| N. miniatus | 30 | 111 | 16 | 48·24 | 56·18 | 53·10 | 4·24 | 25·16 | 38·20 | 35·02 | 3·96 | 39·28 |
| N. elegans | 20 | 37 | 6 | 30·62 | 30·78 | 30·66 | 0·23 | 26·05 | 29·00 | 27·89 | 3·22 | 54·14 |
| N. × alentejanus | 19 | 28 | 4 | 22·92 | 29·84 | 26·54 | 4·12 | 14·03 | 16·78 | 15·52 | 4·22 | 36·15 |
| N. × perezlarae | 29 | 32 | 4 | 39·61 | 46·14 | 43·18 | 4·76 | 26·49 | 29·47 | 28·11 | 4·17 | 39·39 |
2n = Mitotic chromosome number reported for each species (Marques et al., 2010). N = Number of individuals measured per species. Npop = Number of populations measured per species. Min. = Minimum 2C-value recorded considering all populations. Max. = Maximum 2C-value recorded considering all populations.
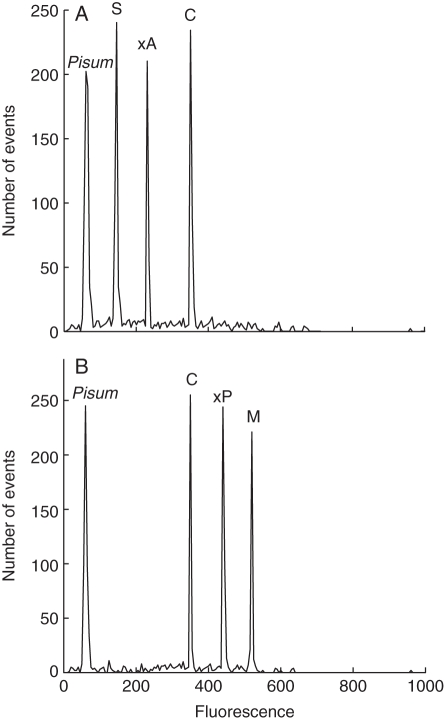
The 2C-values of the two natural hybrids always fell between those of the parental species when estimated both with PI (N. × alentejanus vs. parental species: F2,12 = 61·900, P = 0·0001; N. × perezlarae vs. parental species: F2,12 = 92·895, P = 0·0001) and with DAPI (N. × alentejanus vs. parental species: F2,14 = 83·950, P = 0·0001; N. × perezlarae vs. parental species: F2,15 = 79·589, P = 0·0001). Simultaneous measurements of hybrid samples with their parental species always resulted in separate peaks, the intermediate peak corresponding to the natural hybrid plants (Fig. 2). However, 2C-values of the natural hybrids did not always conform to expectation [(DNA content of parent1 + DNA content of parent2)/2] but depended on the fluorochrome technique. The 2C-values obtained with DAPI were always higher than expected whereas the 2C-values measured with PI did not differ from those expected (Table 2).
Fig. 2.
Fluorescence histogram of propidium iodide-stained (PI) nuclei of parental species and their hybrids analysed in the same sample. (A) Narcissus × alentejanus and parental species. (B) N. × perezlarae and parental species. Abbreviations: C, N. cavanillesii; S, N. serotinus; M, N. miniatus; ×A, N. × alentejanus; ×P, N. × perezlarae.
Table 2.
Comparison of expected and obtained genome size values estimated with PI and DAPI for the two natural hybrids Narcissus × alentejanus and N. × perezlarae
| Natural hybrids | PI |
DAPI |
||||
|---|---|---|---|---|---|---|
| χ2 | d.f. | P | χ2 | d.f. | P | |
| N. × alentejanus | 36·026 | 27 | 0·0054 | 38·897 | 27 | 0·05* |
| N. × perezlarae | 27·200 | 31 | 0·2810 | 38·200 | 31 | 0·01** |
Comparisons were performed with a χ2-test. Significant results are indicated in bold. *P < 0·05, **P < 0·01.
Inter- and intra-population variation in natural hybrids
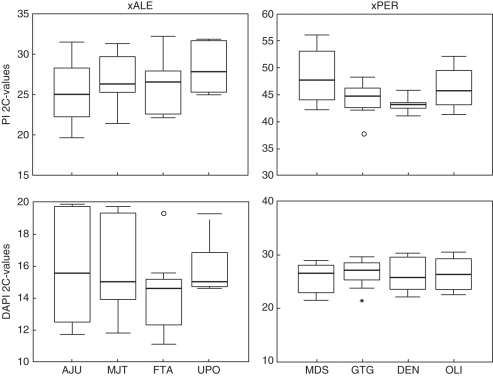
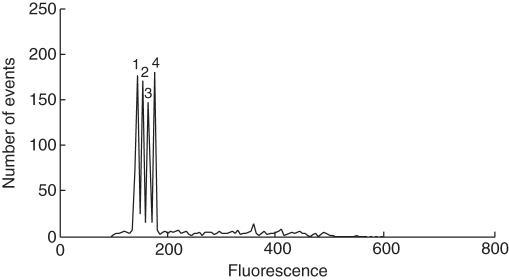
For N. × alentejanus, the 2C-values measured with PI varied between 22·92 pg in population AJU and 29·84 pg in population UPO, with an intra-population variance of 21·32 pg. Estimations with DAPI ranged between 14·03 pg in population FTA and 16·78 pg in population AJU, with an intra-population variance of 10·15 pg (Fig. 3). This high intra-population variance was demonstrated as a clear double peak in the simultaneous measurement of several samples (Fig. 4). Likewise, in N. × perezlarae, genome size measured with PI varied between 39·61 pg in population GTG and 46·14 pg in population MDS, with an intra-population variance of 27·32 pg. The 2C-values measured with DAPI varied between 26·49 pg in population DEN and 29·47 pg in population MDS, with a variance of 27·41 pg (Fig. 3).
Fig. 3.
Intra-population variation of 2C values obtained for the natural hybrids N. × alentejanus (xALE) and N. × perezlarae (xPER). Top: genome size measured with PI. Below: genome size measured with DAPI. Values are expressed in pg. The horizontal line across the box represents the median, and the box represents the interquartile range that contains 50 % of the values. The whiskers are the lines that extend from the box to the highest and lowest values excluding outliers. Asterisks represent extreme values (cases with values more than 3 box lengths from the upper or the lower edge of the box) and open circles represent outliers (cases with values between 1·5 and 3 box lengths from the upper or the lower edge of the box).
Fig. 4.
Differences in genome size among Narcissus × perezlarae samples from four different populations. The nuclei of all individuals were stained with propidium iodide and analysed simultaneously. The peak ratios were 1 : 1·068 : 1·127 : 1·164. The mean 2C-values were 39·61 pg (population 1: GTG), 42·32 pg (population 2: DEN), 44·68 pg (population 3: OLI) and 46·14 pg (population 4: MDS).
No statistically significant differences in 2C-values were found across populations of N. × alentejanus (ANOVA, PI: F3,5 = 5·071, P = 0·054; DAPI: F3,6 = 1·466, P = 0·241) or across populations of N. × perezlarae (PI: F3,5 = 4·583, P = 0·052; DAPI: F3,6 = 1·695, P = 0·561; Fig. 3).
Genome size of synthetic hybrids
Comparing the four classes of experimental crosses, hybrids generated between N. cavanillesii and N. miniatus always had higher 2C-values than hybrids between N. cavanillesii and N. serotinus (Table 3), which is expected based on the genome size of N. miniatus as compared with N. serotinus. Comparing the reciprocal hybrids, 2C-values depended on the fluorochrome used. When assessed with PI, CS hybrids yielded higher 2C-values than SC hybrids while MC hybrids yielded higher 2C-values than CM hybrids (Table 3). In contrast, when genome was estimated with DAPI, no significant differences were found between the reciprocal synthetic hybrids CS and SC and between CM and MC (Table 3).
Table 3.
Results of t-test analyses assessing genome size values (PI and DAPI) obtained for the synthetic hybrids
| Synthetic hybrids | PI |
DAPI |
||||
|---|---|---|---|---|---|---|
| d.f. | t | P | d.f | t | P | |
| CS and SC vs. CM and MC | 19 | 11·230 | 0·0001*** | 19 | 18·082 | 0·0001*** |
| CS vs. SC | 9 | 13·077 | 0·012** | 9 | 11·928 | 0·847 |
| CM vs. MC | 9 | 18·637 | 0·0001*** | 9 | 18·637 | 0·610 |
CS, N. cavanillesii × N. serotinus; SC, N. serotinus × N. cavanillesii; CM, N. cavanillesii × N. miniatus; MC, N. miniatus × N. cavanillesii. The first species of each pair is the ovule donor while the second species corresponds to the pollen donor. Significant differences are indicated in bold type. **P < 0·01, ***P < 0·001.
Comparisons between synthetic and natural hybrids
Estimated with PI, synthetic CS hybrids yielded higher 2C-values than the natural hybrid N. × alentejanus (t = 23·418, d.f. = 32, P = 0·0001) while synthetic CM hybrids had lower 2C-values than the natural hybrid N. × perezlarae (t = 22·789, d.f. = 36, P = 0·015; Fig. 5). The remaining reciprocal hybrids showed no differences from their corresponding natural hybrids (SC vs. N. × alentejanus: t = 1·054, d.f. = 32, P = 0·306; MC vs. N. × perezlarae: t = 1·086, d.f. = 36, P = 0·067; Fig. 5). When stained with DAPI, synthetic F1 hybrids always yielded higher values than their natural hybrid counterparts (N. × alentejanus vs. synthetic hybrids: F2,21 = 21·312, P = 0·0001; N. × perezlarae vs. synthetic hybrids: F2,37 = 21·379, P = 0·0001).
Fig. 5.
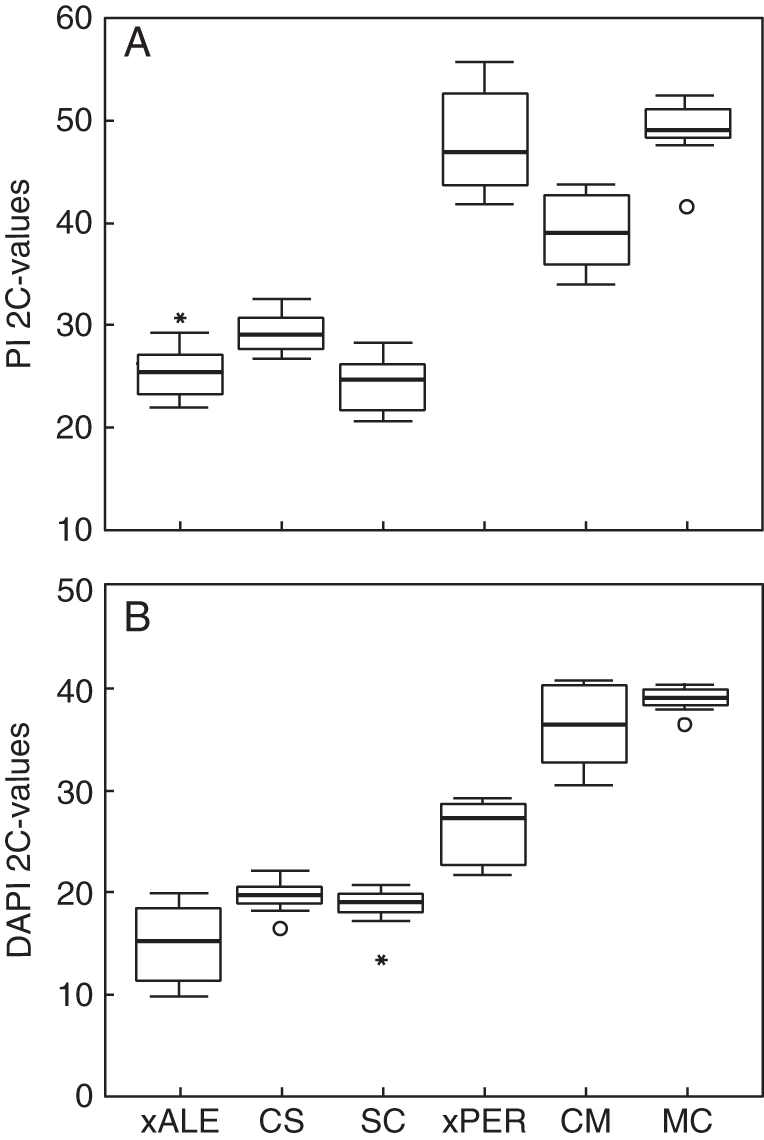
2C-values in natural hybrids, N. × alentejanus (xALE) and N. × perezlarae (xPER) and the equivalent artificial hybrids, CS (N. cavanillesii × N. serotinus), SC (N. serotinus × N. cavanillesii), CM (N. cavanillesii × N. miniatus) and MC (N. miniatus × N. cavanillesii). (A) Genome size obtained with PI. (B) Genome size obtained with DAPI. Values are expressed in pg. For details of the box-and-whisker plots see Fig. 3.
DISCUSSION
The genome size values here obtained are largely congruent with those previously reported in a study of the genus Narcissus as a whole using PI staining (Zonneveld, 2008). However, our extensive sampling provides a more accurate picture of the intraspecific variation of genome size in Narcissus (Table 1). Specifically, our results attest to quantitative and qualitative genome changes found in synthetic F1 interspecific hybrids, corresponding to the unstable phase that is characterized by structural changes and heterochromatic transposon amplification (McClintock, 1984).
A general observation is that the range of genome size values is usually lower for those species not considered of hybrid origin such as N. cavanillesii, N. serotinus and N. elegans as compared with those of hybrid origin such as N. × alentejanus, N. × perezlarae and the allopolyploid N. miniatus where s.d. values fall above 4 (Table 1). This observation is also true for 2C-values obtained with DAPI and therefore calls for an explanation as intraspecific variation in genome size is considered to be rather scarce (Šmarda and Bureš, 2006; Suda et al., 2007; but see Slovák et al., 2008).
A first relevant aspect of our results is the differences found in the 2C-values assessed with PI between the reciprocal F1 hybrids (Fig. 5, Table 3). A maternal effect for genome size in hybrids has been previously reported in synthetic homoploid hybrids of Helianthus (Baack et al., 2005), and in synthetic Brassica polyploids (Song et al., 1995), among other groups. To explain such effects, there is increasing evidence, observed in polyploids, that the maternal cytoplasm produces some selective pressure via seed inviability on the nuclear genome, which discards non-harmonious combinations and thus contributes to stabilize the newly synthesized hybrid (Soltis and Soltis, 1993; Caceres et al., 1998). One possible mechanism is that in many angiosperms the viability of embryos depends on the balance of maternal and paternal genomes in the endosperm. This balance should normally be 2 : 1 because of fusion of the polar nuclei with one sperm nucleus to form the central cell on the maternal side (Martienssen, 2010). Our two hybrids deviate from this ratio because of the divergent parental genomes involved, so we could expect a strong differential maternal effect.
A second relevant aspect of our study is the genome size differences, estimated with PI, found between natural hybrids and some of the synthetic hybrids. Although no significant differences were found between natural hybrids and some of the corresponding synthetic crosses (SC, MC), N. × alentejanus had lower values than synthetic CS hybrids while N. × perezlarae had higher values than synthetic CM hybrids (Fig. 5). One possible explanation for this pattern is that individuals studied from the natural hybrid populations represent late hybrid generations, which would have undergone substantial genome size changes as compared with F1s or are simply a mixture of hybrid genotypes, not necessarily from late hybrid generations. We cannot answer this question conclusively because in previous studies partially contradictory evidence was obtained as to the type of generation the natural hybrids belong to (see Introduction). Regardless, the fact that significant PI 2C-value differences are found in comparisons involving synthetic hybrids in which N. cavanillesii acts as ovule donor (CS, CM) suggests that those differences are representative of natural conditions because N. cavanillesii has been found to be the mother species in most natural hybrids (Marques et al., 2010). Therefore, our results suggest two different trends in the natural hybrids: to increase genome size in N. × perezlarae and to decrease it in N. × alentejanus after the first stages of hybridization.
Another conclusion is that because 2C-values obtained with DAPI were significantly larger in the synthetic F1 hybrids than in the respective natural hybrids (Fig. 4), an increase in GC-rich regions should occur in nature following hybridization. Although there are uncertainties as to the fine interpretation of DAPI values due to nucleotide requirements for the binding of this dye (Barow and Meister, 2002), our results are quite consistent in this respect.
In summary, our results from natural and synthetic hybrids indicate genome size variation and changes of different sorts. These include differences between reciprocal crosses for 2C values estimated with PI as well as differences between natural hybrids and some of the synthetic hybrids, an overall higher variation of genome size estimates in hybrids (both estimated with PI and DAPI), and higher 2C-values obtained with DAPI in synthetic F1 hybrids compared with their natural counterparts. With the exception of the first item already discussed (maternal effects), the others require specific and perhaps also some general underlying mechanisms.
The tendency towards genome size reduction, documented especially in allopolyploids (Rieseberg et al., 1995; Pikaard and Chen, 1998; Bureš et al., 2004; Garnatje et al., 2006), has been explained by rapid elimination of sets of non-coding sequences (Ozkan et al., 2001), and which can even be strongly biased towards one of the progenitor genomes (Levy and Feldman, 2004; Skalická et al., 2005). Although rarer, there are also reports of the opposite trend, of increasing genome size in hybrids (Baack et al., 2005).
In Narcissus, we suggest that chromosome rearrangements and recombination are involved in the genome size changes detected here since they can explain both the variation in genome size and the rate of the changes produced as fast as (but not only) in the F1 generation. In previous chromosome studies of the two hybrids, we observed a high frequency of aberrant meioses with chromosome bridges and lagging chromosomes, sometimes even preventing chromosome counts (Marques et al., 2010). Such aberrant meioses, due to lack of homology between chromosomes, may result in the asymmetric loss and gain of chromosome regions (Song et al., 1995). This is consistent with the extensive cytogenetic pioneering studies of Fernandes (1943, 1951, 1967, 1968, 1975) in Narcissus. He pointed out the importance of chromosome rearrangements in the evolution of Narcissus, specifically in the origin of the two base numbers x = 5 (subgenus Hermione) and x = 7 (subgenus Narcissus) and in the diversification of sections such as Jonquillae or Apodanthae. Recently, karyological polymorphisms that have been detected in N. serotinus and N. miniatus are clearly attributed to chromosome rearrangements caused by hybridization (Díaz Lifante et al., 2009).
Not much can be concluded regarding the functional and evolutionary significance of changes and variation in genome size in hybrids. In a previous study, we found that the natural hybrids studied here exhibit high fitness for vegetative components and are even able to set some seeds albeit at low frequency (Marques et al., 2011). Taking into account the higher range of genome size values in hybrids as well as the frequency of structural changes in Narcissus detected from cytogenetic approaches, it seems that a strong selection operating over a wide variety of genomes has resulted in a number of evolutionary viable genotypes in the two hybrids of Narcissus studied here.
SUPPLEMENTARY DATA
ACKNOWLEDGMENTS
We thank Miguel Serrano for assistance during the performance of this work, João Loureiro, Teresa Garnatje and two anonymous referees for comments on a previous version of this manuscript and Duncan Gilson for linguistic assistance. Seeds were provided by the Institut des Sciences du Végétal, Gif-sur-Yvette, France. This work was supported by a PhD fellowship to I.M. from FCT, Ministério de Ciência e Tecnologia, Portugal (PD/1245/2004), and through projects from CSIC-Comunidad de Madrid, Spain (CCG07-CSIC/AMB-1978), and Spanish Ministerio de Ciencia e Innovación (CGL2007-66516).
LITERATURE CITED
- Baack EJ, Whitney KD, Rieseberg LH. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytologist. 2005;167:623–630. doi: 10.1111/j.1469-8137.2005.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barow M, Meister A. Lack of correlation between AT frequency and genome size in higher plants and the effect of nonrandomness of base sequences on dye binding. Cytometry. 2002;47:1–7. doi: 10.1002/cyto.10030. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Plant genome values: How much do we know? Proceedings of National Academy of Sciences USA. 1998;95:2011–2016. doi: 10.1073/pnas.95.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms – progress, problems and prospects. Annals of Botany. 2005;95:45–90. doi: 10.1093/aob/mci003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureš P, Wang Y-F, Horová L, Suda J. Genome size variation in Central European species of Cirsium (Compositae) and their natural hybrids. Annals of Botany. 2004;94:353–363. doi: 10.1093/aob/mch151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres ME, Pace CD, Mugnozza GTS, Kotsonis P, Ceccarelli M, Cionini PG. Genome size variations within Dasypyrum villosum: correlations with chromosomal traits, environmental factors and plant phenotypic characteristics and behavior in reproduction. Theoretical and Applied Genetics. 1998;96:559–567. [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. Genetic and epigenetic interactions in allopolyploid plants. Plant Molecular Biology. 2000;43:387–399. doi: 10.1023/a:1006480722854. [DOI] [PubMed] [Google Scholar]
- Díaz Lifante Z, Andrés Camacho C, Viruel J, Caballero AC. The allopolyploid origin of Narcissus obsoletus (Alliaceae): identification of parental genomes by karyotype characterization and genomic in situ hybridization. Botanical Journal of the Linnean Society. 2009;159:477–498. [Google Scholar]
- Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM. Rapid elimination of low copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics. 1997;147:1381–1387. doi: 10.1093/genetics/147.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A. Sur la caryo-systématique de la section Autumnales Gay du genre Narcissus L. Boletim da Sociedade Broteriana Série 2. 1943;17:5–54. [Google Scholar]
- Fernandes A. Sur la phylogénie des espèces du genre Narcissus L. Boletim da Sociedade Broteriana Série 2. 1951;25:113–190. [Google Scholar]
- Fernandes A. Contribution à la connaissance de la biosystématique de quelques espèces du genre Narcissus L. Portugalia Acta Biologica Série B. 1967;9:1–44. [Google Scholar]
- Fernandes A. Keys to the identification of native and naturalized taxa of the genus Narcissus L. Daffodil Tulip Year Book. 1968;59:37–66. [Google Scholar]
- Fernandes A. L'évolution chez le genre Narcissus L. Anales del Instituto Botanico Antonio Jose Cavanilles. 1975;32:843–872. [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell-cycle in intact plant-tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- Garnatje T, Garcia S, Vilatersana R, Vallès J. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): systematic implications and additive changes during allopolyploidization. Annals of Botany. 2006;97:461–467. doi: 10.1093/aob/mcj050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. Plant speciation. New York: Columbia University Press; 1981. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RE, Zhao X-P, Islam-Faridi MN, et al. Evolution of interspersed repetitive elements in Gossypium (Malvaceae) American Journal of Botany. 1998;85:1364–1368. [PubMed] [Google Scholar]
- Karrenberg S, Lexer C, Rieseberg LH. Reconstructing the history of selection during homoploid hybrid speciation. American Naturalist. 2007;169:725–737. doi: 10.1086/516758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahulcova A, Krahulec F, Kirschner J. Introgressive hybridization between a native and an introduced species: Viola lutea subsp. sudetica versus V. tricolor. Folia Geobotanica. 1996;31:219–244. [Google Scholar]
- Lai Z, Nakazato T, Salmaso M, et al. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics. 2005;171:291–303. doi: 10.1534/genetics.105.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. Polyploidy in angiosperms. Trends in Plant Science. 1997;2:470–476. [Google Scholar]
- Levy A, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biological Journal of the Linnean Society. 2004;82:607–613. [Google Scholar]
- Liu B, Vega JM, Segal G, Abbo S, Rodova M, Feldman M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops: I. Changes in low-copy non-coding DNA sequences. Genome. 1998a;41:272–277. doi: 10.1139/g98-052. [DOI] [PubMed] [Google Scholar]
- Liu B, Vega JM, Feldman M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops: II. Changes in low-copy DNA sequences. Genome. 1998b;41:535–542. doi: 10.1139/g98-052. [DOI] [PubMed] [Google Scholar]
- Loureiro J, Trávníček P, Rauchová J, et al. The use of flow cytometry in the biosystematics, ecology and population biology of homoploid plants. Preslia. 2010;82:3–21. [Google Scholar]
- Marques I, Draper D. Hibridación natural y el origen de Narcissus perezlarae Font Quer en la Comunidad Valenciana. Valencia: Conselleria de Territori I Habitatge, Servei de Conservació de la Biodiversitat; 2004. [Google Scholar]
- Marques I, Nieto Feliner G, Draper D, Martins-Loução A, Fuertes Aguilar J. Unraveling cryptic reticulate relationships and the origin of orphan hybrid disjunct populations in Narcissus. Evolution. 2010;64:2353–2368. doi: 10.1111/j.1558-5646.2010.00983.x. [DOI] [PubMed] [Google Scholar]
- Marques I, Nieto Feliner G, Martins-Loução A, Fuertes Aguilar J. Fitness in Narcissus hybrids: low fertility is overcome by early hybrid vigour, absence of exogenous selection and high bulb propagation. Journal of Ecology. 2011;99:1508–1519. [Google Scholar]
- Martienssen RA. Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytologist. 2010;186:46–53. doi: 10.1111/j.1469-8137.2010.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Scheid OM, Matzke AJ. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. BioEssays. 1999;21:761–767. doi: 10.1002/(SICI)1521-1878(199909)21:9<761::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Meister A, Barow M. DNA base composition in plant genomes. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Weinheim: Willey-VCH; 2007. pp. 177–216. [Google Scholar]
- Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Chen ZJ. Nucleolar dominance. In: Paule RM, editor. RNA polymerase I: Transcription of eukaryotic ribosomal RNA. Austin, TX: R.G. Landes; 1998. pp. 275–293. [Google Scholar]
- Rieseberg LH, Van Fossen C, Desrochers AM. Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature. 1995;375:313–316. [Google Scholar]
- Salina EA, Ozkan H, Feldman M, Shumny VK. Subtelomeric repeat reorganization in synthesized wheat amphiploids. In: Kolchanov N, Furman D, editors. Proceedings of the Conference on Biodiversity and Dynamics of Ecosystems in North Eurasia. Novosibirsk, Russia: Institute of Cytology and Genetics Press; 2000. pp. 102–105. [Google Scholar]
- Scheid OM, Jakovleva L, Afsar K, Maluszynska J, Paszkowski J. A change in ploidy can modify epigenetic silencing. Proceedings of National Academy of Sciences USA. 1996;93:7114–7119. doi: 10.1073/pnas.93.14.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbach AE, Rieseberg LH. Likely multiple origins of a diploid hybrid sunflower. Molecular Ecology. 2002;11:1703–1715. doi: 10.1046/j.1365-294x.2002.01557.x. [DOI] [PubMed] [Google Scholar]
- Skalická K, Lim KY, Matyasek R, Matzke M, Leitch AR, Kovarik A. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytologist. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Slovák M, Vít P, Urfus T, Suda J. Complex pattern of genome size variation in a polymorphic member of the Asteraceae. Journal of Biogeography. 2008;36:372–384. [Google Scholar]
- Šmarda P, Bureš P. Intraspecific DNA content variability in Festuca pallens on different geographical scales and ploidy levels. Annals of Botany. 2006;98:665–678. doi: 10.1093/aob/mcl150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Molecular data and the dynamic nature of polyploidy. Critical Reviews in Plant Sciences. 1993;12:243–273. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: origins of species and genome evolution. Trends in Ecology and Evolution. 1999;9:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proceedings of National Academy of Sciences USA. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda J, Krahulcová A, Trávnícek P, Rosenbaumová R, Peckert T, Krahulec F. Genome size variation and species relationships in Hieracium subgen. Pilosella (Asteraceae) as inferred by flow cytometry. Annals of Botany. 2007;100:1323–1335. doi: 10.1093/aob/mcm218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium) Proceedings of National Academy of Sciences USA. 1995;92:280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X-P, Si Y, Hanson RE, et al. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Research. 1998;8:479–492. doi: 10.1101/gr.8.5.479. [DOI] [PubMed] [Google Scholar]
- Zonneveld B. The systematic value of nuclear DNA content for all species of Narcissus L. (Amaryllidaceae) Plant Systematics and Evolution. 2008;275:109–132. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.