Abstract
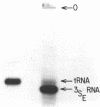
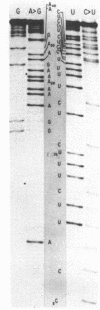
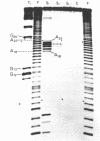
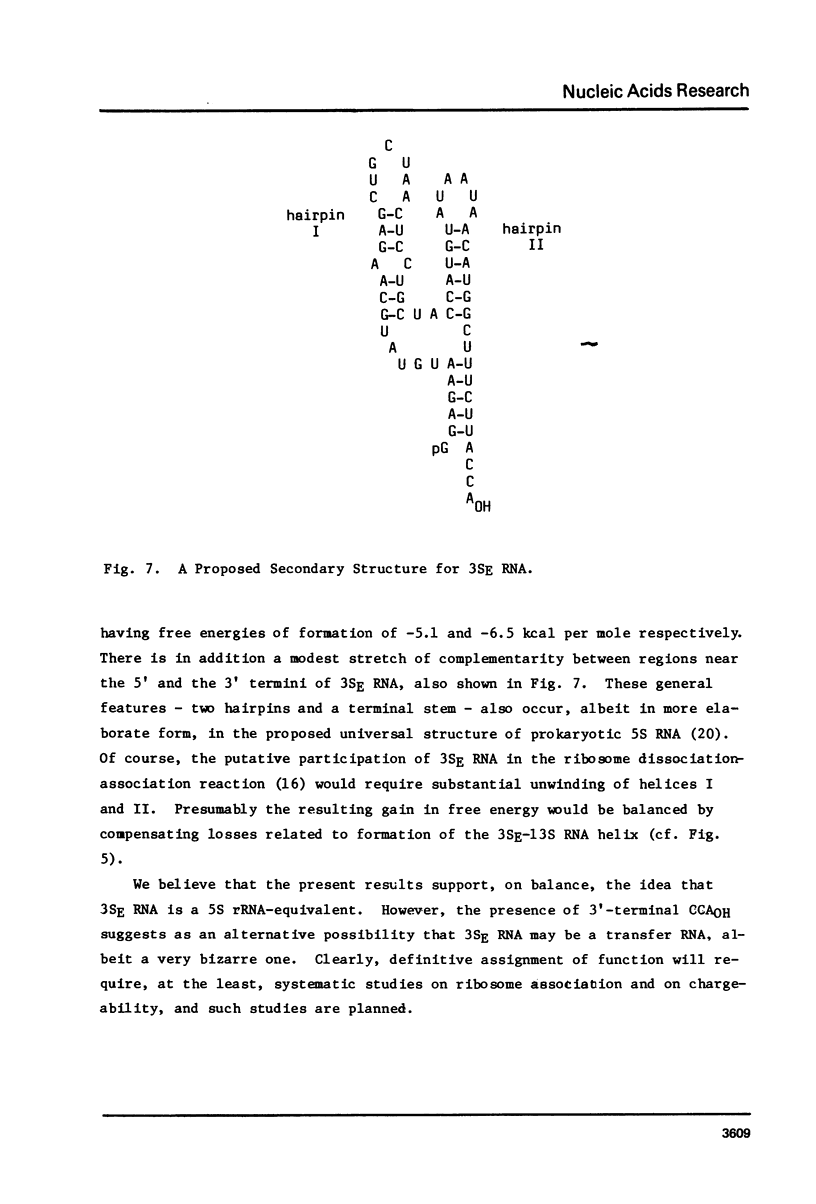
We have sequenced 3SE RNA, an unmodified species from hamster cell mitochondria that may be a 5S rRNA-equivalent. The sequence is [[Formula: see text]. The underlined stretches can form the stems of 2 hairpins whose existence is supported by S1 nuclease analysis. Residues 24 through 34 can also base-pair extensively with a sequence in the 3'-region of the small subunit ("13S") mitochondrial rRNA. These interactions resemble interactions postulated for 5S RNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azad A. A. Intermolecular base-paired interaction between complementary sequences present near the 3' ends of 5S rRNA and 18S (16S) rRNA might be involved in the reversible association of ribosomal subunits. Nucleic Acids Res. 1979 Dec 11;7(7):1913–1929. doi: 10.1093/nar/7.7.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin D. T., Friend D. A. Comparison of cytoplasmic and mitochondrial 4 S RNA from cultured hamster cells: physical and metabolic properties. J Mol Biol. 1972 Nov 14;71(2):163–175. doi: 10.1016/0022-2836(72)90344-0. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Jones T. N., Cleaves G. R. An unmethylated "3 SE" RNA in hamster mitochondria: a 5 S RNA-equivalent? Biochem Biophys Res Commun. 1974 Jan 23;56(2):401–406. doi: 10.1016/0006-291x(74)90856-0. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Montenecourt B. S. Mitochondrial RNA from cultured animal cells. Distinctive high-molecular-weight and 4 s species. J Mol Biol. 1970 Mar 14;48(2):279–295. doi: 10.1016/0022-2836(70)90161-0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Vournakis J. N., Donis-Keller H., Chaconas G., Dougall D. K., Kafatos F. C. End labeling of enzymatically decapped mRNA. Nucleic Acids Res. 1977 Dec;4(12):4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Krupp G., Gross H. J. Rapid RNA sequencing: nucleases from Staphylococcus aureus and Neurospora crassa discriminate between uridine and cytidine. Nucleic Acids Res. 1979 Aug 10;6(11):3481–3490. doi: 10.1093/nar/6.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardi P. M., Luck D. J. Absence of a 5S RNA complnent in the mitochondrial ribosomes of Neurospora crassa. Nat New Biol. 1971 Feb 3;229(5):140–142. doi: 10.1038/newbio229140a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Samols D. R., Hagenbuchle O., Gage L. P. Homology of the 3' terminal sequences of the 18S rRNA of Bombyx mori and the 16S rRNA of Escherchia coli. Nucleic Acids Res. 1979 Nov 10;7(5):1109–1119. doi: 10.1093/nar/7.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]