Abstract
Objectives
To examine the effects of Roux-en-Y gastric bypass (RYGB) surgery with and without laparoscopic removal of omental fat (omentectomy) on the temporal gene expression profiles of skeletal muscle.
Design
Previously reported were the whole-body metabolic effects of a randomized, single-blinded study in patients receiving RYGB surgery stratified to receive or not receive omentectomy. In this follow up study we report on changes in skeletal muscle gene expression in a subset of 21 patients, for whom biopsies were collected preoperatively and at either 6 months or 12 months postoperatively.
Methodology/Principal Findings
RNA isolated from skeletal muscle biopsies of 21 subjects (8 without omentectomy and 13 with omentectomy) taken before RYGB or at 6 and 12 months postoperatively were subjected to gene expression profiling via Exon 1.0 S/T Array and Taqman Low Density Array. Robust Multichip Analysis and gene enrichment data analysis revealed 84 genes with at least a 4-fold expression difference after surgery. At 6 and 12 months the RYGB with omentectomy group displayed a greater reduction in the expression of genes associated with skeletal muscle inflammation (ANKRD1, CDR1, CH25H, CXCL2, CX3CR1, IL8, LBP, NFIL3, SELE, SOCS3, TNFAIP3, and ZFP36) relative to the RYGB non-omentectomy group. Expressions of IL6 and CCL2 were decreased at all postoperative time points. There was differential expression of genes driving protein turnover (IGFN1, FBXW10) in both groups over time and increased expression of PAAF1 in the non-omentectomy group at 12 months. Evidence for the activation of skeletal muscle satellite cells was inferred from the up-regulation of HOXC10. The elevated post-operative expression of 22 small nucleolar RNAs and the decreased expression of the transcription factors JUNB, FOS, FOSB, ATF3 MYC, EGR1 as well as the orphan nuclear receptors NR4A1, NR4A2, NR4A3 suggest dramatic reorganizations at both the cellular and genetic levels.
Conclusions/Significance
These data indicate that RYGB reduces skeletal muscle inflammation, and removal of omental fat further amplifies this response.
Trial Registration
ClinicalTrials.gov NCT00212160
Introduction
Obesity is an important risk factor for prevalent chronic health complications, such as type 2 diabetes (T2D) and cardiovascular disease. At the nexus of obesity-related co-morbidities is insulin resistance, which is characterized by a reduced responsiveness of insulin-sensitive tissues such as skeletal muscle, adipose tissue and liver to insulin-mediated glucose and lipid metabolism. Insulin resistance in skeletal muscle is considered especially pathogenic, as this tissue accounts for the majority of insulin-stimulated glucose disposal [1]. Accumulation of intramuscular lipid and excess plasma free fatty acid levels are considered central to aberrant skeletal muscle insulin signaling [2]. The pro-inflammatory state associated with obesity [3] is also implicated as a factor in skeletal muscle insulin resistance [4], [5].
Expanded visceral fat, as opposed to subcutaneous fat, is more strongly associated with insulin resistance and various comorbidities, especially cardiovascular disease, hypertension and hyperlipidemia [6], [7], [8], [9], [10]; it is also considered an important source of systemic free fatty acid (FFA) overload and inflammation due to enhanced lipolysis, cytokine secretion [11], [12], and macrophage infiltration [13]. Based on the available literature, it is clear that visceral fat is associated with insulin resistance. Our recent findings show that removal of the omentum with RYGB does not impart any additional benefits on hepatic insulin sensitivity or on insulin induced peripheral (muscle) glucose utilization [14]. However, these observations do not rule out any beneficial effects of removal of visceral fat on other muscle-mediated variables that could influence muscle glucose utilization. Recently, Varma et al. provided evidence that pro-inflammatory macrophages infiltrate skeletal muscle of obese, insulin-resistant humans and are activated by fatty acids [15], suggesting that local inflammation might be causative of skeletal muscle insulin resistance. Elevated proinflammatory cytokines such as TNF-α as well as increased proinflammatory pathway activation such as NFκB signaling through IκB kinase (IKK) and JNK-mediated phosphorylation of IRS-1 are also observed in various models of obesity induced insulin resistance [16], [17], [18], [19]. These observations suggest local inflammation in muscle as a possible mechanism by which the effects of insulin may be governed.
RYGB surgery results in 40% weight loss and resolution of T2D in 80% of patients by one year after surgery [20]; additionally, skeletal muscle insulin sensitivity is improved ∼two-fold [14], [21]. Relatively few studies have examined the effect of RYGB on muscle lipid content [22], [23] and insulin signaling proteins [24], [25]. One study revealed a differential expression of genes involved in insulin signaling (growth factor receptor-bound protein 14; GRB14), triglyceride synthesis (glycerol-3-phosphate dehydrogenase 1; GPD1), and muscle mass (myostatin; GDF8) by performing microarray analysis on muscle biopsies from 3 subjects obtained before and 12 months after gastric bypass surgery [26]. In the current study, we report the effects of RYGB combined with omental fat removal on skeletal muscle glucose utilization and on gene expression profiles, especially those related to the inflammatory pathways. Serial skeletal muscle biopsies and hyperinsulinemic-euglycemic clamps were performed preoperatively and at 6 and/or 12 months post-RYGB in 21 obese subjects undergoing RYGB and randomized to omentectomy.
Materials and Methods
Ethics Statement
All subjects provided written, informed consent before participating in this study, which was approved by the Health Sciences Committee of the Vanderbilt University Institutional Review Board.
Subjects
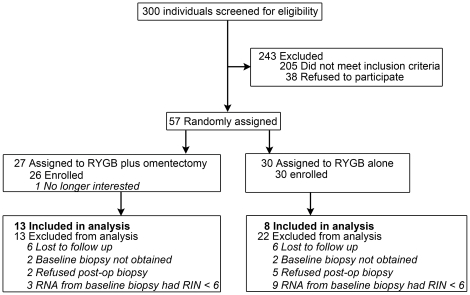
Obese men and women between 18 and 60 years old, with and without T2D, and with physician's approval for RYGB, were recruited from the Center for Surgical Weight Loss at Vanderbilt University Medical Center (Nashville, TN). The cohorts consisted of 13 subjects receiving RYGB surgery plus omentectomy (11 women and 2 men, 44±8 years old, 7 with T2D) and 8 subjects receiving RYGB surgery alone (7 women and 1 man, 40±11 years old, 6 with T2D, see Figure 1 and Table 1 ). Diabetic subjects using oral anti-diabetic agents were asked to discontinue all medications for 5 days prior to each study visit.
Figure 1. CONSORT Diagram.
Table 1. Effect of Roux-en-Y Gastric Bypass surgery with or without omentectomy on body composition and metabolic variables.
| Category | Before surgery | 6 months post-op | 12 months post-op | P value | |||||
| Omentectomy (n) | No (8) | Yes (13) | No (7) | Yes (12) | No (8) | Yes (8) | Group | G×T | Time |
| Gender (F/M) | 7/1 | 11/2 | 6/1 | 10/2 | 7/1 | 6/2 | |||
| Ethnicity (Cau/AfAm) | 6/2 | 9/4 | |||||||
| Diabetes (N/Y) | 2/6 | 6/7 | 2/5 | 5/7 | 2/6 | 3/5 | |||
| Weight (kg) | 129±22 | 137±17 | 97±16 | 98±13 | 87±16 | 89±8 | 0.311 | 0.191 | <0.001 |
| Body mass index (kg/m2) | 46.3±6.2 | 49.6±8.6 | 34.3±4.8 | 34.8±5.7 | 31.1±4.5 | 31.8±4.7 | 0.291 | 0.174 | <0.001 |
| Fat mass (kg) | 62±13 | 62±11 | 42±12 | 40±9 | 33±11 | 34±8 | 0.924 | 0.691 | <0.001 |
| Lean mass (kg) | 64±12 | 67±11 | 53±14 | 54±9 | 50±11 | 52±8 | 0.538 | 0.353 | <0.001 |
| Body fat (%) | 48±5 | 47±5 | 43±7 | 42±6 | 38±8 | 39±7 | 0.661 | 0.478 | <0.001 |
| Glucose (mg/dL) | 141±52 | 133±44 | 93±15 | 96±10 | 92±13 | 107±50 | 0.601 | 0.455 | <0.001 |
| Insulin (mU/L) | 29±14 | 24±12 | 8 ± 5 | 8±3 | 8±4 | 7±4 | 0.239 | 0.504 | <0.001 |
| Glycosylated hemoglobin (%) | 6.3±0.7 | 6.6±0.8 | 5.7±0.8 | 5.8±0.5 | 5.7±0.5 | 6.0±1.4 | 0.706 | 0.843 | <0.001 |
| Free fatty acids (mmol/L) | 0.64±0.12 | 0.70±0.23 | 0.65±0.23 | 0.60±0.15 | 0.53±0.08 | 0.58±0.16 | 0.731 | 0.905 | 0.054 |
| Total triglyceride (mg/dL) | 155±67 | 148±91 | 94±43 | 83±29 | 83±35 | 81±25 | 0.723 | 0.679 | <0.001 |
| Leptin (ng/mL) | 36±13 | 35±10 | 20±12 | 16±5 | 17±12 | 13±9 | 0.701 | 0.507 | <0.001 |
| Total adiponectin (µg/mL) | 7.5±5.0 | 8.1±6.6 | 9.8±3.7 | 11.0±4.3 | 18.6±9.7 | 13.4±8.3 | 0.508 | 0.096 | <0.001 |
| M (mg/kg·min) | 4.4±1.8 | 4.8±1.3 | 6.9±2.0 | 8.6±1.5 | 8.9±1.9 | 9.5±2.2 | 0.287 | 0.917 | <0.001 |
| M/I (mg·mL/kg·min·mU) | 16.0±9.3 | 21.1±13.9 | 32.8±13.5 | 35.3±13.3 | 36.8±14.5 | 37.2±10.4 | 0.367 | 0.595 | <0.001 |
Values are means ± SD; P values for the main effects of group and time and their interaction (G×T) are reported. M, glucose disposal rate per kg of body weight; M/I, insulin sensitivity index (M-value per unit of plasma insulin).
Study Protocol
We conducted a 12-month randomized, controlled trial wherein subjects underwent RYGB with or without omentectomy. Studies were conducted preoperatively and at 6 and 12 months post-RYGB at the Vanderbilt Clinical Research Center (CRC) between March 2005 and December 2008. The protocol for this trial and the CONSORT checklist are available as supporting information (Protocol S1 and Checklist S1). Randomization was conducted using a computer generated code with a permuted block size of four. Separate randomization codes were used for Caucasian females, Caucasian males, African American females, and African American males to achieve equal gender and ethnic distributions in each group. The study coordinator generated the random allocation sequence and notified the surgeon at the beginning of the surgical procedure of the patient's randomization category. All recruited participants and researchers involved in the conduct of the study and sample analysis were blinded to treatment. The primary outcome measure in this study was insulin sensitivity as measured by the hyperinsulinemic euglycemic clamp [14], and a secondary outcome measure was gene expression in skeletal muscle biopsies. Enrollment was concluded when we obtained sufficient power for the primary outcome measure.
Muscle insulin sensitivity was estimated using a hyperinsulinemic euglycemic clamp as the rate of exogenous dextrose (20%) infusion to maintain euglycemia (plasma glucose of 90–100 mg/dl) during a high-dose insulin infusion (2–3 mU/kg.min). The clamp studies were performed in all subjects at three time points: (a) In the preoperative period, at least one week prior to the surgical procedure, and at (b) 6-months and (c) 12 months postoperatively. Muscle biopsies were acquired from the vastus lateralis muscle using a Bergstrom needle as previously described [14]. Basal (prior to RYGB) muscle biopsies were obtained from all subjects (n = 21) at the time of the surgical procedure, immediately after adequate induction of general anesthesia and prior to initiation of the RYGB procedure. The postoperative muscle biopsies were obtained on the CRC on the day of and prior to initiation of the insulin clamp, using 1% Lidocaine solution as local anesthetic. Fourteen subjects had muscle biopsies at the three specified time points (pre-RYGB and at 6 and 12 months post-RYGB); 5 subjects had muscle biopsies obtained during the basal period and at 6 months post-RYGB, and 2 subjects had biopsies performed during the basal period and at 12 months post-RYGB. Biopsy specimen were dissected free of fat and vasculature, blotted free of blood and immediately snap frozen in liquid nitrogen and stored at −80°C until analyzed. The surgical procedure consisted of a laparoscopic RYGB surgery, in which a small gastric pouch (∼25 ml) was created and a Roux-en-Y gastrojejunostomy with a 30- to 50-cm biliopancreatic limb and a 100- to 200-cm Roux limb was constructed. In patients randomized to receive an omentectomy, surgeons resected an average of 0.74 kg of greater omentum [14].
Blood was collected and assays were performed as previously described [14]. The inflammatory cytokines monocyte chemotactic protein-1 (MCP-1), interleukin (IL)-1β, IL-6, IL-8, IL-10, and tumor necrosis factor (TNF)-α were measured by multiplex BD™ Cytokine Bead Array combined with flow cytometry (BD Biosciences, San Diego, California). Fat and lean body mass were assessed with dual-energy x-ray absorptiometry (GE Lunar Prodigy, Madison, WI) using half-body scans[27].
RNA isolation
Skeletal muscle biopsies were chopped into small pieces at −20°C and 1 ml Trizol (Invitrogen, Carlsbad, CA) was added per 100 mg of sample. RNA was extracted using Qiagen's RNase-free DNase Set (Valencia, CA) supplied protocol with the following modification. RNA was precipitated with 0.25 volumes isopropanol (Fisher Scientific, Fairlawn, NJ) and 0.25 volumes of RNA precipitation solution (7% NaCl/21% disodium citrate). DNase treatment was performed by adding 10 µl DNase and 70 µl Buffer RDD (Qiagen) per 100 µl sample. Trizol isolation was repeated once or twice to remove the DNase and any remaining fats or salts. RNA was re-suspended in RNase/DNase free water.RNA quality was assessed by the Vanderbilt Microarray Shared Resource on an Agilent 2100 Bioanalyzer utilizing RNA integrity numbers (RINs), 260/280 absorption and 28 s∶18 s ratios [28]. RNA samples included in this experiment exceeded the recommended RIN of 6, with an average RIN of 7.4.
Generation of cDNA
To minimize costs, initial gene expression profiles of muscle RNA pools were determined using Affymetrix GeneChip Human Exon 1.0 ST arrays. Skeletal muscle RNA pools (pre-RYGB, 6 months post-op, and 12-months post-op) from RYGB subjects with and without omentectomy (6 pools total) were prepared from 42 different skeletal muscle RNA samples. RNA samples were prepared for microarray analysis using the standard Affymetrix protocol (Affymetrix Inc., Santa Clara, CA) and published techniques [29], [30]. Briefly, 200 ng pooled total RNA and poly-A spike in controls, were brought to 5 µl with nuclease free water. First Strand Synthesis Master mix was added to the sample mix and incubated at 42°C for 1 hr; enzymes were heat inactivated 10 minutes at 70°C, followed by a cooling to 4°C for 2 minutes. Subsequently, second strand synthesis master mix was added to the sample mix and incubated at 16°C for 2 hours. The reactions were heat-inactivated by 10 minute incubation at 75°C. The resulting cDNA was then processed in an IVT reaction to generate cRNA that was used in first strand synthesis reactions to generate targets of the correct sense for hybridization to the Affymetrix Exon arrays. The reactions were set up with 10 µg cRNA, random hexamer, and first strand synthesis reagents to make cDNA. The targets were then treated with RNAse to remove template RNA and cleaned up over kit supplied columns, or Agencourt SPRI beads, following manufacturer's protocol. A total of 5.5 µg of the clean cDNA target was then enzymatically fragmented and end-labeled using the Affymetrix kit reagents. The cRNA, cDNA, and fragmented and end-labeled targets were assessed by Agilent bioanalysis to ensure that the amplified targets met the recommended smear range, and that fragmentation and end-labeling were complete.
Microarray Hybridization
The requisite amount of target was then added to hybridization cocktail to give a final target concentration of ∼25 ng/µl in the hybridization cocktail. The targets in hybridization cocktail were heat denatured at 99°C for 5 minutes, cooled to 45°C for 5 minutes, centrifuged and then loaded on the Affymetrix cartridge arrays for a hybridization period of 16 hours at 45°C, with rotation of 60RPM in the Affymetrix Model 645 Hybridization Oven. The cartridge arrays were washed, and stained per standard Affymetrix Protocols using Affymetrix Hybridization Wash and Stain kit reagents. After washing and staining, the arrays were scanned in an Affymetrix 7G plus scanner. The resulting data was acquired by GCOS software.
Validation by Taqman Low Density Array
To validate the expression levels of probes from RNA analyses, the 42 individual RNA samples comprising the pool were analyzed using Taqman Low Density Array (TLDA) [31]. An additional 12 skeletal muscle RNA samples not previously analyzed by microarray (baseline and post-RYGB in seven additional subjects) also were included. Total RNA (500 ng) was reverse transcribed to ds cDNA using random primers, SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the supplied protocol. Following reverse transcription, each 20 µl reaction was diluted with 25 µl of DNase/RNase-free water. Quantitative PCR was then performed using 500 ng of cDNA per reaction well. Thirty-one experimental genes and one control gene were assayed by qPCR. Thirty-two gene specific assays were purchased from Applied Biosystems (Foster City, CA) as custom TLDA microfluidics cards (Format 32) with TLDA probes (Table S3) spotted in triplicate on 384 well plates. All reactions were carried out at the following thermocycling conditions: Step 1: 10 min at 95°C, Step 2: 40 cycles of 15 s at 95°C, and Step 3: 1 min at 60°C. An Applied Biosystems ABI 7900HT unit with automation attachment (Foster City, CA) was used for real-time PCR. This unit is capable of collecting spectral data at multiple points during a PCR run.
Statistical Quality Control, Methods and Bioinformatics
The sample size for the current study was dictated by availability of a baseline and at least one postoperative muscle biopsy collected from the parent study [14]. The study power analysis was completed using the general linear model. A sample size of at least 7 patients per condition per time point, provided at least 90% power to detect an effect size of 4, i.e., 4-fold change, with two-sided false discovery rate (FDR) of 0.05.
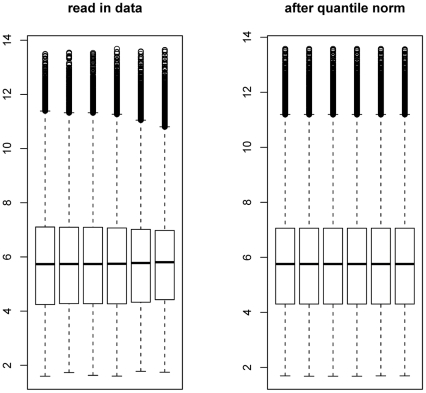
Anthropometric and metabolic parameters were analyzed for an effect of time and group (omentectomy) using linear mixed effects models in IBM SPSS version 18 (Chicago, IL). The overall intensities of all probes across the six microarrays were subjected to box plot analysis ( Figure 2 ). Exon ST 1.0 array data pre-processing was done using GeneSpring v7.3 software (Agilent Technologies, Foster City, CA). Probe-level analysis was performed using the Robust Multichip Analysis (RMA) method [32], but the default quantile across-sample normalization method of RMA was not done in order to avoid over-correction for the small sample size. The expression values were in log2 format after RMA. The change between samples was calculated as the difference between the two expression values. Winner probes for each comparison were selected based on two stringency criteria: a cutoff of 2 fold (2,697 genes, used for global pathway analysis; Table S1) or 4-fold change (103 genes, subset selected for TLDA validation; Table S2). Relative quantitation and standard deviation calculations with TLDA data were performed by the comparative C T method [33]. The 56 cDNA samples were assayed in triplicate for each gene of interest and the control. For each gene in each sample, the average C T and the standard deviation were calculated, and the 18S control average C T was subtracted from the gene average C T. The resulting value is the ΔC T. The standard deviation for the ΔC T was calculated by taking the square root of the sum of the control and gene standard deviations. For comparisons to basal time points, the average ΔC T for the basal period pools was subtracted from the respective average ΔC T of the 6 and 12 months samples generating a ΔΔC T. Fold changes between conditions are given by applying the expression 2−ΔΔCT, and ΔΔC T ± standard deviation applied to this expression gives the range. Correlation analysis was implemented using the “stat” package under R2.12.1 [34] without adjustment for multiple comparisons.
Figure 2. Box plot depicting the difference among microarray expression datasets (chips) before and after normalization by Robust Microarray Averaging (RMA).
Boxes show the 25th and 75th percentiles in the distribution of log-transformed (log base 2) intensities. The median is the horizontal bar in the middle of the box. The whiskers (dotted lines extending from the boxes) illustrate the maximum value or 1.5 times the interquartile range of data (IQR), whichever is smaller. The circles display any points beyond these whiskers.
Results
Participant Flow, Recruitment, and Numbers Analyzed
Fifty-six of 300 screened subjects were enrolled in the trial ( Figure 1 ). Twenty-one subjects completed all study components at all time points (pre-surgery, 6 and 12 month post-operative) and yielded a complement of muscle RNA samples of sufficient quality (average RIN of 7.4) to have expression analysis performed. The first participant was enrolled in March, 2005.
Anthropometric and Metabolic Parameters
Both the omentectomy and non-omentectomy groups exhibited significant decreases in body weight, BMI, fat mass, and lean mass during the 12 months after RYGB; there was neither a group effect nor a group×time interaction ( Table 1 ). Amount of initial weight lost was 28±3% in the first 6 months and 34±6% in 12 months following surgery across both groups. Similarly, fasting levels of glucose, insulin, triglycerides, leptin, and adiponectin were significantly decreased and free fatty acids were marginally decreased over time without a group or group×time effect. Insulin-stimulated glucose uptake in the muscle was significantly increased ∼2-fold by 12 months after RYGB in both groups, without significant differences between groups or group×time interactions.
Circulating Inflammatory Markers
A main effect of time post-RYGB was detected for systemic concentrations of CRP and MCP-1 but not for IL-1β, IL-6, IL-8, IL-10, nor TNF-α. There was no effect of omentectomy or group×time interaction for any inflammatory cytokine ( Table 2 ).
Table 2. Serological measures of inflammation determined before and after RYGB with and without omentectomy.
| Category | Before surgery | 6 months post-op | 12 months post-op | P value | |||||
| Omentectomy | No | Yes | No | Yes | No | Yes | Group | G×T | Time |
| C-reactive protein (µg/mL) | 11.0±9.1 | 9.3±11.0 | 2.4±1.4 | 3.2±3.6 | 2.8±3.8 | 1.0±0.5 | 0.716 | 0.983 | <0.001 |
| IL-1β (pg/ml) | 2.9±4.6 | 18.6±38.5 | 2.2±1.2 | 6.2±10.5 | 1.9±1.4 | 16.2±34.5 | 0.194 | 0.119 | 0.06 |
| IL-6 (pg/ml) | 3.1±2.0 | 62.5±190.5 | 5.4±4.7 | 10.9±21.5 | 4.4±6.6 | 62.1±159.7 | 0.299 | 0.085 | 0.114 |
| IL-8 (pg/ml) | 3.7±2.1 | 18.6±45.4 | 3.3±1.4 | 8.5±9.0 | 3.8±1.7 | 19.6±38.8 | 0.265 | 0.246 | 0.263 |
| IL-10 (pg/ml) | 1.8±1.0 | 3.8±4.4 | 1.9±0.9 | 1.9±1.1 | 1.3±0.5 | 2.0±0.7 | 0.106 | 0.316 | 0.089 |
| MCP-1 (pg/ml) | 85.9±71.1 | 135.5±209.4 | 70.4±66.8 | 59.8±52.9 | 57.4±49.1 | 97.9±175.7 | 0.087 | 0.41 | 0.001 |
| TNF-a (pg/ml) | 1.8±1.7 | 12.1±23.3 | 1.8±1.0 | 4.7±7.4 | 1.2±0.6 | 9.4±15.7 | 0.123 | 0.18 | 0.119 |
Overview of skeletal muscle gene expression patterns after RYGB with and without omentectomy
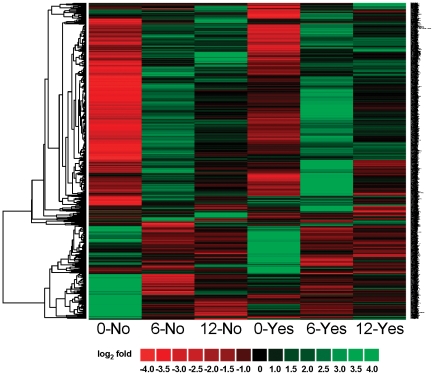
We compared skeletal muscle gene expression patterns between two surgical groups (RYGB surgery with no omentectomy vs. RYGB surgery with omentectomy) over time (6 vs. 0 months, and 12 vs. 0 months) in six skeletal muscle RNA sample pools derived by combining equal amounts of RNA from 7 subjects at each time point. For each time point, the observed gene expression value was compared to an overall average expression value derived from the union of all probe intensities (33,297) across all six Affymetrix Human Exon 1.0 ST Arrays. Probe intensities were then log2 transformed, and those genes expressed greater than 2 fold (log1 = 4 fold) within any time-point comparison were collated. In this manner, not all genes expressed were changed greater than 2-fold among all time-points. Expression levels of the 2,697 probes in each of the 6 microarrays changed greater than 2 fold after hierarchical clustering analysis is displayed graphically in Figure 3 and individual expression values are listed in Table S2. While significance cannot be derived from these microarray hybridizations without experimental replication, general expression were noted. For example, groups without (No) and with (Yes) omentectomy displayed similar expression profiles at 0 months. Additionally, most genes downregulated at 0 month were upregulated at 6 months, and conversely most genes upregulated at 0 month were downregulated at 6 months. At 12 months in both the omentectomy and non-omentectomy groups, the overall abundance of genes changing more than 2-fold was diminished, with no apparent differences as a function of omentectomy.
Figure 3. Heat map displaying hierarchical clustering results from microarray expression profile data derived from skeletal muscle RNA pools.
Samples include 0, 6 and 12 months “No” omentectomy and 0, 6 and 12 months “Yes” omentectomy. The expression of 2,697 genes is represented as a dendrogram after applying filtering criteria for selecting genes with a log2 ratio intensity greater than 1 (ratio>2). Gene expression ratios are displayed by applying progressively brighter shades of red (down-regulated) or green (up-regulated) to log2 ratios that increasingly deviate from zero.
Skeletal muscle gene expression changes without omentectomy
To identify those genes with the greatest change in expression after RYGB we compared genes with greater than 2-fold (log2) changes in expression. Table 3 lists those 25 genes that were differentially expressed comparing 6 vs. 0 months without omentectomy. The AP-1 transcription factors such as FOS, FOSB, JUNB as well as MYC and EGR1 were among those downregulated to the greatest degree at 6 months. These are immediate early genes acting on a specific biological response that directly induces or controls a variety of other genes [35]. HOXC10, a gene associated with muscle satellite cell activation, was the only transcription factor that was upregulated [36]. Genes regulating various cellular processes averaged a greater than 5-fold downregulation: inflammation (ANKRD1, CDR1, CCL2, IL6, MAOB, GADL1, ITLN1); protein turnover (IGFN1, FBXW10); and extracellular matrix remodeling (CYR61, THBS1, THBS4). The small nucleolar RNA C/D box genes SNORD115-44, SNORD25, and SNORD59B were among those genes which were upregulated at 6 months without omentectomy.
Table 3. Genes changed ≥4 fold (log 2 base ≥2) between 6 and 0 months no omentectomy.
| Probe ID | Gene Symbol | Gene Description | Process | Cytoband | mRNA Accession | log2fold |
| 7955858 | HOXC10 | homeobox C10 | transcription factor | 12q13.3 | NM_017409 | 2.05587 |
| 8148317 | MYC | v-myc myelocytomatosis viraloncogene homolog | transcription factor | 8q24.21 | NM_002467 | −2.0047 |
| 8026047 | JUNB | jun B proto-oncogene | transcription factor | 19p13.2 | NM_002229 | −2.0214 |
| 8029693 | FOSB | FBJ murine osteosarcoma viraloncogene homolog B | transcription factor | 19q13.32 | NM_006732 | −2.17407 |
| 8108370 | EGR1 | early growth response 1 | transcription factor | 5q31.1 | NM_001964 | −2.5463 |
| 7975779 | FOS | v-fos FBJ murine osteosarcoma viraloncogene homolog | transcription factor | 14q24.3 | NM_005252 | −3.14739 |
| 7981990 | PWCR1 | Prader-Willi syndrome chromosome region 1 | small nucleolar RNA expression | 15q11.2 | NR_003106 | 2.53618 |
| 7964246 | SNORD59B | small nucleolar RNA, C/D box 59B | small nucleolar RNA expression | 12q13.3 | NR_003046 | 2.49785 |
| 7982094 | SNORD115-44 | small nucleolar RNA, C/D box 115-44 | small nucleolar RNA expression | 15q11.2 | NR_003359 | 2.43735 |
| 7948910 | SNORD25 | small nucleolar RNA, C/D box 25 | small nucleolar RNA expression | 11q13 | NR_002565 | 2.09656 |
| 8012949 | FBXW10 | F-box and WD repeat domaincontaining 10, mRNA | protein turnover | 17p12 | ENST00000308799 | 2.30938 |
| 7908650 | IGFN1 | eEF1A2 binding protein | protein turnover | 1q32.1 | NM_178275 | −2.25232 |
| 8090872 | KY | kyphoscoliosis peptidase | other | 3q22.2 | NM_178554 | 2.06643 |
| 8175311 | CXorf48 | chromosome X open reading frame48 transcript variant 2 | other | Xq26.3 | ENST00000344129 | −2.09328 |
| 8156848 | NR4A3 | nuclear receptor subfamily 4, group A, member 3 | orphan receptor | 9q22 | NM_173198 | −2.75007 |
| 8085972 | GADL1 | glutamate decarboxylase-like 1 | insulin signaling | 3p24.1-p23 | NM_207359 | 2.26062 |
| 7921690 | ITLN1 | intelectin 1 (galactofuranose binding) (omentin-1) | insulin signaling | 1q22-q23.5 | NM_017625 | −2.40258 |
| 8172204 | MAOB | monoamine oxidase B | inflammation | Xp11.23 | NM_000898 | 2.08435 |
| 8131803 | IL6 | interleukin 6 (interferon, beta 2) | inflammation | 7p21 | NM_000600 | −2.27324 |
| 8006433 | CCL2 | chemokine (C-C motif) ligand 2 (MCP-1) | inflammation | 17q11.2-q12 | NM_002982 | −2.3367 |
| 8175531 | CDR1 | cerebellar degeneration-relatedprotein 1, 34kDa | inflammation | Xq27.1-q27.2 | NM_004065 | −2.51038 |
| 7934979 | ANKRD1 | ankyrin repeat domain 1 (cardiac muscle) | inflammation | 10q23.31 | NM_014391 | −3.16087 |
| 7982597 | THBS1 | thrombospondin 1 | extracellular matrix protein | 15q15 | NM_003246 | −2.11267 |
| 8106573 | THBS4 | thrombospondin 4 | extracellular matrix protein | 5q13 | NM_003248 | −2.23557 |
| 7902687 | CYR61 | cysteine-rich, angiogenic inducer, 61 | extracellular matrix protein | 1p31-p22 | NM_001554 | −2.78682 |
Table 4 lists the 36 genes that were most changed at 12 vs. 0 months in the RYGB without omentectomy cohort underscoring the durability of these responses. Several of the genes downregulated at 6 vs. 0 months without omentectomy also were observed at this time point, including the AP-1 transcription factors, markers of satellite cell activation, genes facilitating the inflammatory response, (CCL2, CDR1, and IL6), and the orphan receptor NR4A3, as well as genes regulating extracellular matrix formation and protein turnover. Additionally, over 20 genes regulating small nucleolar RNA (snoRNA) expression, mostly from the C/D box 115 group, were upregulated. SnoRNAs are not transcribed into protein products; they influence the structure of other mRNAs and their encoded proteins. Eighteen microarray probes mapping to fifteen SNORD 115 transcripts derived from cytoband 15q11.2 were upregulated, as were SNORD29, SNORD44, SNORD54 and SNORA73A (from the H/ACA group of snoRNAs).
Table 4. Genes changed ≥4 fold (log 2 base ≥2) between 12 and 0 months no omentectomy.
| Probe ID | Gene Symbol | Gene Description | Process | Cytoband | mRNA Accession | log2fold |
| 7908650 | IGFN1 | eEF1A2 binding protein | protein turnover | 1q32.1 | NM_178275 | −2.55556 |
| 8012949 | FBXW10 | F-box and WD repeat domaincontaining 10 | protein turnover | 17p12 | ENST00000308799 | 2.06496 |
| 7942476 | PAAF1 | proteasomal ATPase-associated factor 1 | protein turnover | 11q13.4 | NM_025155 | 2.01504 |
| 7921690 | ITLN1 | intelectin 1 (galactofuranosebinding) (omentin -1) | insulin signaling | 1q22-q23.5 | NM_017625 | −2.23682 |
| 8108370 | EGR1 | early growth response 1 | transcription factor | 5q31.1 | NM_001964 | −3.05413 |
| 7975779 | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | transcription factor | 14q24.3 | NM_005252 | −4.09474 |
| 8029693 | FOSB | FBJ murine osteosarcoma viraloncogene homolog B | transcription factor | 19q13.32 | NM_006732 | −2.12644 |
| 8026047 | JUNB | jun B proto-oncogene | transcription factor | 19p13.2 | NM_002229 | −2.38627 |
| 8148317 | MYC | v-myc myelocytomatosis viraloncogene homolog | transcription factor | 8q24.21 | NM_002467 | −2.0096 |
| 7955858 | HOXC10 | homeobox C10 | transcription factor | 12q13.3 | NM_017409 | 2.0778 |
| 8006433 | CCL2 | chemokine (C-C motif) ligand 2 | inflammation | 17q11.2-q12 | NM_002982 | −2.09334 |
| 8175531 | CDR1 | cerebellar degeneration-relatedprotein 1, 34kDa | inflammation | Xq27.1-q27.2 | NM_004065 | −3.36346 |
| 8086344 | CX3CR1 | chemokine (C-X3-C motif) receptor 1 | inflammation | 3p21|3p21.3 | NM_001337 | 2.12768 |
| 8131803 | IL6 | interleukin 6 (interferon, beta 2) | inflammation | 7p21 | NM_000600 | −2.57781 |
| 8156848 | NR4A3 | nuclear receptor subfamily 4,group A, member 3 | orphan receptor | 9q22 | NM_173198 | −2.72922 |
| 7899480 | SNORA73A | small nucleolar RNA, H/ACA box 73A | small nucleolar RNA expression | 1p36.1 | NR_002907 | 2.48545 |
| 7982038 | SNORD115-1 | small nucleolar RNA, C/D box 115-1 | small nucleolar RNA expression | 15q11.2 | NR_001291 | 2.68633 |
| 7982078 | SNORD115-11 | small nucleolar RNA, C/D box 115-11 | small nucleolar RNA expression | 15q11.2 | NR_003303 | 2.45315 |
| 7982024 | SNORD115-12 | small nucleolar RNA, C/D box 115-12 | small nucleolar RNA expression | 15q11.2 | NR_003304 | 2.54694 |
| 7982050 | SNORD115-12 | small nucleolar RNA, C/D box 115-12 | small nucleolar RNA expression | 15q11.2 | NR_003304 | 2.53634 |
| 7982084 | SNORD115-12 | small nucleolar RNA, C/D box 115-12 | small nucleolar RNA expression | 15q11.2 | NR_003304 | 2.03653 |
| 7982008 | SNORD115-13 | small nucleolar RNA, C/D box 115-13 | small nucleolar RNA expression | 15q11.2 | NR_003305 | 2.68633 |
| 7982032 | SNORD115-16 | small nucleolar RNA, C/D box 115-16 | small nucleolar RNA expression | 15q11.2 | NR_003308 | 2.68633 |
| 7982046 | SNORD115-20 | small nucleolar RNA, C/D box 115-20 | small nucleolar RNA expression | 15q11.2 | NR_003312 | 2.56052 |
| 7982052 | SNORD115-23 | small nucleolar RNA, C/D box 115-23 | small nucleolar RNA expression | 15q11.2 | NR_003315 | 3.03821 |
| 7982056 | SNORD115-25 | small nucleolar RNA, C/D box 115-25 | small nucleolar RNA expression | 15q11.2 | NR_003342 | 2.77049 |
| 7982058 | SNORD115-26 | small nucleolar RNA, C/D box 115-26 | small nucleolar RNA expression | 15q11.2 | NR_003343 | 2.46435 |
| 7982028 | SNORD115-43 | small nucleolar RNA, C/D box 115-43 | small nucleolar RNA expression | 15q11.2 | NR_003358 | 2.45315 |
| 7982094 | SNORD115-44 | small nucleolar RNA, C/D box 115-44 | small nucleolar RNA expression | 15q11.2 | NR_003359 | 2.75753 |
| 7982016 | SNORD115-5 | small nucleolar RNA, C/D box 115-5 | small nucleolar RNA expression | 15q11.2 | NR_003297 | 2.54694 |
| 7982018 | SNORD115-6 | small nucleolar RNA, C/D box 115-6 | small nucleolar RNA expression | 15q11.2 | NR_003298 | 2.30491 |
| 7982090 | SNORD115-6 | small nucleolar RNA, C/D box 115-6 | small nucleolar RNA expression | 15q11.2 | NR_003298 | 2.27704 |
| 7982020 | SNORD115-7 | small nucleolar RNA, C/D box 115-7 | small nucleolar RNA expression | 15q11.2 | NR_003299 | 3.00484 |
| 7982092 | SNORD115-9 | small nucleolar RNA, C/D box 115-9 | small nucleolar RNA expression | 15q11.2 | NR_003301 | 2.45315 |
| 7948902 | SNORD29 | small nucleolar RNA, C/D box 29 | small nucleolar RNA expression | 11q13 | NR_002559 | 2.34649 |
| 7922410 | SNORD44 | small nucleolar RNA, C/D box 44 | small nucleolar RNA expression | 1q25.1 | NR_002750 | 2.10829 |
| 8150877 | SNORD54 | small nucleolar RNA, C/D box 54 | small nucleolar RNA expression | 8q12 | NR_002437 | 2.06555 |
| 7902687 | CYR61 | cysteine-rich, angiogenic inducer, 61 | extracellular matrix | 1p31-p22 | NM_001554 | −2.6069 |
| 8106573 | THBS4 | thrombospondin 4 | extracellular matrix | 5q13 | NM_003248 | −2.07002 |
| 8042788 | ACTG2 | actin, gamma 2, smooth muscle, enteric | other | 2p13.1 | NM_001615 | −2.1841 |
| 7931832 | AKR1C2 | aldo-keto reductase family 1, member C2 | other | 10p15-p14 | NM_205845 | 2.20199 |
| 7924821 | gm127 | similar to RIKEN 2610020C11 | other | 1q42.13 | AF387613 | −2.183 |
| 8048014 | RPE | ribulose-5-phosphate-3-epimerase | other | 2q32-q33.3 | NM_199229 | 2.17662 |
Skeletal muscle gene expression changes with omentectomy
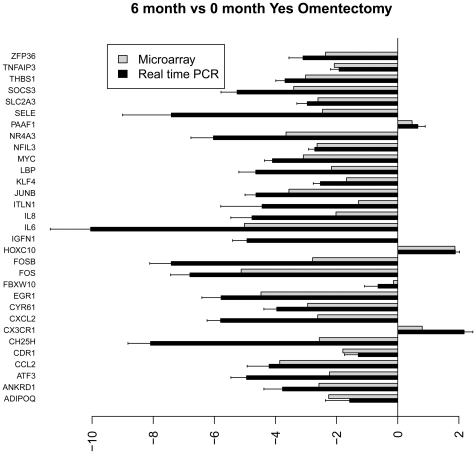
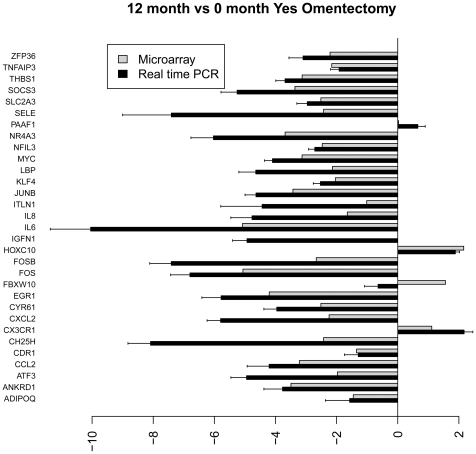
At 6 vs. 0 months in the RYGB with omentectomy group, the number of genes differentially expressed in muscle was nearly double that observed in the RYGB without omentectomy cohort at the same time point. Table 5 lists the 46 genes that expressed greater than a 4-fold change. Genes known to be involved in facilitating the inflammatory response were particularly responsive. In addition to ANKRD1, CCL2, and IL6 observed as being greater than 4-fold downregulated in the RYGB without omentectomy group, the inflammatory genes CH25H, CXCL2, SOCS3, IL8, LBP, NFIL3, SELE and TNFAIP3 also were downregulated. All of the transcription factors differentially expressed at 6 months in the non-omentectomy group were also downregulated in the 6 months with omentectomy group, as was ATF3. Adiponectin (ADIPOQ) and the glucose transporter (SLC2A3) genes along with the extracellular matrix proteins ADAM metallopeptidase 1 and 4 (ADAMTS1 and ADAMTS4), CYR61, CD54, THBD and THBS1 were all downregulated; however a handful of genes with varied functions (ATRX, DLEU2, ZNF780B, YIPF7, RWDD3, ANGPT1) were upregulated. All 3 members of the nuclear orphan nuclear receptor family 4 (NRFA1-3) recently identified as controlling skeletal muscle metabolism [37], [38] were downregulated; in particular NR4A3 which was downregulated over 12 fold. Similar phenomena were observed in the 12 vs. 0 months with omentectomy group as were found at 6 vs. 0 months with omentectomy group. A subset of other proteins with varying functions complete the list of genes with differential expression values greater than 4 fold; in total 36 genes were observed ( Table 6 ).
Table 5. Genes changed ≥4 fold (log 2 base ≥2) between 6 and 0 months with omentectomy.
| Probe ID | Gene Symbol | Gene Description | Process | Cytoband | mRNA Accession | log2fold |
| 7908650 | IGFN1 | eEF1A2 binding protein | protein turnover | 1q32.1 | NM_178275 | −3.10661 |
| 8108370 | EGR1 | early growth response 1 | transcription factor | 5q31.1 | NM_001964 | −4.47459 |
| 7975779 | FOS | v-fos FBJ murine osteosarcoma viraloncogene homolog | transcription factor | 14q24.3 | NM_005252 | −5.12292 |
| 8029693 | FOSB | FBJ murine osteosarcoma viraloncogene homolog B | transcription factor | 19q13.32 | NM_006732 | −2.78581 |
| 8026047 | JUNB | jun B proto-oncogene | transcription factor | 19p13.2 | NM_002229 | −3.56001 |
| 8148317 | MYC | v-myc myelocytomatosis viraloncogene homolog (avian) | transcription factor | 8q24.21 | NM_002467 | −3.08841 |
| 7909610 | ATF3 | activating transcription factor 3 | transcription factor | 1q32.3 | NM_001040619 | −2.23226 |
| 8084710 | ADIPOQ | adiponectin, C1Q and collagen domain containing | insulin signaling | 3q27 | NM_004797 | −2.25931 |
| 7960865 | SLC2A3 | solute carrier family 2 (facilitated glucose transporter), 3 | insulin signaling | 12p13.3 | NM_006931 | −2.60975 |
| 7934979 | ANKRD1 | ankyrin repeat domain 1 (cardiac muscle) | inflammation | 10q23.31 | NM_014391 | −2.57698 |
| 8006433 | CCL2 | chemokine (C-C motif) ligand 2 (MCP-1) | inflammation | 17q11.2-q12 | NM_002982 | −3.85999 |
| 7934916 | CH25H | cholesterol 25-hydroxylase | inflammation | 10q23 | NM_003956 | −2.56701 |
| 8100994 | CXCL2 | chemokine (C-X-C motif) ligand 2 (MIP-2a) | inflammation | 4q21 | NM_002089 | −2.61796 |
| 8131803 | IL6 | interleukin 6 (interferon, beta 2) | inflammation | 7p21 | NM_000600 | −5.01826 |
| 8018864 | SOCS3 | suppressor of cytokine signaling 3 | inflammation | 17q25.3 | NM_003955 | −3.40317 |
| 8095680 | IL8 | interleukin 8 | inflammation | 4q13-q21 | NM_000584 | −2.02015 |
| 8062461 | LBP | lipopolysaccharide binding protein | inflammation | 20q11.23-q12 | NM_004139 | −2.16848 |
| 8162276 | NFIL3 | nuclear factor, interleukin 3 regulated | inflammation | 9q22 | NM_005384 | −2.63846 |
| 7922229 | SELE | selectin E (endothelial adhesion molecule 1) | inflammation | 1q22-q25 | NM_000450 | −2.46165 |
| 8122265 | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | inflammation | 6q23 | NM_006290 | −2.0769 |
| 8028652 | ZFP36 | zinc finger protein 36, C3H type,homolog (mouse) | inflammation | 19q13.1 | NM_003407 | −2.36152 |
| 7955589 | NR4A1 | nuclear receptor subfamily 4, group A,member 1 | orphan receptor | 12q13 | NM_002135 | −2.36834 |
| 8055952 | NR4A2 | nuclear receptor subfamily 4, group A,member 2 | orphan receptor | 2q22-q23 | NM_006186 | −2.11225 |
| 8156848 | NR4A3 | nuclear receptor subfamily 4, group A,member 3 | orphan receptor | 9q22 | NM_173198 | −3.65489 |
| 7929816 | SCD | stearoyl-CoA desaturase (delta-9-desaturase) | fatty acid metabolism | 10q23-q24 | NM_005063 | −2.69181 |
| 7920873 | SNORA42 | small nucleolar RNA, H/ACA box 42 | small nucleolar RNA expression | 1q22 | NR_002974 | −2.13869 |
| 8069676 | ADAMTS1 | ADAM metallopeptidasew/thrombospondin type 1 motif, 1 | extracellular matrix protein | 21q21.2 | NM_006988 | −2.77417 |
| 7921821 | ADAMTS4 | ADAM metallopeptidasew/thrombospondin type 1 motif, 4 | extracellular matrix protein | 1q21-q23 | NM_005099 | −2.82585 |
| 7902687 | CYR61 | cysteine-rich, angiogenic inducer, 61 | extracellular matrix protein | 1p31-p22 | NM_001554 | −2.94737 |
| 8025601 | ICAM1 | intercellular adhesion molecule 1 (CD54) | extracellular matrix protein | 19p13.3-p13.2 | NM_000201 | −2.25818 |
| 8065353 | THBD | thrombomodulin | extracellular matrix protein | 20p11.2 | NM_000361 | −2.80578 |
| 7982597 | THBS1 | thrombospondin 1 | extracellular matrix protein | 15q15 | NM_003246 | −3.01553 |
| 8152297 | ANGPT1 | angiopoietin 1 | other | 8q22.3-q23 | NM_001146 | 2.01173 |
| 8176276 | ATRX | alpha thalassemia/mental retardationsyndrome X-linked | other | Xq13.1-q21.1 | NM_138270 | 2.62294 |
| 8086330 | AXUD1 | AXIN1 up-regulated 1 | other | 3p22 | NM_033027 | −2.06593 |
| 8119088 | CDKN1A | cyclin-dependent kinase inhibitor 1A(p21, Cip1) | other | 6p21.2 | NM_078467 | −2.02657 |
| 7971653 | DLEU2 | deleted in lymphocytic leukemia, 2 | other | 13q14.3 | NR_002612 | 2.22215 |
| 8024485 | GADD45B | growth arrest and DNA-damage-inducible, beta | other | 19p13.3 | NM_015675 | −2.811 |
| 7954090 | EMP1 | epithelial membrane protein 1 | other | 12p12.3 | NM_001423 | −2.17339 |
| 7995806 | MT1A | metallothionein 1A | antioxidant | 16q13 | AY028617 | −2.5602 |
| 7995787 | MT1M | metallothionein 1M | antioxidant | 16q13 | NM_176870 | −2.49124 |
| 7903171 | RWDD3 | RWD domain containing 3 | other | 1p21.3 | NM_015485 | 2.03845 |
| 8135069 | SERPINE1 | other | 7q21.3-q22 | NM_000602 | −2.67627 | |
| 8070665 | SNF1LK | SNF1-like kinase | other | 21q22.3 | NM_173354 | −2.38371 |
| 8100076 | YIPF7 | Yip1 domain family, member 7 | other | 4p13 | NM_182592 | 2.07083 |
| 8036813 | ZNF780B | zinc finger protein 780B | other | 19q13.2 | NM_001005851 | 2.1765 |
Table 6. Genes changed ≥4 fold (log 2 base ≥2) between 12 and 0 months with omentectomy.
| Probe ID | Gene Symbol | Gene Description | Process | Cytoband | mRNA Accession | log2fold |
| 7908650 | IGFN1 | eEF1A2 binding protein | protein turnover | 1q32.1 | NM_178275 | −2.33443 |
| 8108370 | EGR1 | early growth response 1 | transcription factor | 5q31.1 | NM_001964 | −4.20485 |
| 7975779 | FOS | v-fos FBJ murine osteosarcoma viral oncogenehomolog | transcription factor | 14q24.3 | NM_005252 | −5.064 |
| 8029693 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B | transcription factor | 19q13.32 | NM_006732 | −2.66602 |
| 8026047 | JUNB | jun B proto-oncogene | transcription factor | 19p13.2 | NM_002229 | −3.42952 |
| 8148317 | MYC | v-myc myelocytomatosis viral oncogenehomolog (avian) | transcription factor | 8q24.21 | NM_002467 | −3.13093 |
| 7955858 | HOXC10 | homeobox C10 | transcription factor | 12q13.3 | NM_017409 | 2.15884 |
| 7902687 | CYR61 | cysteine-rich, angiogenic inducer, 61 | insulin signaling | 1p31-p22 | NM_001554 | −2.51451 |
| 8163002 | KLF4 | Kruppel-like factor 4 (gut) | insulin signaling | 9q31 | NM_004235 | −2.04104 |
| 7960865 | SLC2A3 | solute carrier family 2 (facilitated glucosetransporter) member3 | insulin signaling | 12p13.3 | NM_006931 | −2.51532 |
| 7934979 | ANKRD1 | ankyrin repeat domain 1 (cardiac muscle) | inflammation | 10q23.31 | NM_014391 | −3.4932 |
| 8006433 | CCL2 | chemokine (C-C motif) ligand 2 | inflammation | 17q11.2-q12 | NM_002982 | −3.21861 |
| 7934916 | CH25H | cholesterol 25-hydroxylase | inflammation | 10q23 | NM_003956 | −2.42635 |
| 8100994 | CXCL2 | chemokine (C-X-C motif) ligand 2 | inflammation | 4q21 | NM_002089 | −2.2466 |
| 8162276 | NFIL3 | nuclear factor, interleukin 3 regulated | inflammation | 9q22 | NM_005384 | −2.47028 |
| 7922229 | SELE | selectin E (endothelial adhesion molecule 1) | inflammation | 1q22-q25 | NM_000450 | −2.42547 |
| 8018864 | SOCS3 | suppressor of cytokine signaling 3 | inflammation | 17q25.3 | NM_003955 | −3.36272 |
| 8122265 | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | inflammation | 6q23 | NM_006290 | −2.15921 |
| 8028652 | ZFP36 | zinc finger protein 36, C3H type, homolog(mouse) | inflammation | 19q13.1 | NM_003407 | −2.21499 |
| 8131803 | IL6 | interleukin 6 (interferon, beta 2) | inflammation | 7p21 | NM_000600 | −5.07812 |
| 8062461 | LBP | lipopolysaccharide binding protein | inflammation | 20q11.23-q12 | NM_004139 | −2.13523 |
| 8025828 | LDLR | low density lipoprotein receptor (familial hypercholesterolemia) | receptors | 19p13.3 | NM_000527 | −2.22451 |
| 7955589 | NR4A1 | nuclear receptor subfamily 4, group A, member 1 | receptors | 12q13 | NM_002135 | −2.04616 |
| 8156848 | NR4A3 | nuclear receptor subfamily 4, group A, member 3 | receptors | 9q22 | NM_173198 | −3.6856 |
| 8069676 | ADAMTS1 | ADAM metallopeptidase with thrombospondintype 1 motif, 1 | extracellular matrix | 21q21.2 | NM_006988 | −3.41834 |
| 7921821 | ADAMTS4 | ADAM metallopeptidase with thrombospondintype 1 motif, 4 | extracellular matrix | 1q21-q23 | NM_005099 | −2.4919 |
| 8065353 | THBD | thrombomodulin | extracellular matrix | 20p11.2 | NM_000361 | −2.61527 |
| 7982597 | THBS1 | thrombospondin 1 | extracellular matrix | 15q15 | NM_003246 | −3.12862 |
| 8086330 | AXUD1 | AXIN1 up-regulated 1 | other | 3p22 | NM_033027 | −2.08176 |
| 8084206 | B3GNT5 | UDP-GlcNAc:betaGal β-1,3-N-acetylglucosaminyltransferase 5 | other | 3q28 | NM_032047 | −2.25232 |
| 7954090 | EMP1 | epithelial membrane protein 1 | other | 12p12.3 | NM_001423 | −2.14602 |
| 8024485 | GADD45B | growth arrest and DNA-damage-inducible, beta | other | 19p13.3 | NM_015675 | −2.47919 |
| 8086538 | LOC644714 | hypothetical protein LOC644714 | other | 3p21.31 | BC047037 | −2.13574 |
| 7995806 | MT1A | metallothionein 1A | other | 16q13 | AY028617 | −2.47681 |
| 7995787 | MT1M | metallothionein 1M | other | 16q13 | NM_176870 | −2.24145 |
| 8135069 | SERPINE1 | serpin peptidase inhibitor, E1 (plasminogenactivator inhibitor type 1) | other | 7q21.3-q22 | NM_000602 | −2.56719 |
Effects of omentectomy evaluated by global pathway analysis
To identify pathways in skeletal muscle that were regulated by RYGB with and without omentectomy we performed two complimentary analyses. For these analyses those genes detected as being changed greater than 2-fold (2,697 genes; Table S1) were analyzed. First, the gene list was analyzed by the FuncAssociate Gene Ontology program (http://llama.mshri.on.ca/funcassociate/) to statistically identify overrepresented gene categories [39]. Cadmium ion binding (GO:0046870), positive regulation of leukocyte binding (GO:0002687) and cellular macromolecule catabolic processes (GO:0044265) were top ranked. Second, we annotated our expression data with the wiring diagrams of molecular interactions, reactions, and relations maintained at the Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY Database. As reported in Table S4, coverage of 201 pathways by our experimental datasets revealed that metabolic pathways, cytokine-cytokine receptor interaction, MAPK signaling pathways, chemokine signaling pathways, and spliceosome were top ranked.
Validation of microarray expression profiles with TLDA
As the significance of gene expression changes determined with microarray could not be determined empirically due to the low number of array replicates, and because a single outlier sample within an RNA pool could in theory skew results, we elected to validate the expression changes of select genes in each of the individual RNA samples comprising the RNA pools using Taqman Low Density Arrays (TLDA). In addition to the RNA samples used to prepare the RNA pools, we also included RNA samples prepared from additional participants lacking a single study (either 6 or 12 months) time point RNA sample. Using a spot checking strategy, expression of 31 genes was used to validate differential expression data discovered during the microarray study (Table S2). This approach is admittedly restrictive; however, we focused on genes involved in pathways such as inflammation, transcriptional regulation, protein turnover and insulin signaling [40], [41]. Gene selection was guided based on literature reviews of the effects of weight loss on skeletal muscle, our recent report that omentectomy concurrent with RYGB did not improve muscle insulin sensitivity [14], and recent observations that altered intramuscular lipid metabolism, circulating cytokines and macrophage infiltration may act synergistically to coordinate insulin sensitivity [40], [41].
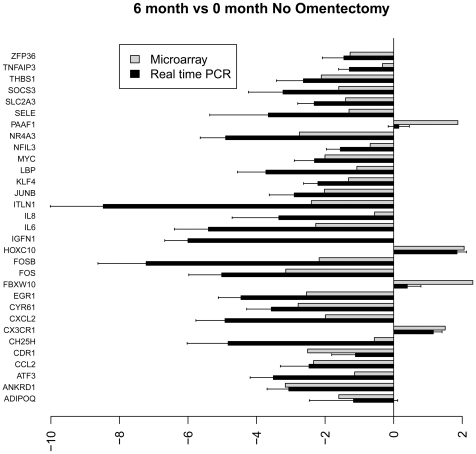
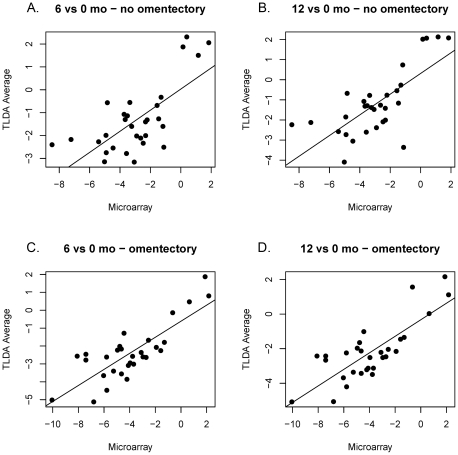
Figures 4 – 7 show the relative average differential expression values determined by microarray (light grey) and TLDA (dark grey) for 31 genes, normalized to an 18S RNA control, when analyzed in triplicate at 0, 6 and 12 months. Of the 5,184 Taqman analyses performed to validate select microarray-derived expression values, only 3 reactions did not amplify. In general, there was good directional (increased or decreased) concordance between expression values derived from analysis of pooled RNA samples and the average of each RNA sample analyzed independently. In most instances, fold expression change values determined by exon array in pooled RNA samples were greater than the averages of individual expression values obtained by TLDA (e.g. FOSB, EGR and IL6 in Figures 4 – 7 ). We examined the relationship between gene expression values revealed by exon array and TLDA profiling. In all group comparisons, there were strong positive relationships: 1) 6 vs. 0 months without omentectomy rho = 0.559 ( Figure 8A ); 12 vs. 0 months without omentectomy rho = 0.720 ( Figure 8B ); 6 vs. 0 months with omentectomy rho = 0.646 ( Figure 8C ); and 12 vs. 0 months with omentectomy rho = 0.640 ( Figure 8D ).
Figure 4. Bar graph illustrating the fold change of individual genes at 6 vs. 0 months without omentectomy as determined by microarray (grey) and TLDA profiling (black).
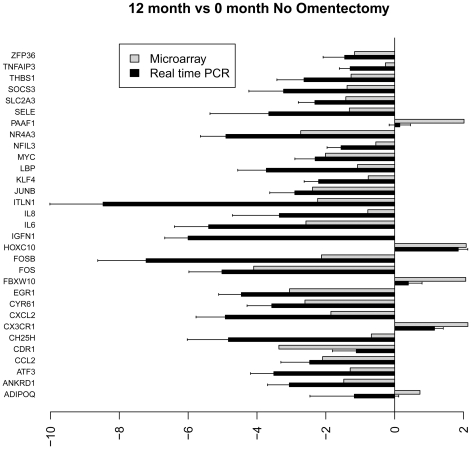
Figure 5. Bar graph illustrating the fold change of individual genes at 12 vs. 0 months without omentectomy as determined by microarray (grey) and TLDA profiling (black).
Figure 6. Bar graph illustrating the fold change of individual genes at 6 vs. 0 months with omentectomy as determined by microarray (grey) and TLDA profiling (black).
Figure 7. Bar graph illustrating the fold change of individual genes at 12 vs. 0 months with omentectomy as determined by microarray (grey) and TLDA profiling (black).
Figure 8. Correlation in fold change (log2) of gene expression as determined by Affymetrix Exon microarray and TLDA profiling.
A) 6 vs. 0 months, no omentectomy, Pearson correlation coefficient rho = 0.559, p<0.0001; B) 12 vs. 0 months, no omentectomy, Pearson correlation coefficient rho = 0.646, p = 0.0002; C) 6 vs. 0 months, yes omentectomy, Pearson correlation coefficient rho = 0.649, p = 0.0002; D) 12 vs. 0 months, yes omentectomy, Pearson correlation coefficient rho = 0.640, p = 0.0002.
Discussion
This is the first high-throughput study to explore the temporal changes in human skeletal muscle gene expression after weight loss through RYGB surgery with tandem laparoscopic omentectomy. In addition, randomization of subjects to removal of the omentum at the time of RYGB allowed us to explore the interplay between visceral adipose and skeletal muscle. While the effects of bariatric surgery on gene expression in muscle have been reported for a few targets [42], [43], [44], [45], the influence of the omentum (visceral adipose) on global skeletal muscle phenotypes (such as insulin resistance) has been understudied. Visceral adipose is the source of 14–20% of the total FFAs delivered to skeletal muscle in obese individuals [46], [47] and numerous studies have linked circulating FFA levels and muscle triglyceride accumulation with peripheral insulin sensitivity [48], [49], [50], [51]. Visceral adipose tissue is also the site of production of several cytokines thought to initiate and maintain pro-inflammatory states both locally and in other tissues [52]. Infiltration and local activation of macrophages in skeletal muscle also has been described [40], [41]. Our group recently reported, in the same subjects examined in the present study, that laparoscopic removal of the greater omentum at the time of RYGB did not significantly enhance weight loss-induced improvements in peripheral insulin sensitivity beyond those observed with RYGB alone [14]. These findings were unanticipated in light of the numerous studies associating increased visceral adiposity with insulin resistance and T2D [53], [54], [55]. Despite these findings, it remained possible that omentectomy produced local changes at the level of the muscle that may not be totally reflected with associated improvements in peripheral glucose utilization. The Affymetrix Exon arrays and TLDA approaches utilized in this study provided a robust and reproducible technique for quantifying gene expression in thousands of independent genes concurrently in RNA samples isolated from multiple tissue samples. This approach represents a significant advance in multivariate gene analysis that was less time- and labor-intensive than individually analyzing single genes by quantitative RT-PCR.
Our major finding was a broad-based downregulation of inflammatory-related pathways in skeletal muscle after RYGB surgery. This response was amplified in the cohort receiving omentectomy, as evidenced by a greater number of genes with at least a 4-fold decrease at 6 or 12 months (at least 4 genes in the RYGB non-omentectomy cohort compared to 11 in the RYGB with omentectomy cohort). Most of the observed inflammatory genes that were only decreased in the RYGB with omentectomy cohort are reportedly associated with macrophage activation (CH25H, CXCL2, SOCS3, LBP, NFIL3, ZFP36) [56], [57], [58], [59], [60], consistent with the systemic decrease in MCP-1, or associated with inflammatory events surrounding sarcopenia (ANKRD1, LBP) [61], [62]. Certain genes such as IL6 and CCL2 were decreased in both groups at both time points relative to pre-surgery levels, but the reductions were greater in the omentectomy group. Changes in IL6 expression levels at 6 and 12 months in the RYGB with omentectomy cohort were the greatest of any gene detected. IL6 is a pleiotropic cytokine described to impact insulin action [63], [64]. IL-6 infusion reportedly increased fatty acid oxidation in human skeletal muscle but not adipose tissue, suggesting a role for this cytokine in stimulating skeletal muscle lipolysis specifically [65]. In the current study, despite glucose utilization (M) being significantly improved at both time-points, omentectomy did not have a differential effect. Interestingly, systemic levels of IL-6 did not change after RYGB. In contrast, other cytokines such as CX3CR1 [66] were equally diminished at 6 and 12 months in both groups after RYGB. Circulating monocytes with high levels of CX3CR1 (frataxylin), are present in systemic disease, such as sepsis or HIV infection, while the macrophage subset that invades tissues during acute localized inflammation expresses lower levels of CX3CR1. These data provide evidence that RYGB reduces the local inflammatory response in muscle which is enhanced by omentectomy, and the physiological consequences warrant further study.
The expression of several transcription factors involved in proliferation control, (FOS, FOSB, JUNB, MYC and EGR1) was reduced at 6 and 12 months after RYGB in both RYGB cohorts with and without omentectomy. These five genes were among those that showed the greatest drop in expression post-RYGB, and their decrease was greater in the omentectomy cohort at both time-points. FOS, FOSB and JUNB are intermediate early genes comprising the AP-1 basic leucine zipper transcription factor family that act on specific biological responses to regulate gene expression. Each has been shown to be increased immediately after exercise [35], but this study is the first to associate them with changes in skeletal muscle after RYGB. Adipose-specific overexpression of ΔFOSB, a naturally occurring alternatively spliced variant of FosB that is antagonistic to normal FosB-mediated gene transcription, was recently shown to increase energy expenditure in muscle and increase insulin sensitivity [67]. Expression of EGR1, a GC box-binding transcription factor, has been shown to regulate TGF-β expression which, in turn increases myostatin expression [68]. The expression of EGR-1 was decreased over 4-fold in the omentectomy cohorts compared to the 2- and 3-fold reductions observed in the non-omentectomy cohort at 6 and 12 months, respectively. EGR-1 binding sites are prevalent in the promoter binding sites for genes such as peroxisomal gamma coactivator-1α (PGC-1α), that are important regulators of mitochondrial biogenesis [69]. The nuclear phosphoprotein MYC was decreased 3-fold at both 6 and 12 month time points in the RYGB with omentectomy cohort but only 2-fold at the same time points in the group without omentectomy. In contrast, HOXC10, a homeoprotein associated with muscle satellite cell activation was upregulated over 2-fold at 12 months in both cohorts, and at 6 months in the RYGB with omentectomy group. The protein product of this gene, together with Pax7, Asb5 and IgSF4, was shown to be markers for skeletal muscle satellite cells [70]. In the present study, increased HOXC10 expression at all time points in both cohorts implies that muscle satellite cell activation and deactivation of transcriptional responses are an important phenomenon in muscle remodeling, particularly after weight loss.
Genes facilitating extracellular matrix (ECM) remodeling such as CYR61, THBS1 and THBS4 were downregulated, consistent with an improvement in inflammation. High levels of muscle expression of ECM, inflammatory, and immediate early genes in obesity could indicate a continual, unsuccessful attempt to repair muscle damage caused by fat overload. A role for the ECM in insulin resistance has been described but is largely unappreciated. Infusion of FFA in subjects with normal glucose tolerance has been shown to increase the expression of ECM genes in muscle coincident with a decrease in insulin sensitivity [71]; furthermore, animal studies indicate that ECM expansion contributes to insulin resistance [72]. These studies in concert with our current results indicate that further studies connecting obesity, the ECM, and insulin resistance are warranted.
Three genes annotated as regulating protein turnover were significantly altered upon RYGB in both surgical groups. The gene encoding eEF1A2 binding protein (IGFN1 or Dkfzp434B1231) plays a key role in protein translation, carrying each aminoacyl-transfer RNA complex to the A site of the ribosome after codon-anticodon recognition [73]. In cultured myotubes, IGFN1 expression inhibited apoptosis and promoted cell proliferation in hyper-catabolic trauma patients [74] and was required for the muscle-specific expression of utrophin A, a sarcomellar protein causal in the fatal neuromuscular disease Duchenne Muscular Dystrophy [75]. In our studies IGNF1 expression was reduced greater than 4-fold, a pattern consistent with muscle catabolism known to occur in this population. Other genes involved in protein turnover such as FBXW10, a component of the E3 ubiquitin ligase system that determines substrate specificity during proteasomal degradation, demonstrated an increased expression pattern primarily at the 12-month time point. It seems likely that as a result of RYGB the regulation of muscle protein maintenance is upset. Future studies in this population examining muscle protein synthesis and muscle protein breakdown using state-of-the-art isotopic tracer methodologies may delineate mechanisms contributory to this observation.
At 6 and 12 months after RYGB in the non-omentectomy group, several small nucleolar RNAs from the C/D box group (SNORDs) were among the few genes that were upregulated in skeletal muscle. SNORDs are a class of small RNA non-protein coding micro RNAs that molecules that guide chemical modification, particularly 2′-O-methylation [76] and alternative splicing [77] of other RNAs. Interestingly, the absence of small nucleolar RNA C/D box 115 gene expression units have been implicated as causal in Prader-Willi Syndrome [78], the most common form of monogenetic obesity. Previous studies, however, indicate that expression of HBII-52 is restricted to the brain [78]; thus, the relevance of the observed expression of SNORD 115 variants in muscle and the increased expression after RYGB is highly unclear.
We have identified a set of genes, which were significantly and differentially altered after RYGB with and without omentectomy; however, there are some important limitations to this study. First, while the utmost effort was placed on ensuring the quality of the muscle biopsies used for RNA extraction both before and after surgery, it remains possible that some intercalated adipose, adventitia and/or microvasculature may have been present in some tissues. This phenomenon was more likely in biopsies obtained prior to RYGB surgery, compared to those at either time point after surgery when significant weight loss occurred, and may lead to significant gene expression changes that are not exclusive to skeletal muscle. For example, the observed changes in adiponectin (ADIPOQ) and omentin-1 (ITLN1) might be reflective of intercalated adipose tissue as this is their primary site of production, although muscle specific expression has been reported [79]. Thirdly, we elected to use Robust Multi-array Averaging, a common normalization method, but did not perform the default quantile across sample normalization method of RMA to avoid over-correction for the small sample size. The data were also analyzed without adjustments for multiplicity, which could alter differential expression estimates [80]. Nevertheless, our validation efforts focused primarily on those genes affected 4 fold or greater (log2 – 2 fold) and, as such, any misinterpretations based on this should be minimal. Finally, we are unable to account for the extrinsic factors, other than those reported in Tables 1 and 2 that can affect end-organ metabolic response. As RYGB surgery is a procedure known for its systemic and interconnected effects on hormonal, neuronal, and biochemical impulses, the discrimination of a muscle-specific response due solely to the removal of omentum remains a challenge.
With regard to our overall study design, there are some limitations that should be mentioned. The population studied was reflective of Nashville, TN and the surrounding counties with an ethnic distribution (Caucasians to African Americans) of 4∶1. While both the omentectomy and non-omentectomy group contained participants with and without T2D, the proportion of subjects with T2D was unequal between groups. There were also no Hispanics in the studied cohorts, based on our geographic locale, and the subjects studied were also primarily female reflective of the patient population seeking this surgical approach; therefore, our findings may be gender and ethnically limited.
Supporting Information
Consort 2010 checklist for randomized trial.
(DOC)
Trial protocol.
(DOC)
Expression values for genes changed greater than 2 fold between 6 vs. 0 months, no omentectomy; 12 vs. 0 months, no omentectomy; 6 vs. 0 months with omentectomy; and 12 vs. 0 months, with omentectomy.
(PDF)
Expression values for genes changed greater than 4 fold between 6 vs. 0 months, no omentectomy; 12 vs. 0 months, no omentectomy; 6 vs. 0 months with omentectomy; and 12 vs. 0 months, with omentectomy.
(PDF)
TLDA probes utilized in this study.
(PDF)
Average gene expression values for those molecular interaction members within the indicated molecular pathway. Pathways are ranked highest to lowest in terms of the number of genes detected per pathway.
(PDF)
Acknowledgments
We thank Erik N. Hansen, MD, MPH, James M. Isbell, MD, Jabbar Saliba, MD, Marcy Buckley, RN, Joan Kaiser, RN, MS, and Kareem Jabbour, BS, for acquisition of data, and Julia P. Dunn, MD, and Nadine Saliba, BS, for technical support. We also thank the volunteers and patients who took part in these studies. We are grateful for Dr. Lillian Nanney, Alonda Pollins, and Andrew Judson Hill IV for isolating and qualifying muscle RNA.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: All microarray experiments were performed in the Vanderbilt Functional Genomics Shared Resource, which is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Digestive Disease Center (P30 DK58404) and the Vanderbilt Vision Center (P30 EY08126). Funding support was from the following National Institutes of Health grants: R01 DK70860 to NNA; UL1 RR024975 (Vanderbilt Clinical and Translational Science Award), P30 DK020593 (Vanderbilt Diabetes Research and Training Center) and P30 DK058404 (Vanderbilt Digestive Disease Research Center). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Defronzo RA. Banting Lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58:773–795. doi: 10.2337/db09-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdul-Ghani MA, DeFronzo RA. Pathogenesis of insulin resistance in skeletal muscle. J Biomed Biotechnol. 2010;2010:476279. doi: 10.1155/2010/476279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 2004;25:4–7. doi: 10.1016/j.it.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Bastard JP, Maachi M, Van Nhieu JT, Jardel C, Bruckert E, et al. Adipose tissue IL-6 content correlates with resistance to insulin activation of glucose uptake both in vivo and in vitro. J Clin Endocrinol Metab. 2002;87:2084–2089. doi: 10.1210/jcem.87.5.8450. [DOI] [PubMed] [Google Scholar]
- 5.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 6.Chiba Y, Saitoh S, Takagi S, Ohnishi H, Katoh N, et al. Relationship between visceral fat and cardiovascular disease risk factors: the Tanno and Sobetsu study. Hypertension research : official journal of the Japanese Society of Hypertension. 2007;30:229–236. doi: 10.1291/hypres.30.229. [DOI] [PubMed] [Google Scholar]
- 7.McLaughlin T, Lamendola C, Liu A, Abbasi F. Preferential Fat Deposition in Subcutaneous Versus Visceral Depots Is Associated with Insulin Sensitivity. The Journal of clinical endocrinology and metabolism. 2011 doi: 10.1210/jc.2011-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosito GA, Massaro JM, Hoffmann U, Ruberg FL, Mahabadi AA, et al. Pericardial fat, visceral abdominal fat, cardiovascular disease risk factors, and vascular calcification in a community-based sample: the Framingham Heart Study. Circulation. 2008;117:605–613. doi: 10.1161/CIRCULATIONAHA.107.743062. [DOI] [PubMed] [Google Scholar]
- 9.Veilleux A, Caron-Jobin M, Noel S, Laberge PY, Tchernof A. Visceral adipocyte hypertrophy is associated with dyslipidemia independent of body composition and fat distribution in women. Diabetes. 2011;60:1504–1511. doi: 10.2337/db10-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Despres JP. Intra-abdominal obesity: an untreated risk factor for Type 2 diabetes and cardiovascular disease. J Endocrinol Invest. 2006;29:77–82. [PubMed] [Google Scholar]
- 11.Ibrahim MM. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes Rev. 2010;11:11–18. doi: 10.1111/j.1467-789X.2009.00623.x. [DOI] [PubMed] [Google Scholar]
- 12.Lafontan M, Girard J. Impact of visceral adipose tissue on liver metabolism. Part I: heterogeneity of adipose tissue and functional properties of visceral adipose tissue. Diabetes Metab. 2008;34:317–327. doi: 10.1016/j.diabet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Harman-Boehm I, Bluher M, Redel H, Sion-Vardy N, Ovadia S, et al. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. J Clin Endocrinol Metab. 2007;92:2240–2247. doi: 10.1210/jc.2006-1811. [DOI] [PubMed] [Google Scholar]
- 14.Fabbrini E, Tamboli RA, Magkos F, Marks-Shulman PA, Eckhauser AW, et al. Surgical removal of omental fat does not improve insulin sensitivity and cardiovascular risk factors in obese adults. Gastroenterology. 2010;139:448–455. doi: 10.1053/j.gastro.2010.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varma V, Yao-Borengasser A, Rasouli N, Nolen GT, Phanavanh B, et al. Muscle inflammatory response and insulin resistance: synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am J Physiol Endocrinol Metab. 2009;296:E1300–1310. doi: 10.1152/ajpendo.90885.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, et al. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 17.Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nature medicine. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 18.Itani SI, Ruderman NB, Schmieder F, Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IkappaB-alpha. Diabetes. 2002;51:2005–2011. doi: 10.2337/diabetes.51.7.2005. [DOI] [PubMed] [Google Scholar]
- 19.Yuan M, Konstantopoulos N, Lee J, Hansen L, Li ZW, et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 20.Buchwald H, Estok R, Fahrbach K, Banel D, Jensen MD, et al. Weight and type 2 diabetes after bariatric surgery: systematic review and meta-analysis. Am J Med. 2009;122:248–256 e245. doi: 10.1016/j.amjmed.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 21.Pereira JA, Lazarin MA, Pareja JC, de Souza A, Muscelli E. Insulin resistance in nondiabetic morbidly obese patients: effect of bariatric surgery. Obes Res. 2003;11:1495–1501. doi: 10.1038/oby.2003.200. [DOI] [PubMed] [Google Scholar]
- 22.Gray RE, Tanner CJ, Pories WJ, MacDonald KG, Houmard JA. Effect of weight loss on muscle lipid content in morbidly obese subjects. Am J Physiol Endocrinol Metab. 2003;284:E726–732. doi: 10.1152/ajpendo.00371.2002. [DOI] [PubMed] [Google Scholar]
- 23.Houmard JA, Tanner CJ, Yu C, Cunningham PG, Pories WJ, et al. Effect of weight loss on insulin sensitivity and intramuscular long-chain fatty acyl-CoAs in morbidly obese subjects. Diabetes. 2002;51:2959–2963. doi: 10.2337/diabetes.51.10.2959. [DOI] [PubMed] [Google Scholar]
- 24.Friedman JE, Dohm GL, Leggett-Frazier N, Elton CW, Tapscott EB, et al. Restoration of insulin responsiveness in skeletal muscle of morbidly obese patients after weight loss. Effect on muscle glucose transport and glucose transporter GLUT4. J Clin Invest. 1992;89:701–705. doi: 10.1172/JCI115638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pender C, Goldfine ID, Tanner CJ, Pories WJ, MacDonald KG, et al. Muscle insulin receptor concentrations in obese patients post bariatric surgery: relationship to hyperinsulinemia. Int J Obes Relat Metab Disord. 2004;28:363–369. doi: 10.1038/sj.ijo.0802565. [DOI] [PubMed] [Google Scholar]
- 26.Park JJ, Berggren JR, Hulver MW, Houmard JA, Hoffman EP. GRB14, GPD1, and GDF8 as potential network collaborators in weight loss-induced improvements in insulin action in human skeletal muscle. Physiol Genomics. 2006;27:114–121. doi: 10.1152/physiolgenomics.00045.2006. [DOI] [PubMed] [Google Scholar]
- 27.Tataranni PA, Ravussin E. Use of dual-energy X-ray absorptiometry in obese individuals. Am J Clin Nutr. 1995;62:730–734. doi: 10.1093/ajcn/62.4.730. [DOI] [PubMed] [Google Scholar]
- 28.Pollins AC, Friedman DB, Nanney LB. Proteomic investigation of human burn wounds by 2D-difference gel electrophoresis and mass spectrometry. The Journal of surgical research. 2007;142:143–152. doi: 10.1016/j.jss.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brem H, Stojadinovic O, Diegelmann RF, Entero H, Lee B, et al. Molecular markers in patients with chronic wounds to guide surgical debridement. Mol Med. 2007;13:30–39. doi: 10.2119/2006-00054.Brem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dasu MR, Barrow RE, Herndon DN. Gene expression changes with time in skeletal muscle of severely burned children. Ann Surg. 2005;241:647–653. doi: 10.1097/01.sla.0000157266.53903.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greco JA, 3rd, Pollins AC, Boone BE, Levy SE, Nanney LB. A microarray analysis of temporal gene expression profiles in thermally injured human skin. Burns. 2010;36:192–204. doi: 10.1016/j.burns.2009.06.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. ISBN 3-900051-07-0, URL. R website. 2010. Available: http://www.R-project.org/. Accessed 2011 Nov 15.
- 35.Puntschart A, Wey E, Jostarndt K, Vogt M, Wittwer M, et al. Expression of fos and jun genes in human skeletal muscle after exercise. Am J Physiol. 1998;274:C129–137. doi: 10.1152/ajpcell.1998.274.1.C129. [DOI] [PubMed] [Google Scholar]
- 36.Gabellini D, Colaluca IN, Vodermaier HC, Biamonti G, Giacca M, et al. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 2003;22:3715–3724. doi: 10.1093/emboj/cdg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearen MA, Myers SA, Raichur S, Ryall JG, Lynch GS, et al. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 2008;149:2853–2865. doi: 10.1210/en.2007-1202. [DOI] [PubMed] [Google Scholar]
- 38.Pearen MA, Ryall JG, Maxwell MA, Ohkura N, Lynch GS, et al. The orphan nuclear receptor, NOR-1, is a target of beta-adrenergic signaling in skeletal muscle. Endocrinology. 2006;147:5217–5227. doi: 10.1210/en.2006-0447. [DOI] [PubMed] [Google Scholar]
- 39.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kewalramani G, Bilan PJ, Klip A. Muscle insulin resistance: assault by lipids, cytokines and local macrophages. Curr Opin Clin Nutr Metab Care. 2010;13:382–390. doi: 10.1097/MCO.0b013e32833aabd9. [DOI] [PubMed] [Google Scholar]
- 41.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 42.Adami GF, Parodi RC, Papadia F, Marinari G, Camerini G, et al. Magnetic resonance spectroscopy facilitates assessment of intramyocellular lipid changes: a preliminary short-term study following biliopancreatic diversion. Obesity surgery. 2005;15:1233–1237. doi: 10.1381/096089205774512483. [DOI] [PubMed] [Google Scholar]
- 43.Greco AV, Mingrone G, Giancaterini A, Manco M, Morroni M, et al. Insulin resistance in morbid obesity: reversal with intramyocellular fat depletion. Diabetes. 2002;51:144–151. doi: 10.2337/diabetes.51.1.144. [DOI] [PubMed] [Google Scholar]
- 44.Johansson L, Roos M, Kullberg J, Weis J, Ahlstrom H, et al. Lipid mobilization following Roux-en-Y gastric bypass examined by magnetic resonance imaging and spectroscopy. Obesity surgery. 2008;18:1297–1304. doi: 10.1007/s11695-008-9484-0. [DOI] [PubMed] [Google Scholar]
- 45.Leichman JG, Wilson EB, Scarborough T, Aguilar D, Miller CC, 3rd, et al. Dramatic reversal of derangements in muscle metabolism and left ventricular function after bariatric surgery. The American journal of medicine. 2008;121:966–973. doi: 10.1016/j.amjmed.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein S. The case of visceral fat: argument for the defense. The Journal of clinical investigation. 2004;113:1530–1532. doi: 10.1172/JCI22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen S, Guo Z, Johnson CM, Hensrud DD, Jensen MD. Splanchnic lipolysis in human obesity. The Journal of clinical investigation. 2004;113:1582–1588. doi: 10.1172/JCI21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bajaj M, Suraamornkul S, Romanelli A, Cline GW, Mandarino LJ, et al. Effect of a sustained reduction in plasma free fatty acid concentration on intramuscular long-chain fatty Acyl-CoAs and insulin action in type 2 diabetic patients. Diabetes. 2005;54:3148–3153. doi: 10.2337/diabetes.54.11.3148. [DOI] [PubMed] [Google Scholar]
- 49.Cameron-Smith D, Burke LM, Angus DJ, Tunstall RJ, Cox GR, et al. A short-term, high-fat diet up-regulates lipid metabolism and gene expression in human skeletal muscle. The American journal of clinical nutrition. 2003;77:313–318. doi: 10.1093/ajcn/77.2.313. [DOI] [PubMed] [Google Scholar]
- 50.Hegarty BD, Cooney GJ, Kraegen EW, Furler SM. Increased efficiency of fatty acid uptake contributes to lipid accumulation in skeletal muscle of high fat-fed insulin-resistant rats. Diabetes. 2002;51:1477–1484. doi: 10.2337/diabetes.51.5.1477. [DOI] [PubMed] [Google Scholar]
- 51.Heron-Milhavet L, Haluzik M, Yakar S, Gavrilova O, Pack S, et al. Muscle-specific overexpression of CD36 reverses the insulin resistance and diabetes of MKR mice. Endocrinology. 2004;145:4667–4676. doi: 10.1210/en.2003-1543. [DOI] [PubMed] [Google Scholar]
- 52.Donohoe CL, Doyle SL, Reynolds JV. Visceral adiposity, insulin resistance and cancer risk. Diabetology & metabolic syndrome. 2011;3:12. doi: 10.1186/1758-5996-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colberg SR, Simoneau JA, Thaete FL, Kelley DE. Skeletal muscle utilization of free fatty acids in women with visceral obesity. The Journal of clinical investigation. 1995;95:1846–1853. doi: 10.1172/JCI117864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macor C, Ruggeri A, Mazzonetto P, Federspil G, Cobelli C, et al. Visceral adipose tissue impairs insulin secretion and insulin sensitivity but not energy expenditure in obesity. Metabolism: clinical and experimental. 1997;46:123–129. doi: 10.1016/s0026-0495(97)90288-2. [DOI] [PubMed] [Google Scholar]
- 55.Coker RH, Williams RH, Yeo SE, Kortebein PM, Bodenner DL, et al. Visceral fat and adiponectin: associations with insulin resistance are tissue-specific in women. Metabolic syndrome and related disorders. 2009;7:61–67. doi: 10.1089/met.2008.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park K, Scott AL. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. Journal of leukocyte biology. 2010;88:1081–1087. doi: 10.1189/jlb.0610318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolpe SD, Sherry B, Juers D, Davatelis G, Yurt RW, et al. Identification and characterization of macrophage inflammatory protein 2. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:612–616. doi: 10.1073/pnas.86.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nature reviews Immunology. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 59.Grube BJ, Cochane CG, Ye RD, Green CE, McPhail ME, et al. Lipopolysaccharide binding protein expression in primary human hepatocytes and HepG2 hepatoma cells. The Journal of biological chemistry. 1994;269:8477–8482. [PubMed] [Google Scholar]
- 60.Hubal MJ, Rubinstein SR, Clarkson PM. Mechanisms of variability in strength loss after muscle-lengthening actions. Medicine and science in sports and exercise. 2007;39:461–468. doi: 10.1249/01.mss.0000247007.19127.da. [DOI] [PubMed] [Google Scholar]
- 61.Badi I, Cinquetti R, Frascoli M, Parolini C, Chiesa G, et al. Intracellular ANKRD1 protein levels are regulated by 26S proteasome-mediated degradation. FEBS letters. 2009;583:2486–2492. doi: 10.1016/j.febslet.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Oropeza M, Petersen B, Carnwath JW, Lucas-Hahn A, Lemme E, et al. Transgenic expression of the human A20 gene in cloned pigs provides protection against apoptotic and inflammatory stimuli. Xenotransplantation. 2009;16:522–534. doi: 10.1111/j.1399-3089.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 63.Franckhauser S, Elias I, Rotter Sopasakis V, Ferre T, Nagaev I, et al. Overexpression of Il6 leads to hyperinsulinaemia, liver inflammation and reduced body weight in mice. Diabetologia. 2008;51:1306–1316. doi: 10.1007/s00125-008-0998-8. [DOI] [PubMed] [Google Scholar]
- 64.Holmes AG, Mesa JL, Neill BA, Chung J, Carey AL, et al. Prolonged interleukin-6 administration enhances glucose tolerance and increases skeletal muscle PPARalpha and UCP2 expression in rats. The Journal of endocrinology. 2008;198:367–374. doi: 10.1677/JOE-08-0113. [DOI] [PubMed] [Google Scholar]
- 65.Wolsk E, Mygind H, Grondahl TS, Pedersen BK, van Hall G. IL-6 selectively stimulates fat metabolism in human skeletal muscle. American journal of physiology Endocrinology and metabolism. 2010;299:E832–840. doi: 10.1152/ajpendo.00328.2010. [DOI] [PubMed] [Google Scholar]
- 66.Teran-Garcia M, Rankinen T, Koza RA, Rao DC, Bouchard C. Endurance training-induced changes in insulin sensitivity and gene expression. American journal of physiology Endocrinology and metabolism. 2005;288:E1168–1178. doi: 10.1152/ajpendo.00467.2004. [DOI] [PubMed] [Google Scholar]
- 67.Rowe GC, Choi CS, Neff L, Horne WC, Shulman GI, et al. Increased energy expenditure and insulin sensitivity in the high bone mass DeltaFosB transgenic mice. Endocrinology. 2009;150:135–143. doi: 10.1210/en.2008-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Irrcher I, Hood DA. Regulation of Egr-1, SRF, and Sp1 mRNA expression in contracting skeletal muscle cells. Journal of applied physiology. 2004;97:2207–2213. doi: 10.1152/japplphysiol.00388.2004. [DOI] [PubMed] [Google Scholar]
- 69.Freyssenet D, Irrcher I, Connor MK, Di Carlo M, Hood DA. Calcium-regulated changes in mitochondrial phenotype in skeletal muscle cells. American journal of physiology Cell physiology. 2004;286:C1053–1061. doi: 10.1152/ajpcell.00418.2003. [DOI] [PubMed] [Google Scholar]
- 70.Seale P, Ishibashi J, Scime A, Rudnicki MA. Pax7 is necessary and sufficient for the myogenic specification of CD45+:Sca1+ stem cells from injured muscle. PLoS biology. 2004;2:E130. doi: 10.1371/journal.pbio.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richardson DK, Kashyap S, Bajaj M, Cusi K, Mandarino SJ, et al. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases the expression of extracellular matrix genes in human skeletal muscle. The Journal of biological chemistry. 2005;280:10290–10297. doi: 10.1074/jbc.M408985200. [DOI] [PubMed] [Google Scholar]
- 72.Kang L, Ayala JE, Lee-Young RS, Zhang Z, James FD, et al. Diet-induced muscle insulin resistance is associated with extracellular matrix remodeling and interaction with integrin alpha2beta1 in mice. Diabetes. 2011;60:416–426. doi: 10.2337/db10-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murray LM, Thomson D, Conklin A, Wishart TM, Gillingwater TH. Loss of translation elongation factor (eEF1A2) expression in vivo differentiates between Wallerian degeneration and dying-back neuronal pathology. Journal of anatomy. 2008;213:633–645. doi: 10.1111/j.1469-7580.2008.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bosutti A, Scaggiante B, Grassi G, Guarnieri G, Biolo G. Overexpression of the elongation factor 1A1 relates to muscle proteolysis and proapoptotic p66(ShcA) gene transcription in hypercatabolic trauma patients. Metabolism: clinical and experimental. 2007;56:1629–1634. doi: 10.1016/j.metabol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 75.Miura P, Coriati A, Belanger G, De Repentigny Y, Lee J, et al. The utrophin A 5′-UTR drives cap-independent translation exclusively in skeletal muscles of transgenic mice and interacts with eEF1A2. Human molecular genetics. 2010;19:1211–1220. doi: 10.1093/hmg/ddp591. [DOI] [PubMed] [Google Scholar]
- 76.Galardi S, Fatica A, Bachi A, Scaloni A, Presutti C, et al. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Molecular and cellular biology. 2002;22:6663–6668. doi: 10.1128/MCB.22.19.6663-6668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet. 19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang RZ, Lee MJ, Hu H, Pray J, Wu HB, et al. Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action. American journal of physiology Endocrinology and metabolism. 2006;290:E1253–1261. doi: 10.1152/ajpendo.00572.2004. [DOI] [PubMed] [Google Scholar]
- 80.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Consort 2010 checklist for randomized trial.
(DOC)
Trial protocol.
(DOC)
Expression values for genes changed greater than 2 fold between 6 vs. 0 months, no omentectomy; 12 vs. 0 months, no omentectomy; 6 vs. 0 months with omentectomy; and 12 vs. 0 months, with omentectomy.
(PDF)
Expression values for genes changed greater than 4 fold between 6 vs. 0 months, no omentectomy; 12 vs. 0 months, no omentectomy; 6 vs. 0 months with omentectomy; and 12 vs. 0 months, with omentectomy.
(PDF)
TLDA probes utilized in this study.
(PDF)
Average gene expression values for those molecular interaction members within the indicated molecular pathway. Pathways are ranked highest to lowest in terms of the number of genes detected per pathway.
(PDF)