Abstract
The Diels-Alder reaction is a cornerstone in organic synthesis, forming two carbon-carbon bonds and up to four new stereogenic centers in one step. No naturally occurring enzymes have been shown to catalyze bimolecular Diels-Alder reactions. We describe the de novo computational design and experimental characterization of enzymes catalyzing a bimolecular Diels-Alder reaction with high stereoselectivity and substrate specificity. X-ray crystallography confirms that the structure matches the design for the most active of the enzymes, and binding site substitutions reprogram the substrate specificity. Designed stereoselective catalysts for carbon-carbon bond forming reactions should be broadly useful in synthetic chemistry.
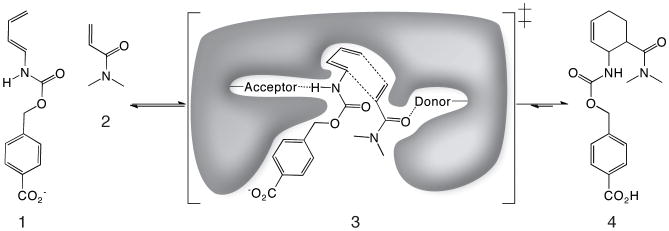
Intermolecular Diels-Alder reactions are important in organic synthesis (1-3), and enzyme Diels-Alder catalysts could be invaluable in increasing rates and stereoselectively. No naturally occurring enzyme has been demonstrated (16) to catalyze an intermolecular Diels-Alder reaction (1, 2), although catalytic antibodies have been generated for several Diels-Alder reactions (3, 16). We have previously used the Rosetta computational design methodology to design novel enzymes (4, 5) that catalyze bond breaking reactions. However, bimolecular bond forming reactions present a greater challenge, since both substrates must be bound in the proper relative orientation in order to accelerate the reaction and induce stereoselectivity. Also, previous successes with computational enzyme design have involved general acid-base catalysis and covalent catalysis, but the Diels-Alder reaction instead can be primarily influenced by modulation of molecular orbital energies (6). To investigate the feasibility of designing intermolecular Diels-Alder enzyme catalysts we chose to focus on the well-studied model Diels-Alder reaction between 4-carboxybenzyl trans-1,3-butadiene-1-carbamate and N,N-dimethylacrylamide (Fig. 1, substrates 1 and 2, respectively) (10, 11).
Figure 1. The Diels-Alder reaction.
Diene (1) and dienophile (2) undergo a pericyclic [4+2] cycloaddition (3) to form a chiral cyclohexene ring (4). Also shown in (3) is a schematic of the design target active site, with hydrogen bond acceptor and donor groups activating the diene and dienophile and a complementary binding pocket holding the two substrates in an orientation optimal for catalysis.
The first step in de novo enzyme design is to decide on a catalytic mechanism and an associated ideal active site. For normal-electron-demand Diels–Alder reactions, Frontier Molecular Orbital Theory dictates that the interaction of the highest occupied molecular orbital (HOMO) of the diene with the lowest unoccupied molecular orbital (LUMO) of the dienophile is the dominant interaction in the transition state (6). The reaction can be catalyzed by bringing the energies of the HOMO and LUMO closer together. This can be accomplished by positioning a hydrogen bond acceptor to interact with the carbamate NH of the diene, (thus raising the energy of the HOMO energy and stabilizing the positive charge accumulating in the transition state), and a hydrogen bond donor to interact with the carbonyl of the dienophile, (lowering the LUMO energy and stabilizing the negative charge accumulating in the transition state) (7). Quantum mechanical (QM) calculations predict that these hydrogen bonds can stabilize the transition state by up to 4.7 kcal/mol. (Fig. S0). In addition to electronic stabilization, binding of the two substrates in a relative orientation optimal for the reaction is expected to produce a large increase in rate through entropy reduction (8). Thus, a protein with a binding pocket (Fig. 1) that positions the two substrates in the optimal relative orientation, and has appropriately placed hydrogen bond donors and acceptors, is expected to be an effective Diels-Alder catalyst.
We used the Rosetta methodology to design in silico enzyme models containing active sites with the desired properties (Fig. 1). The design methodology starts from three-dimensional atomic models of minimal active sites (theozymes) consisting of the reaction transition state and protein functional groups involved in binding and catalysis. We chose the carbonyl oxygen from a glutamine or asparagine to hydrogen bond with the N-H of the diene carbamate, and the hydroxyl from a serine, threonine, or tyrosine to hydrogen bond with the carbonyl oxygen of the dienophile amide moiety (Fig. 1). QM calculations were carried out to determine the geometry of the lowest free energy barrier transition state between substrates and product in the presence of these hydrogen bonding groups. Starting from these coordinates, a diverse ensemble of 1019 distinct minimal active sites was then generated by systematically varying the identity and rotameric state of the catalytic side chains, the hydrogen bonding geometry between these residues and the transition state, and the internal degrees of freedom of the transition state (Fig. S3-S4).
Using RosettaMatch (9), a set of 207 stable protein scaffolds was searched for backbone geometries that allow the two catalytic residues and the two substrates, oriented as in one of the minimal active sites, to be placed without making significant steric clashes with the protein backbone. A hashing technique allows efficient searching through the very large number of distinct sites (16). From the set of 1019 possible active site configurations, approximately 106 could be matched in a stable protein scaffold. Each match was then optimized using RosettaDesign (10) to maximize transition state binding while not clashing with bound substrates or product (16). These designs were filtered based on satisfaction of catalytic geometry, transition state binding energy, and shape complementarity between designed pocket and the transition state (16). A total of 84 designs were selected for experimental validation.
Genes encoding these 84 designs were synthesized with a C-terminal 6-histidine affinity tag and expressed in E. coli. 50 of the designed proteins were soluble; these were purified using affinity chromatography and Diels-Alder activity was monitored using a liquid chromatography-tandem mass spectrometry assay in a phosphate buffered saline (PBS) solution at pH 7.4, 298K (16). Two designs (DA_20_00, DA_42_00) were found to have Diels-Alderase activity. The active design DA_20_00 was created from a 6-bladed β-propeller scaffold (PDB-ID 1E1A; a diisopropylfluorophosphatase from Loligo Vulgaris, 13 mutations, Fig. S5A). As observed for many native β-propeller enzymes, the functional groups that play key roles in catalysis – a glutamine carbonyl group and a tyrosine hydroxyl group that provide the activating hydrogen bonds – are located in the middle of one side of the propeller. The rest of the pocket is lined with hydrophobic residues that form a tight shape-complementary surface (Fig. 2A). The active design DA_42_00 was created from the ketosteroid isomerase scaffold (PDB-ID 1OHO, 14 mutations, Fig. S5B). The active site is quite different than that of DA_20_00, in that only the carbon-carbon bond forming portion of the diene and dienophile are actually buried within the protein.
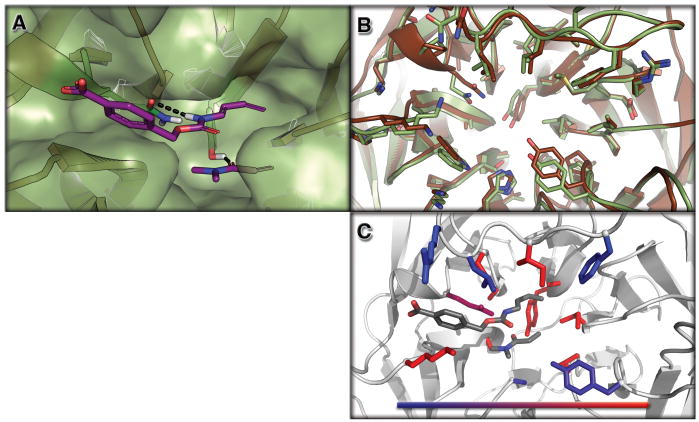
Figure 2. Structure of a Designed Diels-Alderase.
Top panel (2A): surface view of the design model (DA_20_00, green) bound to the substrates (diene and dienophile, purple). The catalytic residues making the designed hydrogen bonds are depicted as sticks. Middle panel (2A): overlay of the design model (DA_20_00, green) and the apo enzyme crystal structure of DA_20_00_A74I (brown). Bottom panel (2C): Contribution of designed residues to catalysis assessed through reversions back to the native amino acid at each position individually. Colors indicate the reduction in activity upon reversion to the native amino acid; 2 fold (blue) to > 10 fold (red). The figure was generated with PyMol (15).
To further improve the catalytic activity of DA_20_00 and DA_42_00, residues that were in direct contact with the transition state in each designed enzyme were mutated individually to sets of residues that were predicted to retain or improve transition state binding and bolster the two catalytic residues. A set of six mutations (A21T, A74I, Q149R, A173C, S271A, and A272N) was found to increase the overall catalytic efficiency of DA_20_00 by over 100-fold relative to the original design model (Table I; we refer to the DA_20_00 protein with these six additional mutations as DA_20_10, Fig. S5C). Three of the mutations improve the packing around the transition state (A74I, A21T) and the catalytic glutamine (A173C). Two of the mutations likely improve the overall electrostatic complementarity with the bound substrates: Q149R hydrogen bonds to the carboxylate on the diene, and S271A makes the dienophile environment more non-polar. The last mutation (A272N) reverts a designed alanine residue back to the native asparagine: molecular dynamics simulations (16) suggested that the catalytic tyrosine can flip into an alternative conformation not positioned to activate the dienophile, and a larger residue at 272, such as the native asparagine, was predicted to hold the tyrosine in the conformation required for catalysis.
Table 1. Kinetic Parameters for DA_20_00, DA_20_10, and DA_42_04.
Reactions for DA_20_00, DA_20_10, and DA_42_04 were carried out at 298K (16). The errors represent the calculated 95% confidence interval. Kinetic parameters for catalytic antibodies 7D4 and 4D5 at 310K were taken from (11,12). The kuncat for the Diels-Alder reaction at 298K was found to be 2.44 × 10−2 M−1 hr−1, in good agreement with the previously reported value at 310K of 4.29 × 10−2 M−1 hr−1.
| Catalyst |
kcat (hr−1) |
KM-diene (mM) |
KM-dienophile (mM) |
kcat/KM-diene (s−1 M−1) |
kcat/KM-dienophile (s−1 M−1) |
kcat/(KM-diene × KM-dienophile) (s−1 M−1 M−1) |
|---|---|---|---|---|---|---|
| DA_20_00 | 0.10 ± 0.02 | 3.5 ± 1.5 | 146.0 ± 2.5 | 0.008 | 0.0002 | 0.06 |
| DA_20_10 | 2.13 ± 0.24 | 1.3 ± 0.1 | 72.8 ± 5.1 | 0.455 | 0.0081 | 6.23 |
| DA_42_04 | 0.03 ± 0.01 | 0.5 ± 0.1 | 16.2 ± 3.2 | 0.017 | 0.0005 | 1.03 |
| mAb 7D4 | 0.21 | 1.0 | 1.7 | 0.058 | 0.0343 | 20.18 |
| mAB 4D5 | 0.21 | 1.6 | 5.9 | 0.036 | 0.0099 | 6.19 |
For DA_42_00, a set of four mutations (Q58R, L61M, A99N, V101I) was found to increase the observed catalytic activity roughly 20-fold over the original design (Table I; we refer to the DA_42_00 protein with four additional mutations as DA_42_04, Fig. S5D). As in the case of DA_20_00, all of these mutations increase the size of the amino acid and either improve packing or electrostatic interactions with the ligand.
To investigate the contributions of the two catalytic residues in DA_20_10 to catalysis, glutamine 195 was mutated into a glutamate (Q195E) and tyrosine 121 was mutated into a phenylalanine (Y121F). We had originally incorporated a glutamine rather than a glutamate at position 195, despite the fact that the carboxylate is more effective than the amide at increasing the energy of the diene HOMO, because we were concerned about the unfavorable contribution of carboxylate desolvation to substrate binding. Furthermore, QM calculations predict that the amide group of glutamine can simultaneously interact with the diene and dienophile, resulting in a 2 kcal/mol lower activation barrier than if glutamate was used as a catalytic residue (Fig S0C). Indeed, the Q195E mutation showed almost complete loss of activity (450-fold), illustrating the sensitivity of the enzyme to the details of the designed active site. The Y121F mutation decreases catalytic activity 27-fold, consistent with the removal of a hydrogen bond that contributes to dienophile binding and a lowering of its LUMO.
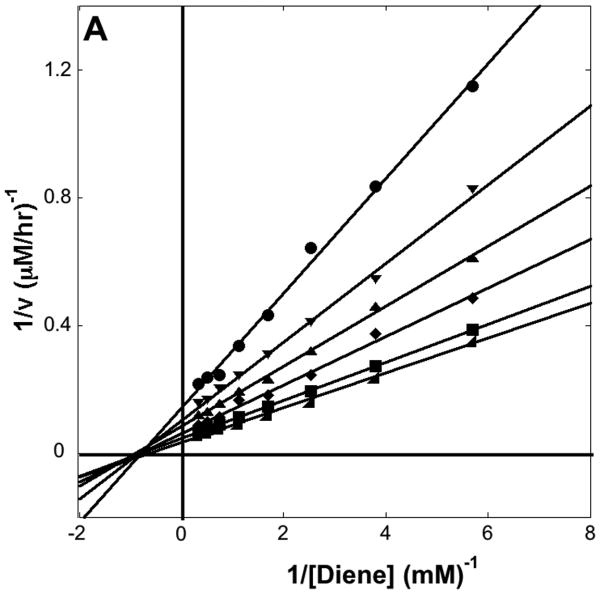
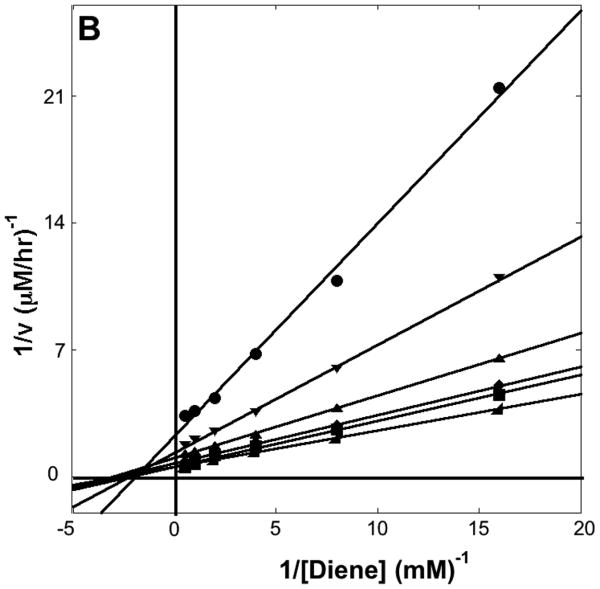
The kinetic parameters of the DA_20_00, DA_20_10, and DA_42_04 catalyzed reactions were determined by measuring the dependence of the reaction velocity on the concentration of both diene and dienophile (16). The kinetic parameters are summarized in Table I, and double reciprocal plots for DA_20_10 and DA_42_04 are shown in Figure 3A and 3B. DA_20_10 has an effective molarity (kcat/kuncat = 89 M) 20 fold greater than those of the catalytic antibodies 7D4 (11) and 4D5 (12) previously elicited for the same reaction. DA_42_04 binds both the diene and the dienophile more tightly (significantly lower KM) than DA_20_10, but the kcat is 100-fold lower suggesting that the orientation of the two substrates relative to each other and/or to the catalytic groups is not optimal.
Figure 3. Kinetic Characterization.
(A) Dependence of reaction velocity for DA_20_10 on diene concentration for different fixed dienophile concentrations. The diene concentration was varied from 3.0 to 0.18 mM with a fixed concentration of 100 mM (◢), 66 mM (■), 44 mM (◆), 30 mM(▲), 20 mM(▼), or 13 mM (●) dienophile (B) Dependence of reaction velocity for DA_42_04 at varying diene concentrations for different fixed dienophile concentrations. The diene concentration was varied from 2.0 to 0.06 mM with a fixed concentration of 100 mM (◢), 50 mM (■), 25 mM (◆), 13 mM(▲), 7 mM(▼), or 3 mM (●) dienophile. Reaction conditions are described in the supplementary material.
At high substrate concentrations DA_20_10 proceeds for more than 30 turnovers with some loss of activity over time due to aggregation (16). At high enzyme concentrations more than 80% of the diene substrate is converted to product (Fig. S7). These properties suggest that de novo designed enzymes could be useful as catalysts in production level chemical syntheses.
Some Diels-Alder reactions can be accelerated by binding within a non specific hydrophobic pocket (13). This, however, does not appear to be the case for the reaction studied here: E. coli cell lysate, cyclodextrins, and BSA have either no effect or actually inhibit the reaction (Table S2). The importance of the active site binding geometry is highlighted by a comparison of DA_42_04 and DA_20_10: DA_42_04 binds the substrates much more tightly but has a much lower kcat. To further probe the sensitivity of DA_20_10 catalysis to the details of the active site geometry, each of the 15 residues constituting the active site was reverted one at a time to its identity in the original scaffold. Remarkably, nine of the reversions completely abolished activity; the other six mutations decreased activity by 1.5-fold to 10-fold (Fig. 2C, Table S5). The reversions that significantly reduced the catalytic activity of DA_20_10 are primarily in the core of the binding site, while mutations that had a less significant effect on activity are closer to the active site rim. Similar levels of sensitivity were observed for mutations that disrupt binding in DA_42_04 (16). Thus, while the catalytic efficiencies of the computationally designed Diels-Alderases are small in comparison with those of native enzymes, they exhibit similar levels of sensitivity to the details of the active site, and clearly provide much more than a general hydrophobic environment.
To determine how well the structure of DA_20_00 matched the design model, we solved the crystal structure of one of the active variants of DA_20_00 (harboring the A74I mutation; see Fig. 2B). The crystal structure solved to 1.5Å resolution (Table S4, Fig 2B) shows atomic level agreement with the design model, with an all-atom RMSD of 0.5Å. The major deviation between the crystal structure and the design model is in a surface loop, which appears to be pulled back from the predicted active site (RMSD on residues 32 to 46, 0.93Å). The conformations of the sidechains at the active site in the crystal structure are close to those in the design model; taken together with the reversion data described above, and the complete lack of activity observed for the starting scaffold (Fig S7), these results strongly suggest that the experimentally observed activity is generated by the designed active site.
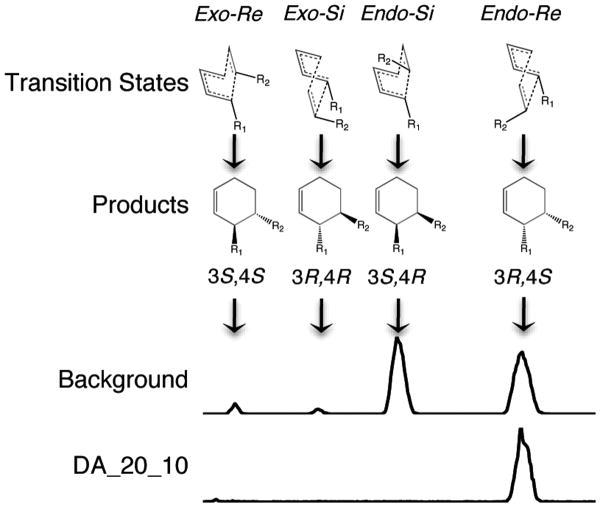
The Diels-Alder reaction studied here can, in principle, produce eight different isomeric products, four of which are experimentally observed in the reaction in solution (11). The computational design was directed at the transition state that yields the 3R,4S endo product, which only comprises 47% of the total product mixture formed in the uncatalyzed reaction. To determine the stereoselectivity of DA_20_10, a previously described liquid chromatography-tandem mass spectrometry assay with a chiral column was used (14). Consistent with the design, DA_20_10 only catalyzes the formation of the expected 3R,4S product (>97%, Fig. S8).
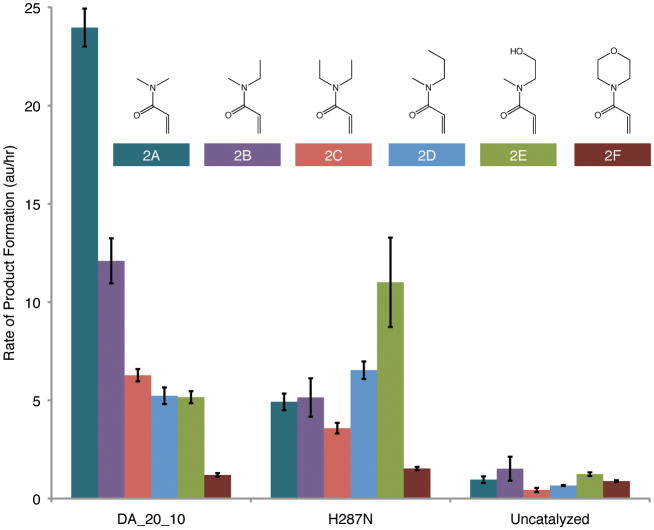
Besides stereoselectivity, the level of control over a chemical reaction by a designed enzyme is reflected by its substrate specificity. To investigate the substrate specificity of DA_20_10, we characterized product formation with six different dienophiles that share the same acrylamide core but have different nitrogen substituents (Fig. 5). The catalytic activity against each of the substrates was measured using a liquid-chromatography mass spectrometry assay (16). DA_20_10 was observed to strongly favor the substrate for which it was designed. Even slight changes, such as adding a methyl group to the N,N-dimethylacrylamide (Fig 5, 2A vs. 2B), significantly decreased the activity of DA_20_10, consistent with the tight packing of the active site around the two substrates.
Figure 5. Designed Control of Substrate Specificity.
Reactions were carried out with 0.2 mM diene 1 (Figure 1), and 10 mM of one of the six dienophiles depicted above in PBS at 298K in the absence or presence of 60 μM DA_20_10 or DA_20_10_H287N. The values in the figure are the mean (colored bars) and standard deviation (error bars) of four independent measurements of the product peak area (arbitrary units) formed per hour (16).
In addition to the ability to catalyze new reactions with high substrate specificity and stereoselectivity, one of the promises of de novo enzyme design is that once an initial active enzyme is engineered it can be easily modified to catalyze similar reactions with alternate substrates. To explore this possibility, we mutated histidine 287 on one side of the dienophile binding pocket in DA_20_10 to asparagine and several other residues. The H287N mutation has a substrate specificity profile different from DA_20_10; in particular there is a 13-fold switch in specificity for dienophile 2E relative to 2A, while the selectivity against 2F is maintained (Fig. 5). The specificity switch may have two origins: the histidine in the crystal structure clashes with the larger substrates, and the amino group on the asparagine can hydrogen bond with the hydroxyl in 2F.
While we have succeeded in computationally designing an enzyme that catalyzes an enantio- and diastereo- selective intermolecular reaction, there is clearly much room for improvement in our computational design methods. Only two of the fifty designed enzymes tested had measurable activity, and a much higher success rate and higher overall activities are clearly desirable. The differences in kcat in the two designed enzymes suggest that precise orientation of the two substrates relative to one another and to the catalytic residues is critical; more precise control over the designed ligand binding orientation is clearly desirable. On the experimental side, by analogy with our previous results with computationally designed Kemp eliminases (4), it should be possible to increase the activity of these enzymes by directed evolution.
The agreement between the designed and the experimentally observed substrate specificity and stereoselectivity of DA_20_10 is notable given the importance of selectivity in organic chemistry reactions. The capability to rationally control both substrate specificity and stereoselectivity via designed enzymes opens up new avenues of research in both basic and applied chemistry.
Supplementary Material
Figure 4. Absolute stereoselectivity of DA_20_10.
The transition states which lead to the four possible ortho-stereoisomers are shown above the reaction chromatograms. Background reaction: 2 mM diene and 70 mM dieneophile in a PBS solution for 24 hours at 298 K. DA_20_10 reaction: 50 μM protein, 0.5 mM diene, and 10 mM dieneophile in a PBS solution for 48 hours at 298K (16).
Acknowledgments
This work was supported by the Defense Advances Research Projects Agency (DARPA), the Howard Hughes Medical Institute (HHMI), a Molecular Biophysics traineeship from the National Institutes of Health for J.B.S, and the LLNL Lawrence Scholars program for G.K. We thank Dr. Miguel Toscano (ETH) and Carolyn Rosewall (UW) for chemical synthesis. Finally we would like to thank Dr. Brock Siegel and Dr. Y.-h. Lam for helpful comments on the manuscript. The x-ray crystallographic coordinates have been deposited in the Protein Data Bank with accession ID 3I1C.
References
- 1.Ose T, et al. Nature. 2003;422:185. doi: 10.1038/nature01454. [DOI] [PubMed] [Google Scholar]
- 2.Serafimov JM, Gillingham D, Kuster S, Hilvert D. Journal of the American Chemical Society. 2008;130:7798. doi: 10.1021/ja8017994. [DOI] [PubMed] [Google Scholar]
- 3.Kim SP, Leach AG, Houk KN. The Journal of Organic Chemistry. 2002;67:4250. doi: 10.1021/jo011180d. [DOI] [PubMed] [Google Scholar]
- 4.Rothlisberger D, et al. Nature. 2008 May 8;453:190. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 5.Jiang L, et al. Science. 2008 Mar 7;319:1387. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boger DL. Modern Organic Synthesis: Lecture Notes. TSRI Press; 1999. pp. 213–272. [Google Scholar]
- 7.Blake JF, Lim D, Jorgensen WL. The Journal of Organic Chemistry. 2002;59:803. [Google Scholar]
- 8.Page MI, Jencks WP. P Natl Acad Sci USA. 1971 August;68:1678. doi: 10.1073/pnas.68.8.1678. 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanghellini A, et al. Protein Sci. 2006 December 1;15:2785. doi: 10.1110/ps.062353106. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhlman B, Baker D. Proc Natl Acad Sci USA. 2000 Sep 12;97:10383. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouverneur VE, et al. Science. 1993 October 8;262:204. doi: 10.1126/science.8211138. 1993. [DOI] [PubMed] [Google Scholar]
- 12.Yli-Kauhaluoma JT, et al. Journal of the American Chemical Society. 1995;117:7041. [Google Scholar]
- 13.Breslow R, Dong SD. Chemical Reviews. 1998;98:1997. doi: 10.1021/cr970011j. [DOI] [PubMed] [Google Scholar]
- 14.Cannizzaro CE, Ashley JA, Janda KD, Houk KN. Journal of the American Chemical Society. 2003;125:2489. doi: 10.1021/ja020879d. [DOI] [PubMed] [Google Scholar]
- 15.W. L. DeLano. (DeLano Scientific LLC, Palo Alto, CA, 2008).
- 16.Supporting Online Material
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.