Abstract
Methyltransferases that use S-adenosylmethionine (AdoMet) as a cofactor to catalyse 5-methyl uridine (m5U) formation in tRNAs and rRNAs are widespread in Bacteria and Eukaryota, and are also found in certain Archaea. These enzymes belong to the COG2265 cluster, and the Gram-negative bacterium Escherichia coli possesses three paralogues. These comprise the methyltransferases TrmA that targets U54 in tRNAs, RlmC that modifies U747 in 23S rRNA and RlmD that is specific for U1939 in 23S rRNA. The tRNAs and rRNAs of the Gram-positive bacterium Bacillus subtilis have the same three m5U modifications. However, as previously shown, the m5U54 modification in B. subtilis tRNAs is catalysed in a fundamentally different manner by the folate-dependent enzyme TrmFO, which is unrelated to the E. coli TrmA. Here, we show that methylation of U747 and U1939 in B. subtilis rRNA is catalysed by a single enzyme, YefA that is a COG2265 member. A recombinant version of YefA functions in an E. coli m5U-null mutant adding the same two rRNA methylations. The findings suggest that during evolution, COG2265 enzymes have undergone a series of changes in target specificity and that YefA is closer to an archetypical m5U methyltransferase. To reflect its dual specificity, YefA is renamed RlmCD.
INTRODUCTION
Ribothymidine (m5U) is present in tRNAs from the three domains of life (1–3) and is additionally found in the rRNAs of all Bacteria (4; http://www.ecosal.org) and certain Archaea (5). The RNAs of the best-studied bacterium, Escherichia coli, contain three m5U's. Two of these are in 23S rRNA and one in tRNA, and these modifications are conserved to varying degrees in other bacteria. The most highly conserved m5U is at position 54 in the T-loop of tRNA (6) and in E. coli this modification is added by the methyltransferase TrmA, which was formerly termed RumT (7). In 23S rRNA, m5U at nucleotide 1939 is present in all bacterial ribosomes characterized to date (8), and its formation is catalysed by the E. coli enzyme RlmD (formerly RumA) (9,10). The third m5U, at 23S rRNA position 747, is the least conserved of the three and is found mainly in Gram-negative Beta-, Epsilon- and Gamma Proteobacteria (3) and, in E. coli, this modification is added by the methyltransferase RlmC (formerly RumB) (10). All three E. coli m5U methyltransferases use S-adenosyl-l-methionine (AdoMet) as the methyl group donor, and their sequence similarity indicates that they share a common evolutionary origin (3). The sequences cluster within the family of orthologous groups COG2265 (11), which contains homologues from phylogenetically distant organisms such as the tRNA m5U54 methyltransferases Trm2p, found in the yeast Saccharomyces cerevisiae (12), and PAB0719 in the hyperthermophilic archaeon Pyrococcus abyssi (3).
The sites of m5U modification in the RNAs of Bacillus subtilis are similar to those in E. coli—the tRNAs of B. subtilis contain m5U54, and our earlier provisional studies on B. subtilis 23S rRNA indicated that nucleotides U747 and U1939 (E. coli numbering system) were also methylated. However, this similarity in m5U location obscured the fact that there are fundamental differences in the modification mechanisms in B. subtilis and E. coli. It had already been shown that in B. subtilis, the tRNA m5U54 modification is added by a folate-dependent methyltransferase (TrmFO) that bears no relationship to the COG2265 family members (13), and this discovery begged the question as to how m5U's might be added to the rRNA. Our searches of the B. subtilis genome revealed no paralogues of TrmFO, although two COG2265-type genes, yefA and yfjO, were evident. The protein products YefA and YfjO showed sufficient sequence similarity to each other, and to the E. coli RlmC and RlmD enzymes, to rank them as the most likely candidates for catalysis of m5U modification in B. subtilis rRNA.
In this study, we have used Matrix Assisted Laser Desorption/Ionization (MALDI) mass spectrometry (MS) to demonstrate conclusively that m5U's are present at nucleotides 747 and 1939 in B. subtilis 23S rRNA. The rRNAs were then analysed after inactivation of yefA or yfjO to establish whether the protein products of these genes were responsible for the m5U modifications. Enzyme function was re-established by expressing plasmid-encoded recombinant protein in the B. subtilis null-mutant and also in an E. coli mutant in which all three trmA, rlmC and rlmD genes had been deleted. Our findings show that compared to the E. coli RlmC and RlmD counterparts, a functional shift has occurred during the evolution of the B. subtilis YefA and YfjO methyltransferases.
MATERIALS AND METHODS
Bacillus subtilis and E. coli strains
Bacillus subtilis 168 (trpC2) was used here as the reference strain with a wild-type rRNA modification pattern. The B. subtilis knockout strains, BG12826 (yefA::pMutin2) and BG12911 (yfjO::pMutin2), originated from the collection of the Japanese/European B. subtilis Functional Analysis Program, and were created by a standard single crossover-based protocol using PCR-amplified fragments of target genes cloned in the pMUTIN2 vector (14). The ΔyefA::Cm deletion mutant was derived from the B. subtilis 168 strain. The upstream 891 bp and downstream 919 bp fragments of the yefA gene were PCR amplified from the genomic DNA using the primers yefA-up1 (5′-GCGCTCGGAATCCCACGCGAGG) together with yefA-up2 (5′-CGACCTGCAGGCATGCAAGCTCTATAATCACCTCTAGTTTTAAGCGAAAG), as well as yefA-down1 (5′-GCTCGAATTCACTGGCCGTCGTAAATAGTCAAAAATCTATGCTGATGG) together with yefA-down2 (5′-TAGTCCCTCGCAGCTTGCCCGC). The sequences underlined in the yefA-up2 and yefA-down1 primers are homologous to the ends of the pUC19-cloned 727-bp chloramphenicol resistance cassette (15) that was amplified using the PCR primers Cm5 (5′-CGACGGCCAGTGAATTCGAGC) together with Cm3 (5′-AGCTTGCATGCCTGCAGGTCG). The three PCR products were mixed in equimolecular amounts and joined in a subsequent PCR. The resulting 2.5-kb DNA fragment was used to transform competent B. subtilis 168 cells prior to selection at 37°C on LB rich medium containing chloramphenicol (5 µg/ml). Chromosomal DNA was extracted from chloramphenicol-resistant clones and yefA gene deletion was verified by PCR using the upstream out5 (5′-GAGTTTGACCTGATCATTGCGGC-3′) and downstream out3 (5′-GAATCTTTTCAGAATATATTGCTGG-3′) primers. One of the correct strains was dubbed 168 ΔyefA::Cm and was used for further study. The B. subtilis strain BFS2838 carrying an inactivated trmFO (gid-0) gene was kindly provided by S. Serror (European functional analysis project of B. subtilis; http://genome.jouy.inra.fr/cgi-bin/micado/fonct_analysis.cgi).
Escherichia coli strains used in this work are derivatives of BW 25113 [F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514] and were obtained from the Coli Genetic Stock Center. The BW 25113 ΔtrmA/ΔrlmC/ΔrlmD strain was obtained by sequential P1-mediated transductions of the ΔtrmA753::kan, ΔrlmD783::kan and ΔrlmC760::kan alleles (16). After each transduction, the kanamycin cassette was excised by transient expression of the specific FLP recombinase (17). Deletion of the three genes was verified by PCR using primers adjacent to the inactivated sequences.
Plasmids
The 1.4-kb yefA ORF was amplified from genomic DNA with the PCR primers atg_Hind (5′CCCAAGCTTATGAAAATGAAACCCCCAGTAG-3′) and stop_Xba (5′-CTAGTCTAGATTATTCTTTTAATTTTATCAACACACAG-3′) containing HindIII and XbaI sites, respectively. The PCR fragment was cloned into the SmaI-digested pUC19 vector to form pUC-yefA and was then digested with HindIII and XbaI before cloning the yefA sequence into these sites in plasmid pDG148 (18) forming pDG-yefA. In this vector, expression of yefA was under control of the Pspac promoter, and was induced with 0.5–1 mM IPTG. The pDG148 and pDG-yefA vectors were shuttled into competent 168 ΔyefA::Cm cells, and transformants were selected on LB plates containing chloramphenicol (5 µg/ml) and kanamycin (6 µg/ml).
The pBAD-yefA vector was obtained by digesting pUC-yefA with EcoRI and XbaI, and cloning the yefA fragment into the same sites in the pBAD24 vector bringing yefA expression under the control of the arabinose promoter, which was induced with 0.2% arabinose. Electrocompetent ΔtrmA/ΔrlmC/ΔrlmD cells were transformed with the pBAD-yefA vector and selected on LB plates containing ampicillin (100 µg/ml). All recombinants were verified by sequencing.
Preparation of B. subtilis and E. coli rRNA
The E. coli and B. subtilis strains were grown at 37°C to an OD600 of 0.6 in 200 ml LB medium containing the appropriate antibiotics. For strains complemented with recombinant copies of yefA and yfjO, IPTG was added at this stage to a final concentration of 1 mM to induce protein expression. Cells were kept at 37°C for three more hours, and were then harvested by centrifugation, washed and resuspended on ice in 3 ml of 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM NH4Cl before being lysed by sonication. Cell debris was removed by centrifugation and ribosomal particles were pelleted by ultracentrifugation (19) leaving bulk tRNA in the supernatant. The tRNA and rRNA fractions were extracted with phenol and chloroform, the RNAs were recovered by ethanol precipitation and dissolved in H2O.
MALDI mass spectrometry analyses of rRNA
RNA was extracted from the ribosomal particles of the B. subtilis 168 wild-type, yefA and yfjO knockouts, and from the knockouts complemented with active copies of yefA and yfjO; rRNAs were additionally extracted from the E. coli ΔrlmC/ΔrlmD/ΔtrmA triple knockout and from this strain expressing an active copy of yefA or yfjO. The sequence from G725 to G772 within domain II of the B. subtilis 23S rRNA (Figure 1A) was isolated by hybridization to a complementary 48-mer deoxyoligonucleotide, 5′-CCCACACCTCATCCCCGCACTTTTCAACGTGCGTGGGTTCGGGCCTCC, while the sequence from C1914 to C1961 within domain IV of the B. subtilis 23S rRNA (Figure 1B) was hybridized to the 48-mer, 5′GTCGGAACTTACCCGACAAGGAATTTCGCTACCTTAGGACCGTTATAG. The corresponding regions in E. coli rRNA were isolated using similar oligonucleotides adjusted for the sequence variations shown in Figure 1. In each case, 100 pmol of total rRNA was hybridized to 500 pmol of deoxyoligonucleotide; unprotected regions of the rRNAs were digested away with mung bean nuclease and RNase A; and the rRNA sequences paired to the deoxyoligonucleotide were separated by gel electrophoresis and extracted (20,21). The rRNA sequences of around 48 nt were digested with either RNase A or RNase T1 in aqueous solution containing 3-hydroxypicolinic acid and analysed by MALDI-MS (Voyager Elite, Perseptive Biosystems) recording in reflector and positive ion mode (22). Spectra were analysed using the program m/z (Proteometrics Inc). Tandem mass spectra were recorded in positive ion mode on a MicroMass MALDI Q-TOF Ultima mass spectrometer (23).
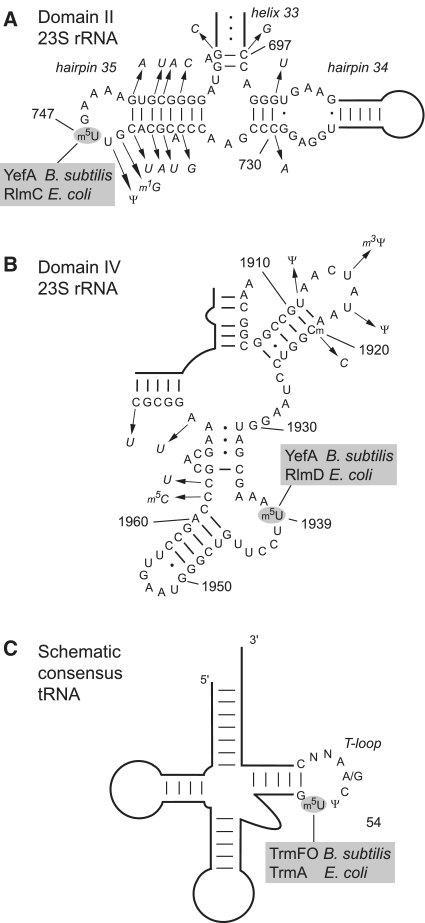
Figure 1.
The three characterized sites of m5U modification in B. subtilis and E. coli RNAs. The 23S rRNA secondary structures are based on the B. subtilis sequence and nucleotide differences in E. coli are indicated by the arrows pointing to the italicized nucleotides. (A) Portion of domain II of 23S rRNA, containing hairpins 34 and 35 and part of helix 33, showing the location of m5U747 modified by YefA in B. subtilis rRNA and RlmC in E. coli. The m1G745 modification is seen in E. coli rRNA but is absent in B. subtilis; the presence of pseudouridine Ψ746 could not be tested by mass spectrometry. (B) Region from domain IV of 23S rRNA containing m5U 1939 that is modified by YefA in B. subtilis rRNA, and by RlmD in E. coli. The B. subtilis rRNA is methylated on the ribose of nucleotide C1920 presumably by an orthologue of the enzyme TlyA (42) and this modification is missing in E. coli; the m5C1962 modification is present in the E. coli rRNA (47) but was not detected in B. subtilis; the pseudouridines Ψ1911, Ψ1915 and Ψ1917 could not be verified by mass spectrometry. (C) Schematic consensus of tRNA structures indicating the U54 target of the E. coli TrmA (48) and the structurally unrelated enzyme TrmFO that modifies the same nucleotide in B. subtilis using a tetrahydrofolate cofactor as the methyl donor (13,25).
Analyses of RNAs by HPLC and ion-trap electrospray mass spectrometry
The supernatant fractions containing tRNAs and other soluble RNAs were passed through Microcon YM-100 columns (Millipore) to reduce the amount of impurities. The supernatant samples were digested to completion to form nucleosides (24), and the pellet samples containing the rRNAs were similarly treated. The resultant nucleosides were analysed using a HPLC/ES-MS ion-trap system. Briefly, the loading pump of an Agilent 1100 HPLC was run at 5 μl/min; the column was a Hypercarb 150 × 0.3 mm, 5 µm PGC with a pore size of 250 (Thermo Scientific), linked to an Agilent XCT Ultra 6340 ion trap. Separation was achieved on gradients of 0–90% acetonitrile in 0.1% formic acid. ES-MS spectra were recorded in negative ion mode scanning initially in the m/z range 220–400 for intact nucleosides and then in lower m/z ranges after fragmentation (24). Nucleosides from the tRNAs were additionally analysed using reversed phase HPLC as previously described (24).
RESULTS
Nucleotide modifications m5U747 and m5U1939 are present in B. subtilis 23S rRNA
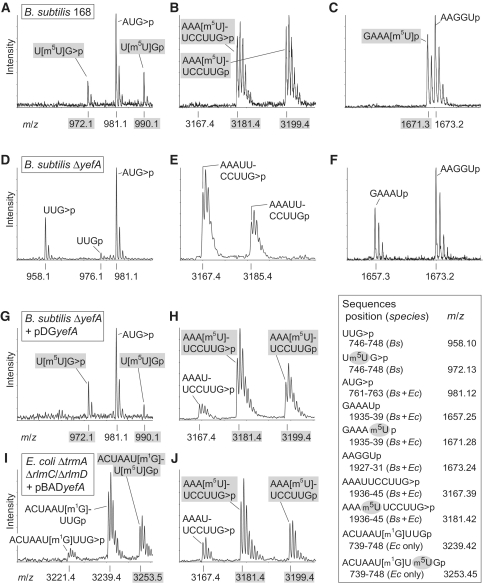
Isolation of the B. subtilis 23S rRNA domain II region (Figure 1A) followed by RNase T1 digestion gives rise to a unique UUGp fragment containing nucleotide U747. In the MALDI-MS spectra of the rRNA from the wild-type B. subtilis strain, this fragment produced peaks at m/z 990 and m/z 972, respectively, for the linear and cyclic phosphate forms of UUGp (Figure 2A), corresponding to the trimer plus 14 Da extra mass of one methyl group. There was no peak at m/z 976 (or at m/z 958 for the cyclic phosphate form), indicating that methylation of the sequence was stoichiometric. Tandem MS of this fragment and a longer partially digested RNase T1 fragment (CACGUUGp) showed that the methyl group resided on nucleotide U747 (Supplementary Figure S1).
Figure 2.
MALDI-MS analyses of m5U methylation sites in 23S rRNA. The upper row of spectra were generated from 23S rRNA of the B. subtilis wild-type strain 168 and show sequences containing m5U nucleotides (shaded in grey) at (A) U747 (in an RNase T1 oligo), (B) U1939 (RNase T1 oligo) and (C) U1939 (RNase A oligo). The corresponding spectral regions from the B. subtilis ΔyefA::Cm strain are shown in panels (D–F). The rRNA spectra from the B. subtilis ΔyfjO strain (data not shown) were identical to the wild-type rRNA. The rRNA analysed in spectra (G and H) was from B. subtilis ΔyefA::Cm strain complemented with an active copy of the yefA gene. (I and J) are spectra from the same rRNA regions of the E. coli ΔrlmC/ΔrlmD/ΔtrmA triple mutant expressing an active copy of yefA (note the E. coli m1G745 is resistant to RNase T1 digestion, producing a longer fragment). Before complementation with yefA, the rRNA from the E. coli ΔrlmC/ΔrlmD/ΔtrmA mutant produced none of the peaks containing m5U (data not shown). The table shows relevant fragments (sequences 5′–3′) generated from digestion of B. subtilis (Bs) and/or E. coli (Ec) rRNAs with RNase A (leaving a 3′-U or -C) and RNase T1 (3′-G). RNase T1 produced a mixture of fragments with a 2′–3′-cyclic phosphate (>p) and a linear 3′-phosphate (p), the latter having an 18 Da larger mass; the theoretical monoisotopic mass/charge values (m/z) are given for the predominant peaks.
In domain IV of the rRNA from the wild-type B. subtilis strain (Figure 1B), RNase T1 digestion produced a unique decamer AAAUUCCUUGp containing nucleotide U1939. The MS peak at m/z 3199 (and at m/z 3181 with a cyclic phosphate) corresponds to the decamer plus a single methyl group (Figure 2B). The lack of any peak at m/z 3185 (and at m/z 3167 with the cyclic phosphate) indicated stoichiometric methylation of this sequence. The RNase A fragment GAAAUp (nucleotides 1935–1939) flew at m/z 1671 (Figure 2C) showing that this overlapping sequence also carried the methyl group. The exact location of the methylation site was pinpointed to nucleotide U1939 by tandem MS analysis of this sequence using the slightly longer fragment GAAAUUp formed by partial RNase A digestion (Supplementary Figure S2).
These regions of the B. subtilis rRNA were analysed by primer extension, which did not reveal any reverse transcriptase stops or pauses at nucleotides U747 and U1939 (data not shown), and ruled out that the methylations could be at the N3 of the bases or the 2′-OH of the riboses (21). These findings were fully consistent with the location of methyl groups added by YefA being at the C5-positions of nucleotides U747 and U1939.
Candidate methyltransferases for the m5U747 and m5U1939 modifications
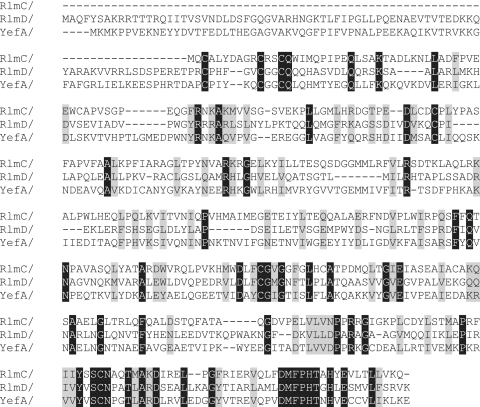
A BLAST search of the B. subtilis genome using the sequence of the tRNA-modifying enzyme TrmFO as the query revealed no paralogue that might function in rRNA modification (25). Similar searches using the E. coli 23S rRNA m5U methyltransferases RlmC and RlmD, revealed two B. subtilis orthologues, YefA and YfjO. These two putative B. subtilis methyltransferases are 43% identical (68% similar) in their amino acid sequences (Supplementary Figure S3) and are clearly paralogues. Comparison with the E. coli methyltransferases revealed that YefA displays 22% and 30% identity (40% and 49% similarity) to RlmC and RlmD, respectively (Figure 3); YjfO is slightly less close to the E. coli sequences and shows corresponding identities of 19% and 24% (42% and 47% similarity). Phylogenetically, all the four proteins YefA, YfjO, RlmC and RlmD fit into the same cluster of orthologous groups COG2265 (11). The yefA and yfjO genes were therefore candidates (and, in fact, the only likely candidates) for encoding enzymes that could catalyse the m5U modifications in B. subtilis 23S rRNA.
Figure 3.
Amino acid sequence alignment of m5U methyltransferases. The E. coli RlmC and RlmD enzymes, which are respectively specific for 23S rRNA nucleotides U747 and U1939, are aligned with the B. subtilis YefA sequence. Identical amino acid residues are highlighted in black, and grey indicates highly similar residues. Full-length sequences are shown; RlmC lacks the N-terminal extension seen in RlmD and YefA. The common core sequences were used for calculating percentages of amino acid identity and similarity.
Identifying the enzyme(s) responsible for the B. subtilis m5U747 and m5U1939 modifications
Strains BG12826 (yefA::pMutin2) and BG12911 (yfjO::pMutin2) were procured and then tested by PCR to established whether gene inactivation had been successful. The BG12911 (yfjO::pMutin2) strain produced PCR fragment patterns that were fully consistent with targeted inactivation of the yfjO gene; however, the (yefA::pMutin2) strain showed anomalies indicating that genes neighbouring yefA on the chromosome might also have been disturbed. We therefore made another yefA knockout (ΔyefA::Cm) in-house using a different procedure; subsequent PCR analysis (data not shown) confirmed that yefA had been inactivated without disruption of any other gene.
Analysis of the rRNA from the ΔyefA::Cm strain by MALDI-MS showed that methylation had been lost at both U747 (Figure 2D) and U1939 (Figure 2E and F). Parenthetically, we note that the initial yefA knockout, BG12826 (yefA::pMutin2), had also lost the methylations at both of these nucleotides (data not shown); however, we continued all recombinant work with our own ΔyefA::Cm strain. Transforming this latter strain with an active copy of yefA on a plasmid restored to a significant degree the methylation at both U747 (Figure 2G) and U1939 (Figure 2H).
The dual methylation function of YefA was additionally tested in a strain of E. coli in which all three m5U methyltransferase genes had been deleted. In this heterologous expression system, methylation was partially restored at both U747 (Figure 2I) and U1939 positions in the E. coli 23S rRNA (Figure 2J) with the latter site being more efficiently methylated. Methylated uridines from E. coli and B. subtilis tRNAs were analysed using an HPLC/ES-MS ion-trap system, which verified that the methyl group was on the C5-position of the uracil base (Supplementary Figure S4). Further HPLC analysis showed that YefA is not involved in m5U modification of tRNAs (Supplementary Figure S5).
Parallel sets of experiments were carried out in attempts to identify the function of the YfjO enzyme. In the BG12911 strain, in which this enzyme is missing, nucleotides m5U747 and m5U1939 remained fully methylated. Furthermore, HPLC analyses of tRNAs from the B. subtilis strains showed that, similar to YefA, YfjO plays no role in tRNA m5U modification (Supplementary Figure S5). At present therefore, the methylation target for the YfjO enzyme remains unknown.
DISCUSSION
The rRNAs of all living organisms are post-transcriptionally modified to improve their performance in protein synthesis (2,26), although there are marked differences in the mechanisms by which rRNA modification is achieved. In Archaea and Eukaryota, most rRNA modifications are uridine isomerizations and 2′-O-ribose methylations (8). These modifications are generally added by enzyme complexes that are guided to their nucleotide targets by complementary sequences in the sRNAs of Archaea (27,28) and the snoRNAs of Eukaryota (29–31), although some non-guided modifications have also been noted (32,33). In the rRNAs of Bacteria, uridine isomerizations and 2′-O-methylations also occur, although base methylation is the most frequent type of modification (4,34,35). None of the bacterial rRNA modifications requires the help of guide RNAs, and the general view has been that each modification is added by a single-specific enzyme that is capable of independently finding its target nucleotide.
On the whole, this view has been substantiated by studies of the numerous rRNA modifying enzymes in E. coli [reviewed in (34)] with only a few amendments being necessary. For instance, a handful of enzymes including the pseudouridine synthase RluD (20,36), the highly conserved m6A dimethyltransferase RsmA/KsgA (37–39) and the m5C methyltransferase RsmF of Thermus thermophilus (40) modify multiple sites that are immediately adjacent (RsmA) or within several nucleotides of each other on the bacterial rRNAs (RluD and RsmF). It could be argued that these enzymes engage in a single binding event with the rRNA during which a second or third nucleotide is accommodated into the active site without the need for the enzyme to dissociate from the substrate. Rare exceptions include the 2′-O-methyltransferase TlyA from Mycobacterium tuberculosis, which in separate reactions modifies nucleotides on both subunits of the ribosome (41), RluC which catalyses pseudouridine formation at distant locations in E. coli 23S rRNA (42), and RluA which forms pseudouridines in both the tRNAs and 23S rRNA of E. coli (43).
The data presented here show that B. subtilis YefA is able to recognize, bind and modify two targets that are at separate locations. From the MS analyses, it can be seen that B. subtilis 23S rRNA possesses the modifications m5U 747 (Figure 2A) and m5U1939 (Figure 2B and C) and that methylation is stoichiometric at each of the two nucleotides. Both methylations are lost upon inactivation of yefA and both are then restored upon complementation with an active copy of this gene. However, as it could formally have been argued that recognition by YefA of the two sites was facilitated by an auxiliary B. subtilis factor or was perhaps due to some peculiarity in the B. subtilis rRNA, a strain of E. coli was constructed in which all three of its m5U methyltransferase genes (rlmC, rlmD and trmA) were deleted. Upon heterologous expression of yefA in this E. coli strain, methylation at U747 and U1939 was regained showing that the specific recognition of dual targets by YefA is a property inherent within the structure of this enzyme. Analyses of the tRNA component of these cells showed that YefA is not involved in tRNA methylation. Thus, in B. subtilis, YefA is specific for the 23S rRNA modifications m5U747 and m5U1939, while modification at m5U54 tRNAs is added exclusively by the folate-dependent methyltransferase TrmFO (13,25).
In wild-type E. coli, the C5 positions of nucleotides U747 and U1939 are stoichiometrically methylated by the independent action of the enzymes RlmC and RlmD (9,10). The requirement for two autonomous m5U rRNA methyltransferases in E. coli, while a single enzyme can do the same job in B. subtilis, raises the question as to how the functions of YefA, RlmC and RlmD evolved. One likely scenario, consistent with phylogenetic analyses (3,5), would be that the contemporary B. subtilis YefA enzyme arose from a more promiscuous multi-site, AdoMet-dependent m5U rRNA methyltransferase that was an earlier (and now perhaps extinct) member of the COG2265 cluster of orthologous genes. Similarly, in E. coli and related species, single-site specificity evolved following duplication of an ancestral COG2265 gene giving rise to the rlmC and rlmD paralogues. The limited amino acid identity between the three enzyme sequences (Figure 3) favours the idea that separation of RlmC, RlmD and YefA was indeed an ancient event. In the context of functional shifts, we note that while YefA stoichiometrically methylates both of its B. subtilis U747 and U1939 targets (Figure 2A and B) and the recombinant YefA methylates with high efficiency at U1939 in B. subtilis (Figure 2H) and E. coli (Figure 2J), the enzyme was clearly less effective at recognizing U747 in E. coli where an appreciable residue of unmethylated nucleotide remained (Figure 2I). A precedence for target shifts in m5U RNA methyltransferases has already been observed in the archaeon Pyrococcus abyssi where two functional RlmD-like paralogues are present, and neither of these modified U1939 (3,5).
Although m5U RNA methyltransferases have changed their modification targets during evolution, the requirement for m5U modification at specific locations has remained remarkably constant. The majority of organisms appear to possess a homologue of either TrmA or TrmFO for m5U54 modification in tRNA (25). Likewise, the majority of bacteria encoded at least one RlmD gene, which fits with the observation that all bacteria studied to date possess the 23S rRNA m5U1939 methylation and many also have the m5U747 modification. The conservation of modified nucleotides at identical positions in rRNA would suggest that they play essential roles. Nevertheless, the E. coli knockout strain grew comfortably (in rich medium) despite the loss of all three of its m5U’s and, even though the strain would be expected to be at a disadvantage if subjected to stresses including antibiotics (44), its resilience was noteworthy. Similarly, the deletion of yefA (or yfjO) in B.subtilis had no major effect on the viability of the strains under standard laboratory growth conditions.
A functional role for the paralogue YfjO remains to be determined. The YfjO enzyme does not play a role in m5U methylation of B. subtilis tRNAs (Supplementary Figure S4). The HPLC and ion-trap-MS approaches (Supplementary Figures S4 and S5) are less well suited to the analysis of m5U nucleotides in the larger rRNAs, where this modification makes up a much smaller a proportion of the total uridine complement and, although we did show by MALDI-MS that YfjO does not modify within the 747 or 1939 regions of 23S rRNA, we have not ruled out that this enzyme might have an rRNA modifying function. In fact, four rlmD orthologues and one trmFO orthologue are evident in the genome of Acholeplasma laidlawii (45), which belongs to the Mollicutes and is distantly related to B. subtilis, and this fits well with an earlier analysis that indicated up to six m5U nucleotides at unidentified locations in the A. laidlawii 23S rRNA (46). The present evidence is consistent with certain members of the Gram-positive Firmicutes (which include Bacillus and Acholeplasma) having, in addition to U747 and U1939, m5U modification(s) in at least one other region of the 23S rRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Research Agency (FNU-rammebevillinger 09064292/10-084554); Nucleic Acid Center of the Danish Grundforskningsfond (to S.D.); H.G. holds a position of Emeritus scientist at Université de Paris-Sud. Funding for open access charge: Danish Research Agency (FNU-rammebevillinger 09064292/10-084554 to S.D.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Jean-Pierre Rousset and Philippe Noirot (INRA, Jouy-en-Josas) for their support of the work in providing laboratory facilities to C.F., D.B. and H.G. Etienne Dervyn (INRA, Jouy-en-Josas) kindly provided the B. subtilis knockout strains BG12826 and BG12911 and assisted with B. subtilis transformation. The authors thank Céline Brochier-Armanet (University of Provence in Marseilles) for discussions on evolutionary aspects of RNA m5U methyltransferases. Anders M. Giessing, Christina S. Sørensen and Mikkel, C. Hansen are thanked for assistance with MS studies, for checking the protein sequence alignments and PCR analyses.
REFERENCES
- 1.Björk GR, Hagervall TG. Transfer RNA modification. In: Böck A, Curtis R, Kaper JB, Neidhardt T, Nyström T, Squires C, editors. Escherichia coli and Salmonella. Washington DC: ASM Press; 2005. p. 4.6.2. [Google Scholar]
- 2.Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin Texas: Landes Biosciences; 2009. [Google Scholar]
- 3.Urbonavičius J, Auxilien S, Walbott H, Trachana K, Golinelli-Pimpaneau B, Brochier-Armanet C, Grosjean H. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol. Microbiol. 2008;67:323–335. doi: 10.1111/j.1365-2958.2007.06047.x. [DOI] [PubMed] [Google Scholar]
- 4.Ofengand J, Del Campo M. Modified nucleotides of E. coli ribosomal RNA. In: Böck A, Curtis R, Kaper JB, Neidhardt T, Nyström T, Squires C, editors. Escherichia coli and Salmonella. Washington DC: ASM Press; 2005. p. 4.6.1. [Google Scholar]
- 5.Auxilien S, Rasmussen A, Rose S, Brochier-Armanet C, Husson C, Fourmy D, Grosjean H, Douthwaite S. Specificity shifts in the rRNA and tRNA nucleotide targets of archaeal and bacterial m5U methyltransferases. RNA. 2011;17:45–53. doi: 10.1261/rna.2323411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Björk GR. Transductional mapping of gene trmA responsible for the production of 5methyluridine in transfer ribonucleic acid of Escherichia coli. J. Bacteriol. 1975;124:92–98. doi: 10.1128/jb.124.1.92-98.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwalla S, Kealey JT, Santi DV, Stroud RM. Characterization of the 23S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 2002;277:8835–8840. doi: 10.1074/jbc.M111825200. [DOI] [PubMed] [Google Scholar]
- 10.Madsen CT, Mengel-Jorgensen J, Kirpekar F, Douthwaite S. Identifying the methyltransferases for m5U747 and m5U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003;31:4738–4746. doi: 10.1093/nar/gkg657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordlund ME, Johansson JO, von Pawel-Rammingen U, Byström AS. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA. 2000;6:844–860. doi: 10.1017/s1355838200992422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urbonavičius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA: m5U methyltransferase in bacteria-evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 15.Trieu-Cuot P, de Cespedes G, Horaud T. Nucleotide sequence of the chloramphenicol resistance determinant of the streptococcal plasmid pIP501. Plasmid. 1992;28:272–276. doi: 10.1016/0147-619x(92)90060-n. [DOI] [PubMed] [Google Scholar]
- 16.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2001. [Google Scholar]
- 19.Douthwaite S, Powers T, Lee JY, Noller HF. Defining the structural requirements for a helix in 23S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 1989;209:655–665. doi: 10.1016/0022-2836(89)93000-3. [DOI] [PubMed] [Google Scholar]
- 20.Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007;425:1–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 22.Kirpekar F, Douthwaite S, Roepstorff P. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA. 2000;6:296–306. doi: 10.1017/s1355838200992148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirpekar F, Krogh TN. RNA fragmentation studied in a matrix-assisted laser desorption/ionisation tandem quadropole/orthogonal time-of-flight mass spectrometer. Rapid Commun. Mass Sp. 2001;15:8–14. doi: 10.1002/1097-0231(20010115)15:1<8::AID-RCM185>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 24.Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbonavičius J, Brochier-Armanet C, Skouloubris S, Myllykallio H, Grosjean H. In vitro detection of the enzyme activity of folate-dependent tRNA(U54, C5)methyltransferase. Methods in Enzymology. 2007;425:103–119. doi: 10.1016/S0076-6879(07)25004-9. [DOI] [PubMed] [Google Scholar]
- 26.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 27.Dennis PP, Omer A, Lowe T. A guided tour: small RNA function in Archaea. Mol Microbiol. 2001;40:509–519. doi: 10.1046/j.1365-2958.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- 28.Tran E, Brown J, Maxwell ES. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem. Sci. 2004;29:343–350. doi: 10.1016/j.tibs.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Bachellerie JP, Cavaille J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 30.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafontaine DL, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 32.Bonnerot C, Pintard L, Lutfalla G. Functional redundancy of Spb1p and a snR52dependent mechanism for the 2′-O-ribose methylation of a conserved rRNA position in yeast. Mol. Cell. 2003;12:1309–1315. doi: 10.1016/s1097-2765(03)00435-0. [DOI] [PubMed] [Google Scholar]
- 33.Decatur WA, Schnare MN. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Mol. Cell Biol. 2008;28:3089–3100. doi: 10.1128/MCB.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YgdE is the 2'-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol. 2009;72:1147–1158. doi: 10.1111/j.1365-2958.2009.06709.x. [DOI] [PubMed] [Google Scholar]
- 35.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ero R, Leppik M, Liiv A, Remme J. Specificity and kinetics of 23S rRNA modification enzymes RlmH and RluD. RNA. 2010;16:2075–2084. doi: 10.1261/rna.2234310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helser TL, Davies JE, Dahlberg JE. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 38.Demirci H, Murphy Ft, Belardinelli R, Kelley AC, Ramakrishnan V, Gregory ST, Dahlberg AE, Jogl G. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010;16:2319–2324. doi: 10.1261/rna.2357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desai PM, Culver GM, Rife JP. Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly. Biochemistry. 2011;50:854–863. doi: 10.1021/bi101005r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demirci H, Larsen LH, Hansen T, Rasmussen A, Cadambi A, Gregory ST, Kirpekar F, Jogl G. Multi-site-specific 16S rRNA methyltransferase RsmF from Thermus thermophilus. RNA. 2010;16:1584–1596. doi: 10.1261/rna.2088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2'-O-methylations in 16S and 23S rRNAs. Mol. Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 42.Conrad J, Sun D, Englund N, Ofengand J. The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23S ribosomal RNA. J. Biol, Chem. 1998;273:18562–18566. doi: 10.1074/jbc.273.29.18562. [DOI] [PubMed] [Google Scholar]
- 43.Wrzesinski J, Nurse K, Bakin A, Lane BG, Ofengand J. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for Ψ746 in 23S RNA is also specific for Ψ32 in tRNAPhe. RNA. 1995;1:437–448. [PMC free article] [PubMed] [Google Scholar]
- 44.Persaud C, Lu Y, Vila-Sanjurjo A, Campbell JL, Finley J, O'Connor M. Mutagenesis of the modified bases, m5U1939 and Ψ2504, in Escherichia coli 23S rRNA. Biochem. Biophys. Res. Commun. 2010;392:223–227. doi: 10.1016/j.bbrc.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 45.Kovaleva GY, Kovaleva GY, Kazanov MD, Malko DB, Vitreschak AG, Sernova NV, Gelfand MS, Lazarev VN, Levitskii SA, Basovskii YI, et al. Acholeplasma laidlawii complete genome. EMBL accession number CP000896. 2007 [Google Scholar]
- 46.Hsuchen CC, Dubin DT. Methylation patterns of mycoplasma transfer and ribosomal ribonucleic acid. J. Bacteriol. 1980;144:991–998. doi: 10.1128/jb.144.3.991-998.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purta E, O'Connor M, Bujnicki J, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 2008;383:641–651. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 48.Ny T, Lindstrom HR, Hagervall TG, Björk GR. Purification of transfer RNA m5U54-methyltransferase from Escherichia coli. Association with RNA. Eur. J. Biochem. 1988;177:467–475. doi: 10.1111/j.1432-1033.1988.tb14396.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.