Abstract
Four members of the mammalian ATP binding cassette (ABC) transporter G subfamily are thought to be involved in transmembrane (TM) transport of sterols. The residues responsible for this transport are unknown. The mechanism of action of ABCG1 is controversial and it has been proposed to act at the plasma membrane to facilitate the efflux of cellular sterols to exogenous high-density lipoprotein (HDL). Here we show that ABCG1 function is dependent on localization to intracellular endosomes. Importantly, localization to the endosome pathway distinguishes ABCG1 and/or ABCG4 from all other mammalian members of this superfamily, including other sterol transporters. We have identified critical residues within the TM domains of ABCG1 that are both essential for sterol transport and conserved in some other members of the ABCG subfamily and/or the insulin-induced gene 2 (INSIG-2). Our conclusions are based on studies in which (i) biotinylation of peritoneal macrophages showed that endogenous ABCG1 is intracellular and undetectable at the cell surface, (ii) a chimeric protein containing the TM of ABCG1 and the cytoplasmic domains of the nonsterol transporter ABCG2 is both targeted to endosomes and functional, and (iii) ABCG1 colocalizes with multiple proteins that mark late endosomes and recycling endosomes. Mutagenesis studies identify critical residues in the TM domains that are important for ABCG1 to alter sterol efflux, induce sterol regulatory element binding protein-2 (SREBP-2) processing, and selectively attenuate the oxysterol-mediated repression of SREBP-2 processing. Our data demonstrate that ABCG1 is an intracellular sterol transporter that localizes to endocytic vesicles to facilitate the redistribution of specific intracellular sterols away from the endoplasmic reticulum (ER).
Keywords: cholesterol, 25-hydroxycholesterol, 27-hydroxycholesterol
The ATP binding cassette (ABC) superfamily of transporters are found in all phyla including bacteria, plants, yeast, flies, fish, and mammals (1, 2). All functional eukaryotic ABC transporters contain two ABCs and two transmembrane (TM) domains, each of which is composed of six TM α-helices, although additional TM helices are present in some transporters. “Full” transporters contain two ABCs and two TM domains on one polypeptide, whereas “half” transporters, such as ABCG1, -G2, -G4, -G5 and -G8, contain one ABC and one TM domain on one polypeptide and thus must form dimers to function. Specific mammalian ABC transporters have been shown to localize to the plasma membrane, endoplasmic reticulum (ER), mitochondria, peroxisomes, and lamellar bodies (3–6). In each case, the two ABC domains are present in the cytoplasm where they interact to form a site that binds and hydrolyzes ATP to facilitate transport of substrates across the membrane bilayer (7).
Early studies showed that overexpression of ABCG1 resulted in increased efflux of cellular cholesterol to exogenously added high-density lipoprotein (HDL) (8, 9). These initial studies led to the suggestion that ABCG1 might be critical for efflux of cholesterol from lipid-loaded macrophages, in the first step of reverse cholesterol transport (10). However, subsequent studies demonstrated that there was little/no specificity for the exogenous acceptor of cholesterol as other lipoproteins, in addition to HDL and protein-free lipid vesicles, could serve as acceptors (9, 11–13). These data suggest that, unlike ABCA1, which effluxes cholesterol and phospholipid after binding exogenous lipid-poor apoproteins, ABCG1 effluxes cellular cholesterol by a process that is not dependent upon interaction with an extracellular protein.
Studies with Abcg1−/−, Abcg1−/−Abcg4−/−, Abcg1−/−Apoe−/−, and Abcg1−/−Abca1−/− mice indicate that loss of ABCG1 has no effect on plasma lipoprotein levels (8, 14–18). Rather, studies indicate that loss of ABCG1 was associated with an inability to control cellular sterol levels, especially in pulmonary macrophages (19, 20). This led to an alternative proposal that the primary function of ABCG1 may be to control intracellular sterol homeostasis, rather than to efflux cellular sterols to HDL (21, 22).
Identification of the cellular location and substrates of ABCG1 might provide important insights into the normal physiologic function of ABCG1. However, there have been conflicting reports on the cellular localization of ABCG1 as it has been reported to be predominantly localized to the cell surface (23, 24) or to be transported to the plasma membrane in response to liver X receptor (LXR) agonists (25) or independent of LXR activation (24). Others have reported that ABCG1 is largely intracellular with little (21) or undetectable levels at the cell surface (22). The reasons for these discrepant conclusions remain unclear.
In the current studies, we use multiple approaches to demonstrate that ABCG1 is associated with endosomal vesicles and is undetectable at the cell surface. We also generated ABCG1–ABCG2 chimeric fusion proteins and demonstrated that the TM domains of ABCG1 are necessary and sufficient to both correctly target ABCG1 to the endocytic pathway and to increase the processing of SREBP-2, presumably by facilitating the movement of sterols away from the ER. Finally, we used a luciferase reporter assay to identify specific amino acids within the TM domains of ABCG1 that are necessary for function, but not for correct membrane targeting.
Results
ABCG1 Localizes to Endosomes.
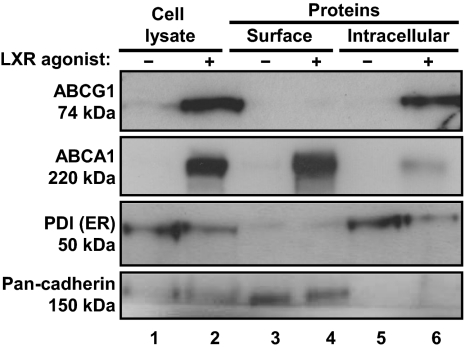
To determine whether endogenous ABCG1 is expressed at the cell surface, we incubated primary mouse peritoneal macrophages for 24 h in the absence or presence of the LXR agonist GW3965. Cells were exposed to biotin before quenching the reaction to prevent modification of intracellular proteins after cell lysis. As expected, Western blot analysis of whole cell lysates showed that GW3965 markedly induced ABCG1 and ABCA1 protein levels, but had no effect on pan-cadherin or protein disulfide isomerase (PDI), markers for the cell surface and endoplasmic reticulum (ER), respectively (Fig. 1, compare lanes 1 and 2).
Fig. 1.
Endogenous ABCG1 is intracellular. Mouse primary peritoneal macrophages were treated with or without LXR agonist GW3965 (1 μM) for 24 h, before biotinylation of cell surface proteins. Total cell lysates (lanes 1 and 2), biotinylated proteins (lanes 3 and 4), and unmodified proteins (lanes 5 and 6) were separated by SDS/PAGE and the indicated proteins identified using specific antibodies.
As expected, Fig. 1 (lanes 3 and 4) shows that ABCA1 and pan-cadherin are biotinylated and precipitated by the streptavidin beads, consistent with the known expression of both proteins at the cell surface. In contrast, ABCG1 and PDI were not biotinylated, and hence they were not precipitated by the streptavidin beads (Fig. 1, lanes 3 and 4), consistent with an intracellular localization of both proteins (Fig. 1, lanes 5 and 6).
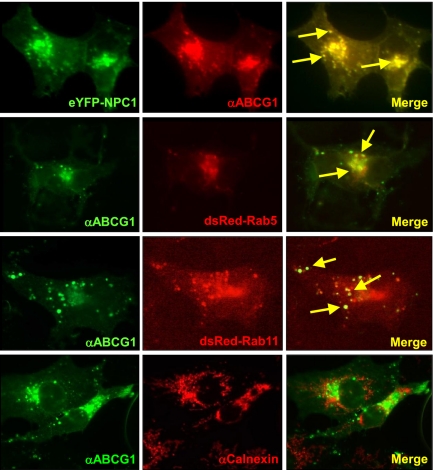
In a complimentary approach, we cotransfected Cos-7 cells with plasmids encoding untagged ABCG1 together with epitope-tagged proteins that serve as markers for early endosomes (Rab5), recycling endosomes (Rab11), or late endosomes/lysosomes Niemann-Pick type C-1 (NPC-1). Immunofluorescence studies demonstrated that ABCG1 colocalized with NPC-1, Rab5, and Rab11 (Fig. 2, yellow arrows), but did not colocalize with calnexin, an ER resident protein (Fig. 2). Importantly, ABCG1 was not detected at the cell surface (Fig. 2), a finding consistent with the data from biotinylation studies of endogenous ABCG1 (Fig. 1) and earlier studies in which ABCG1 was overexpressed in hippocampal neurons (22). Together, these studies demonstrate that ABCG1 localizes to intracellular vesicles of the endosomal pathway.
Fig. 2.
ABCG1 localizes to intracellular vesicles of the endosomal pathway. Cos-7 cells were cotransfected with untagged ABCG1 and either NPC-1-YFP (late endosomes/lysosomes) or DsRed-Rab5 (early endosomes), or DsRed-Rab11 (recycling endosomes). Immunofluorescence was determined after treatment of cells with antibody to ABCG1 or calnexin. Images are at 63× magnification. Representative sites of colocalization are indicated by yellow arrows in the merged images.
ABCG1 Expression Levels Affect an SREBP-2–Dependent Reporter Gene.
We previously reported that overexpression in primary mouse astrocytes of either ABCG1, or the highly homologous ABCG4, resulted in increased processing of the precursor SREBP-2 to form the nuclear/mature SREBP-2 (mSREBP-2) and to increased expression of SREBP-2–regulated genes that included the low density lipoprotein receptor (Ldlr) and 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase (Hmgr) (22). In contrast, expression levels of many SREBP-2 responsive genes were repressed in the brains of Abcg1−/−, Abcg4−/−, or Abcg1−/−Abcg4−/− mice (15) or in macrophages from the lungs of Abcg1−/− mice consistent with intracellular accumulation of sterols and reduced processing of SREBP-2 (19). Given that small changes in ER sterol content are known to affect maturation/nuclear localization of SREBP-2 and subsequent target gene activation (26), these data suggest that ABCG1 may facilitate the movement of sterols away from the ER, thus increasing SREBP-2 processing.
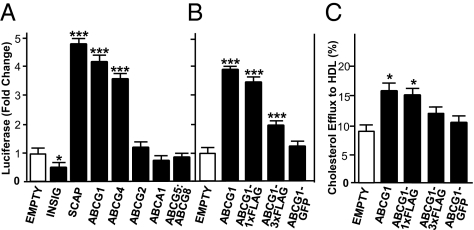
To test this hypothesis, we transfected CHO-K1 cells with a plasmid that encodes a luciferase reporter gene under the control of two SREs from the HMG-CoA synthase promoter (pSynSRE), and is thus activated by mSREBP-2, together with wild-type or epitope-tagged ABCG1, insulin-induced gene 2 (INSIG-2), SREBP cleavage-activating protein (SCAP), or various other ABC transporters. The activity of the luciferase reporter plasmid is completely dependent on endogenous SREBP processing and nuclear localization of mSREBP-2. Such processing of SREBP-2 is partially inhibited when cells are grown in the presence of 10% FBS (Fig. 3 A and B, empty plasmid). Overexpression of INSIG-2 and SCAP were used as controls as they are known to repress or induce SREBP-2 maturation, respectively (27, 28).
Fig. 3.
A high throughput, sensitive luciferase reporter assay measures ABCG1 activity. CHO-K1 cells were cotransfected with pSynSRE, β-galactosidase, and either an empty plasmid (open bars) or myc–INSIG-2, HSV–SCAP, untagged ABCG1, ABCG4, ABCG2, ABCA1, or ABCG5:ABCG8 (A) or untagged ABCG1, ABCG1–1×FLAG, ABCG1–3×FLAG, or ABCG1–GFP (black bars) (B). The cells were then incubated for an additional 24 h in media containing 10% FBS. Normalized luciferase activities are given as fold change relative to cells transfected with pSynSRE and an empty plasmid. (C) CHO-K1 cells were transfected as in A and B. Efflux of 3H-cholesterol to HDL (50 μg/mL) was measured as described (41). The data are representative of three separate experiments, each performed in sextuplet. *P < 0.05; ***P < 0.001.
Fig. 3A shows that overexpression of ABCG1, ABCG4, or SCAP resulted in an approximately fourfold increase in luciferase activity, consistent with increased levels of mSREBP-2 and activation of target genes. Activation of pSynSRE by ABCG1, ABCG4, or SCAP was specific because luciferase activity was unchanged after overexpression of ABCG2, ABCA1, or ABCG5:ABCG8, and, as expected (27), decreased after overexpression of INSIG-2 (Fig. 3A). Fig. 3B shows that the ability of ABCG1 to activate pSynSRE was significantly impaired when three FLAG epitopes or GFP were fused to the COOH terminus. In contrast, ABCG1 containing a single FLAG epitope was nearly as potent as untagged ABCG1 protein (Fig. 3B). All these data suggest that ABCG1 facilitates the movement of sterols away from the ER leading to increased nuclear mSREBP-2 and activation of target genes.
Finally, we show that, whereas overexpression of untagged ABCG1 or ABCG1–1×FLAG stimulated the efflux of radioactive cellular cholesterol to exogenous HDL, addition of larger tags (3×FLAG or GFP) to ABCG1 severely impaired function (Fig. 3C). Importantly, comparison of the data in Fig. 3 A and B vs. C, indicate that the luciferase reporter assay is far more robust and sensitive than the cholesterol efflux assay. As described below, this assay has led to the identification of critical amino acids required for ABCG1 function.
Transmembrane Domains of ABCG1 Are Sufficient for Endosomal Targeting and Sterol Transport.
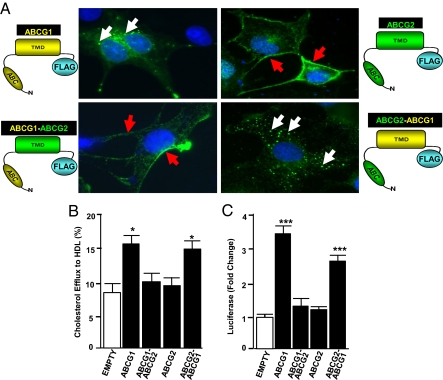
To identify domains of ABCG1 necessary for intracellular targeting and function, we generated plasmids that expressed wild-type ABCG1 or ABCG2 or chimeric fusion proteins comprising the NH2-terminal cytoplasmic domain (∼440 residues) and TM domain (∼220 residues) of ABCG1 and/or ABCG2, each with a single FLAG epitope at the COOH terminal (Fig. 4A).
Fig. 4.
The C-terminal transmembrane domain region of ABCG1 is essential for cellular localization and function. (A) Cos-7 cells were transfected with 1×FLAG-tagged ABCG1, ABCG2, or the indicated chimeric fusion proteins. Confocal images (taken at 63× magnification) are shown. White and red arrows identify immunofluorescent signals in endosomes and the plasma membrane, respectively. (B) CHO-K1 cells were transfected with an empty/control plasmid or plasmids encoding the indicated protein and cholesterol efflux to HDL determined as described in Fig. 3. (C) CHO-K1 cells were cotransfected with pSynSRE and the indicated plasmid before determination of luciferase activity, as described in Fig. 3. Data in B and C are representative of two to three different experiments, each performed in sextuplet. *P < 0.05; ***P < 0.001 vs. cells transfected with pSynSRE and empty plasmid.
The data of Fig. 4A (Upper Left, white arrows) show that overexpressed ABCG1 localizes to intracellular vesicles and is undetectable at the cell surface. The majority of ABCG2 localized to the plasma membrane (Fig. 4A; Upper Right, red arrows) where it can function to efflux drugs from the cell. Overexpression of ABCG1, but not ABCG2, increased both efflux of cellular cholesterol to HDL and the activity of pSynSRE (Fig. 4 B and C). A chimeric fusion protein containing the NH2-terminal domain of ABCG2 and the TM domains of ABCG1 (ABCG2–ABCG1) localized to intracellular vesicles (Fig. 4A; Lower Right, white arrows). Importantly, this fusion protein retained significant functional activity as determined by the activation of pSynSRE and increased efflux of cellular cholesterol to HDL (Fig. 4 B and C). In contrast, a chimeric fusion protein containing the NH2-terminal domain of ABCG1 and the TM domains of ABCG2 (ABCG1–ABCG2) localized to the cell surface (Fig. 4A, Lower Left, red arrows) and failed to activate pSynSRE or the efflux of cellular cholesterol to HDL (Fig. 4 B and C). Taken together, these data demonstrate that the TM domains of ABCG1 are sufficient to correctly target the protein to intracellular vesicles and to stimulate both pSynSRE luciferase activity and cellular cholesterol efflux to HDL and that ABCG1 must be localized to intracellular vesicles to function.
Identification of Critical Residues Required for ABCG1 Function.
To identify critical residues that are required for ABCG1 to activate pSynSRE and/or the efflux of cellular cholesterol to HDL, we aligned the amino acid sequence of the TM domains of mouse ABCG1 with the corresponding sequences from three other members of the murine ABCG subfamily (ABCG4, ABCG5, and ABCG8) that are reported to be involved in sterol flux (Fig. S1). We included INSIG-2, as alanine-scanning mutagenesis studies previously identified five amino acids (asterisks in Fig. S1) that are important for INSIG-2 to bind oxysterols and thus retain the SCAP:SREBP complex in the ER (29). We also included ABCG2 as a negative control because this protein does not affect sterol homeostasis or efflux of cellular sterols (Fig. 3).
We excluded proteins that contained a sterol-sensing domain (SSD) (i.e., SCAP, NPC-1, HMG-CoA reductase) (30–32) because comparison of their amino acid sequences with ABCG1 failed to identify significant regions of amino acid conservation (Fig. S2 A and B).
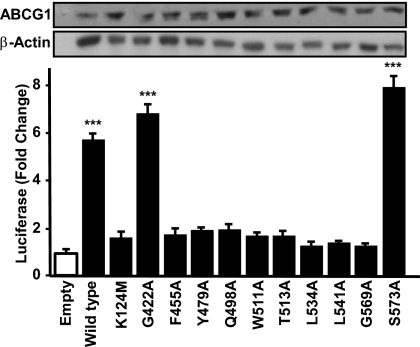
On the basis of the alignment (Fig. S1), we generated a series of ABCG1 mutants wherein a single amino acid was replaced with alanine. We chose to mutate two residues (Q498A and W511A) that are both conserved with INSIG-2 and were previously shown to be important for the interaction of INSIG-2 with oxysterols (29). We also mutated residues that were either conserved with INSIG-2 and at least two other ABCG family members (T513A, G569A, L534A, and L541A) or were found in at least two members of the ABCG subfamily but not INSIG-2 (G422A, Y479A, F455A, and S573A). The positions of the 10 amino acid mutations within the six TM α-helices and the point mutation in the Walker A motif are illustrated in Fig. S3.
CHO-K1 cells were then transfected with untagged wild-type or mutant ABCG1 together with pSynSRE. The data of Fig. 5 show that luciferase activity increased 5.5- to 8-fold after overexpression of wild-type ABCG1 or ABCG1 mutants G422A or S573A, thus indicating that G422 and S573 are not critical for function. In contrast, luciferase activity did not increase significantly following overexpression of ABCG1–K124M that contains a mutation in a critical lysine in the Walker A motif (Fig. 5). This mutant ABCG1 serves as a negative control as mutation of the corresponding lysine in the Walker A motif of many other ABC transporters attenuates ATP hydrolysis and transport function (33). Interestingly, overexpression of the remaining eight ABCG1 constructs failed to induce luciferase activity above the level seen with the ABCG1–K124M mutant (Fig. 5). The finding that S573 is only three residues from G569, a nonfunctional mutant, suggests the effects of the mutations are specific. Thus, we conclude that these eight conserved residues are critical for maintaining ABCG1 function.
Fig. 5.
Alanine-scanning mutagenesis identifies critical residues that are important for ABCG1 function. (A) CHO-K1 cells were cotransfected with pSynSRE and either wild-type ABCG1 or ABCG1 containing the indicated mutation. Luciferase activities are given as fold changes compared with control. (Inset) Western blot analysis of total cell extracts. Data are representative of at least three experiments each performed in sextuplet. ***P < 0.001 vs. cells transfected with pSynSRE plus empty vector.
Western blot analysis of cell lysates showed that wild-type and mutant forms of ABCG1 were all expressed at similar levels (Fig. 5, Inset). Further, immunofluorescence studies of selected mutants showed that wild-type and inactive mutant ABCG1 proteins colocalized with NPC-1 (Fig. S4), suggesting the single point mutations did not result in aberrant targeting or degradation of ABCG1.
To determine whether dimerization was affected by these point mutations, we transfected cells with ABCG1–FLAG together with wild-type or mutant forms of ABCG1 that contained a COOH-terminal HA epitope. Immunoprecipitation followed by Western blot analysis of the precipitated proteins demonstrated that all five of the tested mutant proteins formed dimers in HEK293 cells (Fig. S5). We conclude that the loss in activity of the eight mutant forms of ABCG1 (Fig. 5) is not a result of decreased protein expression, mislocalization, or an inability to form homodimers. Rather, the data suggest that these eight amino acids are critical for ABCG1 to function and alter intracellular sterol flux and SREBP-2 processing.
Inhibition of SREBP-2 Processing by Exogenous Sterols Is Attenuated by ABCG1.
Previous studies by Brown and Goldstein and colleagues demonstrated that processing of SREBP-2 in cultured cells is inhibited following addition of specific sterols to the media (29). Further, Adams et al. demonstrated that exogenously delivered 25-hydroxycholesterol (25-OHC) was more potent than cholesterol in inhibiting SREBP-2 processing (34).
To determine whether overexpression of ABCG1 affects the ability of exogenous sterols to repress SREBP-2 processing, we transfected CHO-K1 cells with pSynSRE, together with either wild-type or mutant ABCG1. Cells were incubated for an additional 24 h in medium supplemented with 5% lipoprotein-deficient serum, 5 μM simvastatin and 50 μM mevalonic acid to activate SREBP-2 processing and induce SREBP-2 target genes, including pSynSRE. As expected, under these conditions overexpression of ABCG1 had a relatively small effect on luciferase activity, consistent with preexisting high levels of nuclear mSREBP-2 (Fig. 6A, compare open bars).
Fig. 6.
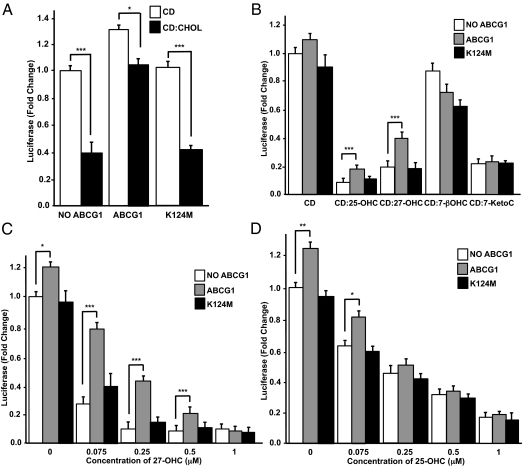
Overexpression of wild-type ABCG1 selectively attenuates the repression of SREBP-2 processing by sterols. (A) CHO-K1 cells were cotransfected with pSynSRE and either wild-type or mutant ABCG1 (ABCG1–K124M). After 5 h transfection media was replaced with medium D (5% lipoprotein-deficient serum, LPDS) containing cyclodextrin (CD) or CD complexed with 25 μM cholesterol (A) or with individual oxysterols (B–D) at 0.5 μM (B) or the indicated concentration (C and D). Luciferase values are normalized to β-galactosidase activity. The values are mean ± SEM from six wells. The data are representative of at least three experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 6A (Left two bars) shows that treatment of the cells with cyclodextrin (CD):cholesterol (25 μM) reduced the activity of pSynSRE by 60%, consistent with the ability of cholesterol to inhibit SREBP-2 processing to mSREBP-2. Importantly, this cholesterol-dependent repression of luciferase activity was attenuated in cells overexpressing wild-type ABCG1, but not the inactive mutant ABCG1–K124M (Fig. 6A). We interpret these data to indicate that wild-type ABCG1 redistributes cholesterol away from the ER, thus allowing maturation of SREBP-2 and activation of pSynSRE. Fig. 6B shows the changes in pSynSRE activity in response to different oxysterols (0.5 μM) in the presence or absence of either wild-type or mutant ABCG1 overexpression. The data show that repression of pSynSRE luciferase activity, in response to 27-hydroxycholesterol (27-OHC), and 25-OHC, was blunted by overexpression of wild-type, but not mutant ABCG1 (Fig. 6B). In contrast, repression of the reporter activity following addition of 7-ketocholesterol, an autooxidation product of cholesterol, to the cells was unaffected by ABCG1 overexpression (Fig. 6B). Addition of 7-β-cholesterol, another autooxidation, nonenzymatic product of cholesterol, resulted in relatively poor repression of pSynSRE and the effect was not impaired by ABCG1 overexpression (Fig. 6B). Finally, we show that the ability of transfected wild-type, but not mutant ABCG1 to limit the oxysterol-mediated repression of pSynSRE luciferase activity was dependent both on oxysterol concentration and structure (Fig. 6 C and D). The data show that wild type, but not mutant ABCG1 is able to blunt the repression of pSynSRE in response to 27-OHC at concentrations below 1 μM (Fig. 6C). In contrast, the ability of ABCG1 to attenuate repression of pSynSRE in response to 25-OHC was muted over the whole concentration range tested (Fig. 6D). The effect of ABCG1 overexpression on oxysterol-mediated inhibition of SREBP-2 processing is specific, as the data in Fig. S6 show that ABCG1 fails to affect the repression of pSynSRE in response to 7-ketocholesterol. Together, these studies demonstrate that ABCG1 modulates the biological effects of specific sterols (cholesterol, 27-OHC, and 25-OHC) on SREBP-2 processing and maturation.
Discussion
Mammalian ABC transporters have been shown to localize to either the plasma membrane, peroxisomes, mitochondria, endoplasmic reticulum (3–5), or lamellar bodies (6). Very recently Sturek et al. (21) showed that >90% ABCG1 was concentrated in secretory granules of pancreatic β-cells. These authors provided evidence that stimulated insulin secretion was dependent upon ABCG1 maintaining appropriate cholesterol levels in the granule membrane (21). To our knowledge, this is the only report demonstrating that a member of the ABC gene superfamily localizes to the endosome/recycling endosome pathway. The conclusion that active ABCG1 localizes to endosomes is supported by data obtained from a number of different experimental approaches, including biotinylation of endogenous ABCG1 in peritoneal macrophages treated with or without an LXR agonist (Fig. 1), immunofluorescent studies involving either untagged or epitope-tagged ABCG1, or hybrid ABCG1–ABCG2 proteins (Figs. 2 and 4), and the ability of the intracellular ABCG1 protein to activate an SRE-luciferase reporter gene (Figs. 3–6). This conclusion is also consistent with a previous observation showing that transiently transfected ABCG1 or ABCG4 localized to intracellular vesicles in primary mouse neurons and astrocytes and to RhoB+ vesicles in Cos-7 cells (22). On the basis of all these data, we hypothesize that one important function of ABCG1 is to maintain normal sterol levels in endocytic vesicles in part by facilitating the flux of specific sterols away from the endoplasmic reticulum.
The current studies also identify important differences between ABCG1 (and the highly homologous transporter ABCG4) (22) and other ABC family members that facilitate “sterol” transport. In contrast to the intracellular localization of ABCG1 and ABCG4 (22), both ABCA1 and the heterodimer ABCG5:ABCG8 localize to the plasma membrane (3, 35). Further, heterodimeric ABCG5:ABCG8 promotes the efflux of cholesterol and sitosterol out of enterocytes or hepatocytes (36), whereas ABCA1 may actually stimulate the transbilayer movement of a phospholipid, with sterol redistributing into the phospholipid-enriched outer leaflet of the plasma membrane, before efflux of both lipids to exogenous apoA-1 (37).
To our knowledge, there are no other reports of alanine-scanning mutagenesis of an ABC sterol transporter. This approach, together with studies using fusion proteins containing domains of ABCG1 and ABCG2, has allowed us to separate the requirements for correct intracellular targeting and function. We show that the TM domains of ABCG1 are sufficient to both target the protein to intracellular vesicles of the endosomal pathway and to alter cellular sterol homeostasis (Fig. 4). However, our mutagenesis studies have led to the identification of eight amino acids within the TM domain that are critical for function, but are themselves not important for ABCG1 targeting.
Interestingly, two of the residues (Q498 and W511) required for ABCG1 function are conserved in INSIG-2, where they are required for interaction with specific oxysterols, including 25-OHC (29). Because we demonstrate that overexpression of ABCG1 partially prevents the inhibitory effects of 25-OHC and 27-OHC on SREBP-2 processing, we suggest that Q498 and W511 are involved in interaction of ABCG1 with specific oxysterols and their subsequent movement away from the ER. However, to date, we have been unable to demonstrate specific binding of these sterols to the polytopic ABCG1 protein.
Only one residue in ABCG1 (G569) is conserved when the amino acids in the TM domains of all six members of the mammalian ABCG family and INSIG-2 are aligned (Fig. S1). Interestingly, a point mutation in this residue in ABCG8 (G574R) has been identified in a patient with sitosterolemia (38). This suggests that this point mutation is sufficient to inactivate the ABCG8:ABCG5 heterodimer. The current studies demonstrate that this same residue is critical for ABCG1 function (Fig. 5; G569). Because ABCG8:ABCG5 functions as a cholesterol transporter (36), the current data suggest that G569 may also be involved in the interaction of ABCG1 with cholesterol to facilitate the transmembrane movement of sterols.
On the basis of biotinylation of HEK293 cells that expressed ABCG1–FLAG in response to mifepristone (23) or studies using sucrose gradient centrifugation (25), it was concluded that ABCG1 localized to the plasma membrane. However, as we have never observed ABCG1 at the cell surface, we are unable to account for these differences.
The current studies were greatly aided by the use of a highly sensitive and robust sterol-sensitive luciferase reporter assay that was responsive to mSREBP levels. In contrast to the 3H-cholesterol efflux assay to measure ABCG1 activity, the pSynSRE assay is more sensitive, reproducible, and can be carried out in normal growth media (10% FBS) and does not depend upon the addition of acyl-CoA:cholesterol acyltransferase (ACAT) inhibitors or culturing cells for prolonged periods in the absence of serum.
On the basis of recent studies that showed that a small decrease or increase in ER-cholesterol levels is sufficient to activate or repress SREBP-2 processing, respectively (26), we have concluded that ABCG1 functions to redistribute intracellular cholesterol and specific oxysterols (27-OHC and 25-OHC) (Fig. 6) away from the ER, thus leading to increased mSREBP-2 levels and activation of pSynSRE (Fig. 6). The finding that ABCG1 coexpression had no effect on the repression of pSynSRE by 7-ketocholesterol (Fig. S6) was unexpected, as it has been reported that ABCG1 protects cells from 7-ketocholesterol–induced toxicity (39). The reason for these differences remains to be identified.
Nonetheless, the observation that repression of SREBP-2 processing by 27- or 25-OHC is attenuated following overexpression of ABCG1 suggested that these oxysterols may accumulate in cells lacking this transporter. Consistent with this proposal, we have previously reported that Abcg1−/− alveolar macrophages accumulate significant amounts of 27-OHC and 25-OHC (17). Further, cholesterol 27-hydroxylase (Cyp27A1) and cholesterol 25-hydroxylase (CH25H) mRNAs, that encode the enzymes that synthesize these two oxysterols, were increased in Abcg1−/− macrophages (Fig. S7).
How can intracellular localization of ABCG1 affect efflux of cellular cholesterol to exogenous HDL? The finding that ABCG1 is present in endosomes and recycling endosomes suggests a mechanism by which ABCG1 transfers sterols to the inner leaflet of these vesicles before their fusion with the plasma membrane. This would result in redistribution of these sterols to the outer leaflet of the plasma membrane such that they can desorb in a nonspecific manner to multiple exogenous lipid acceptors that include HDL, LDL, or nonphysiologically to phospholipid vesicles or CD. Such nonspecific desorption of cholesterol from the plasma membrane of ABCG1-expressing cells has recently been reported (13). In summary, the studies described herein provide important insights into the domains and residues that are important for ABCG1 function and localization. Further studies that use purified ABCG1 protein should provide additional information on possible transporter–substrate interactions.
Materials and Methods
Materials.
Details of materials can be found in SI Materials and Methods.
Preparation of Sterol/Methyl-β-Cyclodextrin Complexes.
Complexes were generated using a modification of the protocol described by Klein et al. (40) Details can be found in SI Materials and Methods.
Further detailed methods can be found in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank our colleagues Dr. Peter Tontonoz and Dr. Thomas Vallim for useful discussions, Lucille Huang for technical assistance, and Dr. T. Osborne (Sanford-Burnham Medical Research Institute) for providing pSynSRE and plasmids expressing INSIG-2 and SCAP. Dr. R. Pagano (deceased) kindly provided pDsRed–Rab5 and pDsRed–Rab11. These studies were supported by Grants from the National Institutes of Health (HL68445) (to P.A.E.) and from the Laubisch Fund (P.A.E.). E.J.T. was supported in part by a postdoctoral fellowship from the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113021108/-/DCSupplemental.
References
- 1.Dean M, Allikmets R. Evolution of ATP-binding cassette transporter genes. Curr Opin Genet Dev. 1995;5:779–785. doi: 10.1016/0959-437x(95)80011-s. [DOI] [PubMed] [Google Scholar]
- 2.Tarr PT, Tarling EJ, Bojanic DD, Edwards PA, Baldan A. Emerging new paradigms for ABCG transporters. Biochim Biophys Acta. 2009;1791:584–593. doi: 10.1016/j.bbalip.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graf GA, et al. Coexpression of ATP-binding cassette proteins ABCG5 and ABCG8 permits their transport to the apical surface. J Clin Invest. 2002;110:659–669. doi: 10.1172/JCI16000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamijo K, Taketani S, Yokota S, Osumi T, Hashimoto T. The 70-kDa peroxisomal membrane protein is a member of the Mdr (P-glycoprotein)-related ATP-binding protein superfamily. J Biol Chem. 1990;265:4534–4540. [PubMed] [Google Scholar]
- 5.Lill R, Kispal G. Mitochondrial ABC transporters. Res Microbiol. 2001;152:331–340. doi: 10.1016/s0923-2508(01)01204-9. [DOI] [PubMed] [Google Scholar]
- 6.Yamano G, et al. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett. 2001;508:221–225. doi: 10.1016/s0014-5793(01)03056-3. [DOI] [PubMed] [Google Scholar]
- 7.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy MA, et al. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005;1:121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Wang N, Lan D, Chen W, Matsuura F, Tall AR. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc Natl Acad Sci USA. 2004;101:9774–9779. doi: 10.1073/pnas.0403506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothblat GH, Bamberger M, Phillips MC. Reverse cholesterol transport. Methods Enzymol. 2003;129:628–644. doi: 10.1016/0076-6879(86)29095-3. [DOI] [PubMed] [Google Scholar]
- 11.Favari E, et al. Small discoidal pre-beta1 HDL particles are efficient acceptors of cell cholesterol via ABCA1 and ABCG1. Biochemistry. 2009;48:11067–11074. doi: 10.1021/bi901564g. [DOI] [PubMed] [Google Scholar]
- 12.Larrede S, et al. Stimulation of cholesterol efflux by LXR agonists in cholesterol-loaded human macrophages is ABCA1-dependent but ABCG1-independent. Arterioscler Thromb Vasc Biol. 2009;29:1930–1936. doi: 10.1161/ATVBAHA.109.194548. [DOI] [PubMed] [Google Scholar]
- 13.Sankaranarayanan S, et al. Effects of acceptor composition and mechanism of ABCG1-mediated cellular free cholesterol efflux. J Lipid Res. 2009;50:275–284. doi: 10.1194/jlr.M800362-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baldán A, et al. Impaired development of atherosclerosis in hyperlipidemic Ldlr-/- and ApoE-/- mice transplanted with Abcg1-/- bone marrow. Arterioscler Thromb Vasc Biol. 2006;26:2301–2307. doi: 10.1161/01.ATV.0000240051.22944.dc. [DOI] [PubMed] [Google Scholar]
- 15.Bojanic DD, et al. Differential expression and function of ABCG1 and ABCG4 during development and ageing. J Lipid Res. 2009;51:169–181. doi: 10.1194/jlr.M900250-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranalletta M, et al. Decreased atherosclerosis in low-density lipoprotein receptor knockout mice transplanted with Abcg1-/- bone marrow. Arterioscler Thromb Vasc Biol. 2006;26:2308–2315. doi: 10.1161/01.ATV.0000242275.92915.43. [DOI] [PubMed] [Google Scholar]
- 17.Tarling EJ, et al. Impaired development of atherosclerosis in Abcg1-/- Apoe-/- mice: Identification of specific oxysterols that both accumulate in Abcg1-/- Apoe-/- tissues and induce apoptosis. Arterioscler Thromb Vasc Biol. 2010;30:1174–1180. doi: 10.1161/ATVBAHA.110.205617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yvan-Charvet L, et al. Combined deficiency of ABCA1 and ABCG1 promotes foam cell accumulation and accelerates atherosclerosis in mice. J Clin Invest. 2007;117:3900–3908. doi: 10.1172/JCI33372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldán A, et al. Deletion of the transmembrane transporter ABCG1 results in progressive pulmonary lipidosis. J Biol Chem. 2006;281:29401–29410. doi: 10.1074/jbc.M606597200. [DOI] [PubMed] [Google Scholar]
- 20.Wojcik AJ, Skaflen MD, Srinivasan S, Hedrick CC. A critical role for ABCG1 in macrophage inflammation and lung homeostasis. J Immunol. 2008;180:4273–4282. doi: 10.4049/jimmunol.180.6.4273. [DOI] [PubMed] [Google Scholar]
- 21.Sturek JM, et al. An intracellular role for ABCG1-mediated cholesterol transport in the regulated secretory pathway of mouse pancreatic beta cells. J Clin Invest. 2010;120:2575–2589. doi: 10.1172/JCI41280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarr PT, Edwards PA. ABCG1 and ABCG4 are coexpressed in neurons and astrocytes of the CNS and regulate cholesterol homeostasis through SREBP-2. J Lipid Res. 2008;49:169–182. doi: 10.1194/jlr.M700364-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Vaughan AM, Oram JF. ABCG1 redistributes cell cholesterol to domains removable by high density lipoprotein but not by lipid-depleted apolipoproteins. J Biol Chem. 2005;280:30150–30157. doi: 10.1074/jbc.M505368200. [DOI] [PubMed] [Google Scholar]
- 24.Xie Q, et al. Cell surface localization of ABCG1 does not require LXR activation. Arterioscler Thromb Vasc Biol. 2006;26:e143–e144, author reply e145. doi: 10.1161/01.ATV.0000245790.47112.b2. [DOI] [PubMed] [Google Scholar]
- 25.Wang N, Ranalletta M, Matsuura F, Peng F, Tall AR. LXR-induced redistribution of ABCG1 to plasma membrane in macrophages enhances cholesterol mass efflux to HDL. Arterioscler Thromb Vasc Biol. 2006;26:1310–1316. doi: 10.1161/01.ATV.0000218998.75963.02. [DOI] [PubMed] [Google Scholar]
- 26.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci USA. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang T, Goldstein JL, Brown MS. Overexpression of membrane domain of SCAP prevents sterols from inhibiting SCAP.SREBP exit from endoplasmic reticulum. J Biol Chem. 2000;275:29881–29886. doi: 10.1074/jbc.M005439200. [DOI] [PubMed] [Google Scholar]
- 29.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millard EE, et al. The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J Biol Chem. 2005;280:28581–28590. doi: 10.1074/jbc.M414024200. [DOI] [PubMed] [Google Scholar]
- 31.Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: Mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 32.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 33.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 34.Adams CM, et al. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 35.Neufeld EB, et al. Cellular localization and trafficking of the human ABCA1 transporter. J Biol Chem. 2001;276:27584–27590. doi: 10.1074/jbc.M103264200. [DOI] [PubMed] [Google Scholar]
- 36.Graf GA, et al. ABCG5 and ABCG8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J Biol Chem. 2003;278:48275–48282. doi: 10.1074/jbc.M310223200. [DOI] [PubMed] [Google Scholar]
- 37.Vedhachalam C, et al. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J Biol Chem. 2007;282:25123–25130. doi: 10.1074/jbc.M704590200. [DOI] [PubMed] [Google Scholar]
- 38.Berge KE, et al. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 39.Terasaka N, Wang N, Yvan-Charvet L, Tall AR. High-density lipoprotein protects macrophages from oxidized low-density lipoprotein-induced apoptosis by promoting efflux of 7-ketocholesterol via ABCG1. Proc Natl Acad Sci USA. 2007;104:15093–15098. doi: 10.1073/pnas.0704602104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein U, Gimpl G, Fahrenholz F. Alteration of the myometrial plasma membrane cholesterol content with beta-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- 41.Venkateswaran A, et al. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc Natl Acad Sci USA. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.