Abstract
The prediction of blood–brain barrier permeation is vitally important for the optimization of drugs targeting the central nervous system as well as for avoiding side effects of peripheral drugs. Following a previously proposed model on blood–brain barrier penetration, we calculated the cross-sectional area perpendicular to the amphiphilic axis. We obtained a high correlation between calculated and experimental cross-sectional area (r = 0.898, n = 32). Based on these results, we examined a correlation of the calculated cross-sectional area with blood–brain barrier penetration given by logBB values. We combined various literature data sets to form a large-scale logBB dataset with 362 experimental logBB values. Quantitative models were calculated using bootstrap validated multiple linear regression. Qualitative models were built by a bootstrapped random forest algorithm. Both methods found similar descriptors such as polar surface area, pKa, logP, charges and number of positive ionisable groups to be predictive for logBB. In contrast to our initial assumption, we were not able to obtain models with the cross-sectional area chosen as relevant parameter for both approaches. Comparing those two different techniques, qualitative random forest models are better suited for blood-brain barrier permeability prediction, especially when reducing the number of descriptors and using a large dataset. A random forest prediction system (ntrees = 5) based on only four descriptors yields a validated accuracy of 88%.
Electronic supplementary material
The online version of this article (doi:10.1007/s10822-011-9478-1) contains supplementary material, which is available to authorized users.
Keywords: Blood–brain barrier, Central nervous system, Membrane permeability, QSAR, LogBB, Random forest
Introduction
The blood–brain barrier (BBB) is a complex system, tightly regulating the transport from and to the central nervous system (CNS) [1]. It separates the systemic bloodstream from the CNS and is therefore important for drug diffusion and transport between them [2]. Drugs targeting the CNS need to be able to pass the BBB to reach their target [3]. In contrast, low BBB permeability reduces the chance of undesirable CNS-related side effects [4, 5]. Therefore an early estimation of BBB permeability would be highly valuable for drug design [6, 7]. The relevance of BBB permeability of therapeutic drugs has been reported in the context of numerous clinical dysfunctions, like dementia [8] and other clinical disorders [9–11].
The most common numeric value describing permeability across BBB is the logBB [12]. It is defined as logarithmic ratio between the concentration of a compound in brain and blood (Eq. 1).
| 1 |
Unfortunately, experiments to measure logBB are time-consuming, laborious and expensive in vitro [13–16] and even more in vivo [17, 18]. So it is not surprising that the number of published experimental values is limited. Experimental methods to assess BBB permeability range from artificial membranes and complex cell culture systems to in vivo methods. The PAMPA assay uses artificial membranes to observe passive (effective) membrane permeability, quantified by Peff [19, 20]. Obviously, those experiments are only able to observe permeability, neglecting the special characteristics of the BBB. Nevertheless, results from these studies support the validity of lipid bilayer systems as strongly simplified representations of the BBB. The main drawback of cell-free methods is that they neglect active transporters acting at the BBB and therefore incorrectly predict substrates to transport systems [21]. Numerous active transport systems and efflux transport systems play an important role at the BBB [22, 23]. One of the most commonly reported transport systems acting at the BBB is P-Glycoprotein (P-Gp) [24–26]. In contrast, in vivo methods, like in situ brain perfusion [17], are able to capture real BBB permeability as given by PS (permeability surface product) or logPS values [27].
Due to these experimental difficulties, it is not surprising, that BBB is frequently addressed via computational approaches. Computer-aided methods applied to this field of interest include multiple linear regression [28–32], bagged regression [33], partial least square analysis [34–37], support vector machines [38–40] and artificial neural networks [39, 41]. These methods are frequently combined with descriptor selection algorithms based on genetic algorithms to name only one [42, 43]. A comprehensive overview of previous models for BBB prediction has been published by Vastag and Keseru [44].
Depending on the size of the dataset, the number of descriptors, and the mathematical approach for prediction range from rough guidelines to quantitative predictions. Complex methods like partial least square analysis and artificial neural networks suffer from the drawback of being hard to interpret, whereas simple methods like multiple linear regression often yield less accurate results or even only rough guidelines [45]. Although different mathematical techniques make it hard to compare the results directly, the performance decreases strongly with larger datasets. High squared correlation coefficients above 0.85 are reported frequently for focused data sets with a size of approximately 50–90 compounds [31]. Predictions based on larger compound collections with a size of over 200 compounds resulted mainly in “rules of thumb” for good BBB permeability [45]. Altogether, these findings clearly show that there is still need for further research on BBB permeability [46].
Summarizing recent work, there is broad agreement on the importance of some molecular properties and descriptors which have been found in numerous publications to influence BBB permeability [45]:
The descriptor most frequently reported with BBB permeability is the polar surface area. The majority of publications report correlation of logBB with the polar surface [30, 47] or a property closely related to it [35]. The sum of oxygen and nitrogen atoms for example is extremely cheap in computation-time, but has still proven to be useful.
There is consensus that BBB permeability is also highly influenced by lipophilicity [48, 49]. One way to quantify lipophilicity is logP, the logarithmic partition coefficient between 1-octanol and water. However, the ability of logP to represent lipophilicity come under discussion recently [50], as octanol is a good hydrogen donor and therefore probably not a typical apolar solvent, even more when used as a calculated in silico descriptor [50, 51]. In addition to that, logP is defined for the neutralized state of a compound. LogP values for ionized (e.g. protonated) compounds are basically not defined [52]. Liu et al. [47] introduced ‘lipoaffinity’ as an easily-accessible descriptor. It is calculated by adding the contributions to the logP values of all but nitrogen and oxygen atoms.
Molecular flexibility has also been reported to influence BBB permeability. This is in agreement with the theory we used in this study (see below), since rigid molecules seem to fit less well to the membrane than more flexible ones (given that both molecules have approximately the same weight) [53]. A simple descriptor representing molecular flexibility would be the number of rotatable bonds, for example [29].
In this study we followed an approach based on physico-chemical properties to address permeation across the BBB, proposed by Fischer et al. [53]. According to this hypothesis, the process of integrating a compound into a membrane can be split into essential steps that can be added up to form the process of membrane permeation:
In the first step the compound needs to be desolvated from the aqueous environment. The process of desolvation is often addressed by molecular dynamics simulations [54]. Simultaneously, a cavity, appropriate for embedding the compound within the membrane is created. The amount of energy required to create this cavity is correlated to the energy needed to insert a molecule into the membrane. Fischer et al. [53] assume that the size of this cavity is crucial for membrane permeation.
In the second step the compound is inserted into the cavity. It is stabilized by electrostatic interactions with the polar headgroup of lipids and hydrophobic interactions with the core region of the lipid bilayer [55].
Finally, the compound needs to resolvate behind the lipid bilayer. This process is similar to the reversion of the solvation process.
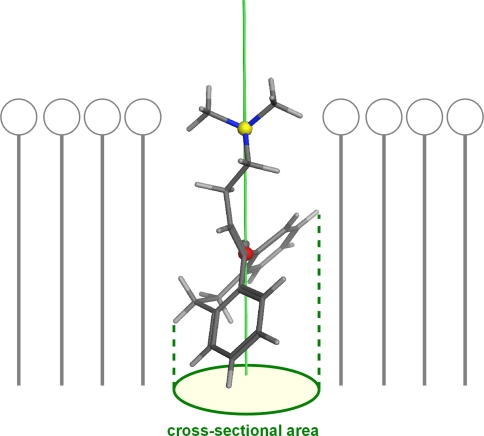
Figure 1 schematically illustrates how a molecule is inserted into a membrane according to this hypothesis. Based on this theory Gerebtzoff and Seelig [56] introduced the cross-sectional area (CSA) of a molecule as a novel descriptor to presumably represent BBB permeability. Because of its well-founded physico-chemical background it promised to achieve good predictability and interpretability, although this descriptor neglects all thermodynamic aspects of desolvation and resolvation. Descriptors based on valid mechanistic models have proven to contribute to the design and optimization of drug molecules [57]. Thus we reproduced this promising molecular descriptor and critically analysed its ability to predict BBB permeability. For this purpose, we compiled a large data set with experimental logBB values from numerous published datasets, instead of using single focused sets.
Fig. 1.
The cross-sectional area (CSA) has been introduced as a measure for the area occupied by a compound after insertion into a lipid membrane. Local polarity of the membrane determines the orientation of the ligand
Methods
Calculation of the CSA
We calculated the amphiphilic axis and CSA, as described in detail in Gerebtzoff and Seelig [56]. Modifications were introduced wherever the description was not clear or the results did not match our expectations. The following section describes and explains these modifications.
The amphiphilic axis is defined by the hydrophobic and hydrophilic center of a molecule. The hydrophilic center was calculated by averaging oxygen and nitrogen atom positions weighted by their contribution to the topological polar surface area (TPSA). Assuming that hydrogen bonds mainly influence BBB permeability [32], we decided to consider only nitrogen and oxygen as hydrophilic atoms and neglect sulphur atoms. The weighting factors were based on TPSA provided by MOE [58]. To emphasize the increased polar character of charged atoms compared to polarized atoms, we assigned a factor of 100 to charged atoms according to Eq. 2, where wf is the weighting factor, z is the charge and w0 is the weighting factor according to the TPSA.
| 2 |
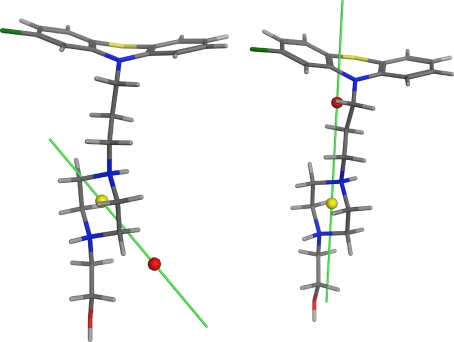
Halogen and carbon atoms were taken into account to place the hydrophobic center. Hydrophobic atom positions were weighted by their contribution to logP prediction by MOE (logP(o/w)) [59]. This fragment-based calculated logP suggests that halogen atoms have a large negative contribution to logP, which results in a displaced hydrophobic center for molecules containing halogen atoms. Thus we removed the logarithm before weighting to avoid negative contributions. Removal of the logarithm resulted in a more intuitive placement of the hydrophobic center (see Fig. 2.)
Fig. 2.
Comparison of two different strategies to calculate the hydrophobic center (red sphere) for compounds with halogen atoms (like perphenazine). On the left side, the hydrophobic center is calculated weighting atom positions by their contribution to logP prediction; on the right side the calculation is done with modifications presented in this study
According to the mechanism outlined by Fischer et al. [53], a molecule inserts into a membrane along the amphiphilic axis. The CSA reflects the area occupied by the molecule when projected to the plane perpendicular to the amphiphilic axis (see Figs. 1, 3). Projecting a molecule onto an area reduces computational efforts from 3D into 2D space, which dramatically increases the calculation speed for larger molecules in contrast to the published procedure [56].
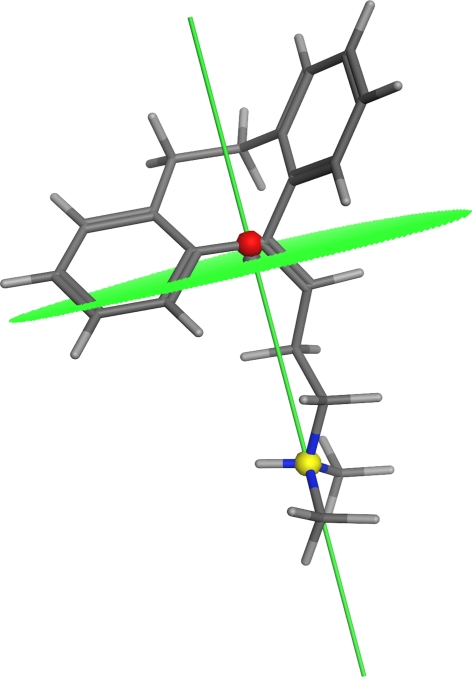
Fig. 3.
Amitriptyline with hydrophilic center (yellow sphere), hydrophobic center (red sphere), amphiphilic axis (green line) and CSA (green dotted area). This BBB-permeable compound illustrates the role of the amphiphilic axis and the CSA
Calculation of amphiphilic axis and CSA were performed with MOE [60] using its scripting language SVL (complete script is available as supplementary information). Partial charges were calculated using MMFF94x forcefield. Protonation states were assigned according to physiological pH of 7.4.
Experimental CSA values
To validate our CSA calculations, we compared our results with experimental CSA values [56]. We obtained all structures as SDF files from PubChem [61]. The reported dataset [56] consists of 32 compounds with experimental CSA values for pH 7.4 and 8. The experimental CSA at pH 7.4 was used, as it represents physiological pH. Carebastine was excluded from the set since the two CSA values for pH 8 and 7.4 differed significantly. We also removed beta-cyclodextrin from the dataset, since it is not a typical drug-like molecule with a molecular weight over 1,000 Da (see Lipinski’s rule-of-5 [62]). For each compounds the most stable conformation according to its conformational energy calculated by Omega (version 2.0) was used to calculate the CSA.
Experimental logBB values
In contrast to the small number of experimental CSA values, various studies containing experimental logBB values have been published. To investigate the ability of CSA for logBB prediction we combined all available published datasets and generated a novel large logBB dataset. Our dataset consisted of 195 compounds from Vilar et al. [63], 119 compounds from Platts et al. [64], 38 compounds from Naranayan and Gunturi [43], 94 compounds from Mente and Lombardo [33], 147 compounds from Zhang et al. [40], 197 compounds from Abraham et al. [65], 168 compounds from Garg and Verma [66], 106 compounds from Guerra et al. [67], 95 compounds from Rose et al. [68], 36 compounds from Kelder et al. [31], 165 compounds from Konovalov et al. [69] and 36 compounds from Zerara et al. [70]. Many compounds were reported multiple times with similar or identical logBB values, especially drugs with CNS-related effects such as antidepressants or neuroleptics. The average of the logBB values was used for identical compounds reported more than once. After removing duplicate structures, we ended up with 362 unique compounds with experimental logBB values ranging from −2.2 to +1.6. 199 logBB values were positive, 163 were negative or zero.
From this set we also wanted to exclude actively transported compounds, since their mechanism of passing the BBB is different to those passively entering CNS. Therefore we searched for substrates of P-Gp, one of the major transport systems acting at the BBB, in three previously published datasets [71–73]. Combining results from these sources we excluded 18 known substrates of P-Gp (bunitrolol, cimetidine, digoxin, domperidone, etoposide, fexofenadine, flunitrazepam, levodopa, loperamide, methotrexate, morphine, nevirapine, phenytoin, quinidine, risperidone, triflupromazine, vincristine, yamatetan). In addition to these 18 compounds, six compounds (chlorpromazine, doxorubicin, nelfinavir, saquinavir, verapamil, vinblastine) are reported ambiguously in the publications, thus we did not exclude them.
To the best of our knowledge, this is to date the largest set of quantitative logBB values, compiled from various resources. This dataset promises to be a very elaborate and refined selection of compounds. The complete dataset can be found in the supplementary material.
Descriptor calculation
We calculated all descriptors provided by MOE 2010.10 [59] and all from ACD/Labs (version 10.0) [74], that could be calculated for all compounds. A complete list of descriptors used is included the supplementary information. In addition, we calculated descriptors reported to be useful in other publications addressing BBB permeability, as far as we were able to reproduce them. Table 1 lists all additional descriptors together with a reference to their original publication. We also implemented size intensive descriptors using molecular weight as a normalizing factor [75]. Finally, our data set comprised over 880 descriptors, ranging from simple atom counts to computationally intensive quantum–mechanical properties.
Table 1.
List of molecular descriptors developed or reproduced in addition to the standard descriptors by ACD/Labs 10.0 and MOE 2010.10
| Descriptor | Description | Reference |
|---|---|---|
| AA | Length of the amphiphilic axis | [56] |
| CSA | Cross-sectional area perpendicular to the amphiphilic axis | [56] |
| LAA | Length along the amphiphilic axis | |
| NOOM | Number of atoms above the hydrophilic center | |
| VOOM | Volume above the hydrophilic center | |
| li | Longest distance from an ionized atom to another atom | |
| mpc | Longest distance from the atom with the highest partial charge | |
| n_COOH | Number of carboxylic acid functions | [76] |
| n_hal | Number of halogen atoms | |
| n_ion | Number of ionized atoms | |
| QMAXneg | Highest negative partial charge | [76] |
| QMAXpos | Highest positive partial charge | [76] |
| QSUMH | Sum of all partial charges on hydrogen atoms | |
| QSUMO | Sum of all partial charges on oxygen atoms | [76] |
| QSUMN | Sum of all partial charges on nitrogen atoms | |
| n_OpN | Sum of nitrogens and oxygen atoms | [35] |
| N_XpC | Sum of halogens and carbon atoms | |
| LA | Lipoaffinity | [47] |
| logP-NO | logP—number of oxygen and nitrogen atoms | [35] |
| I3 | +1 for amines, -1 for acids, otherwise 0 | [30] |
| n_PI | Number of positive ionizable groups | |
| QMAXneg | Highest negative partial charge | [76] |
| QMAXpos | Highest positive partial charge | [76] |
| QSUMH | Sum of all partial charges on hydrogen atoms | |
| QSUMO | Sum of all partial charges on oxygen atoms | [76] |
| QMINN | Lowest partial charge on nitrogen atoms | |
| QSUMN | Sum of all partial charges on nitrogen atoms | |
| QMEANN | Average partial charge in nitrogen atoms | |
| Qamines | Average partial charge on amines | |
| LA | Lipoaffinity | [47] |
| n_pol | Number of polar atoms | [77] |
| n_amines | Number of amines | |
| n_pN | Number of protonated nitrogen atoms at pH 7 |
Quantitative models: beam search and multiple linear regression
A large number of potentially predictive descriptors prompted us to systematically reduce dimensionality (the number of descriptors) used to construct and validate the models. A beam search algorithm (width = number of descriptors = 79) was applied to preselect potentially predictive descriptors [78]. For each combination a bootstrapped multiple linear regression was calculated and the squared correlation coefficient was returned as fitness criterion. We limited the maximum number of generations and subsequently the number of descriptors simultaneously taken into account to 10 and selected the best multiple linear regression model per generation.
Qualitative models: beam search and random forest
To generate qualitative models our dataset was split into BBB permeable (logBB ≥ 0.3, n = 126) and non-permeable (logBB ≤ −0.3, n = 76) compounds. The compounds between the two limits (n = 142) were excluded from the process, as they do not show strong characteristics of BBB permeable or non-permeable, respectively. These limits were adapted from Abraham et al. [65], who assume an experimental error of about 0.3 log units (logBB values range from −2.2 to 1.6). We then performed a beam search from 1 to 5 descriptors (width = number of descriptors = 72). As qualitative model we constructed a random forest model for each combination (ntree = 5, depth = 5), validated by a bootstrapping procedure (sample ratio = 1.0, number of validations = 100). Accuracy was used as the main performance criteria. Again, we captured the best models per generation.
All models were calculated using RapidMiner (version 5.1.1) and the Weka’s implementation of a random forest algorithm. Correlation coefficients were calculated according to Pearson.
Results and discussion
We calculated the CSA for 32 compounds and compared it with experimental values taken from Gerebztoff and Seelig [56]. Similar to the original work we also achieved a good correlation (r = 0.898) to experimental CSA values.
Quantitative models to predict logBB
The main intention of the present study was to investigate a correlation between CSA and BBB permeability, as suggested by Gerebtzoff and Seelig [56]. We therefore constructed multiple linear regression models using a beam search algorithm for feature selection (up to 10 descriptors).
Table 2 shows the squared correlation to increase with respect to the number of descriptors. Simultaneously, the validated squared correlation is constant or even decreases for more than 5 descriptors. Overall, statistical parameters improve only slightly from 5 to 10 descriptors, although the number of descriptors used is doubled. Thus influence of additional descriptors must be questioned. The validated squared correlation increases constantly up to 5 descriptors. So we consider 5 as the maximum number of descriptors to avoid overfitting.
Table 2.
Squared correlation coefficients (raw and bootstrap validated) of the best models with 1–10 descriptors constructed with beam search using multiple linear regression and squared correlation as performance criterion
| natts | Descriptor names | r2 | r2bootstrapping, 100 |
|---|---|---|---|
| 10 | si_TotalFormalCharge, prot_n_pol, neutral_n_pol, a_don, PEOE_VSA_POL, PDist, I3, chi1, logPow-logWeight, n_PI | 0.585 | 0.508 |
| 9 | si_TotalFormalCharge, prot_n_pol, neutral_n_pol, PEOE_VSA_PNEG, PDist, I3, apol, logPow-logWeight, n_PI | 0.577 | 0.534 |
| 8 | si_TotalFormalCharge, prot_n_pol, neutral_n_pol, a_don, PDist, chi1, logPow-logWeight, n_PI | 0.568 | 0.553 |
| 7 | prot_n_pol, neutral_n_pol, a_don, PDist, chi1, logPow-logWeight, n_PI | 0.558 | 0.537 |
| 6 | prot_n_pol, neutral_n_pol, a_acc, PDist, prot_logPow-logWeight, neutral_n_PI | 0.544 | 0.520 |
| 5 | prot_n_pol, neutral_n_pol, a_acc, logPow-logWeight, n_PI | 0.533 | 0.521 |
| 4 | TPSA, I3, logPow-logWeight, n_PI | 0.515 | 0.499 |
| 3 | prot_n_pol, neutral_n_pol, logPow-logWeight | 0.491 | 0.449 |
| 2 | TPSA, logPow-logWeight | 0.431 | 0.459 |
| 1 | TPSA | 0.354 | 0.221 |
In agreement with previous studies, TPSA is highly important for BBB permeability. The number of polar atoms (n_pol) and a descriptor taken from Feher et al. [30] (I3), followed by the number of positive ionisable groups (n_PI) and a descriptor developed for this study (PDist) were also found to influence BBB permeability, as well as the number of hydrogen bond acceptors (a_acc).
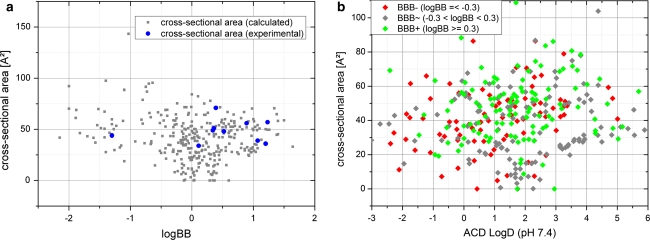
In contrast to our expectations, CSA never appeared in the most predictive models. This leads to the question why CSA does not contribute to BBB prediction as much as TPSA, for example. For BBB-permeable compounds Gerebtzoff and Seelig [56] suggest that there is an upper limit for CSA at 80 Ǻ². Figure 4a shows a scatterplot of logBB versus calculated CSA values for our large dataset, to further analyse this hypothesis. For 11 compounds, both experimental and calculated values for CSA and logBB were available. Overall, this plot does not show a clear correlation between CSA and logBB. As suggested in the original publication, we also investigated our dataset with respect to logD (at pH 7.4) and CSA. In contrast to the original publication Fig. 4b shows no significant separation by logD and CSA. A limit for BBB permeable compounds reflected by the CSA could not be determined.
Fig. 4.
a Experimental logBB plotted against 11 experimental and 362 calculated CSA show no correlation. Blue dots represent experimental CSA values, whereas grey dots are based on calculated CSA values. b Colour coded scatterplot of CSA versus LogD (at pH = 7.4), where green dots represent BBB permeable, red dots represent non-BBB permeable and gray dots represent unclassified compounds
Correlation between CSA and number of atoms
Searching for structural and chemical information covered by CSA, we tested its correlation with all other descriptors. Overall, various descriptors correlate remarkably well with the CSA. Table 3 lists the correlation with prominent other descriptors, including those from the models listed in Table 2. The majority of those are based on properties, easily obtainable from the structure. Remarkably, CSA is highly correlated to numerous simple descriptors that are easier to calculate, such as the number of atoms (see Fig. 5). A good correlation (r = 0.959) between those two properties suggest that CSA can be seen as derivative of the number of atoms. A high correlation of approximately 0.9 has also been found with the molecular weight. It was reported previously that molecular weight contributes to bioavailability in general [77]. Therefore we doubt that CSA provides more information with respect to BBB permeability than the number of atoms or molecular weight.
Table 3.
Various commonly-known descriptors correlate well with the CSA
| Descriptor | Correlation coefficient |
|---|---|
| a_count | 0.959 |
| b_count | 0.957 |
| apol | 0.955 |
| a_heavy | 0.929 |
| Weight | 0.897 |
| WeinerPath | 0.877 |
| mpc | 0.710 |
| n_pol | 0.645 |
| TPSA | 0.590 |
| I3 | 0.317 |
| n_PI | 0.264 |
CSA may therefore be regarded as an expensive replacement for much simpler descriptors. All descriptors are either standard MOE descriptors or explained in Table 1
Fig. 5.
CSA plotted against the number of atoms (a_count) reveals a remarkably high correlation (r = 0.959)
Number of descriptors, dataset size and accuracy
Whenever we tried to construct multiple linear regression models for logBB prediction on our large dataset, we failed to achieve results comparable to those reported by others using smaller data sets. To benchmark this relationship, linear regression models were built based on a single descriptor, but varying the composition of the training set. As descriptor TPSA was chosen, since its impact on BBB permeability has not only been demonstrated by the models presented here but also by other researchers, for example by Kelder et al. [31]. In their study a set of 45 compounds was used to construct a regression model. Similarly, we constructed subsets from our dataset consisting of 50–350 compounds and calculated the squared correlation coefficient for each model. Each subset size was tested 500 times using different random seeds to cover different selection of compounds. Figure 6 illustrates that small sets show a large variability with respect to the squared correlation. Although the number of possible subsets is much lower for the large subsets, those are less likely to suffer from arbitrary correlations. This underlines the need for large datasets like the one we present here.
Fig. 6.
Different training sets with 50–350 examples all selected from our dataset (n = 362) show that the size of the training set highly influences the performance given by squared correlation, even when constructed with exactly the same descriptor (TPSA) and the same procedure
Qualitative models
Focusing on a small number of descriptors, we were not able to obtain simple models with high performance using quantitative techniques. Thus we also calculated qualitative models to predict BBB permeability using a random forest. Again, we could compare our results to various published studies [28, 38, 56].
For the qualitative models we used the same dataset as for the quantitative models, but converted logBB values into three bins. Compounds with a logBB ≥ 0.3 comprise the set of BBB permeable compounds, whereas compounds with a logBB ≤ 0.3 are considered as not BBB permeable. The remaining compounds are excluded from the qualitative modelling. This left us with a set of 202 compounds for the training set. From the initial set of 886 descriptors, only 72 descriptors remained after preselection. Similar to the quantitative approach we aimed to obtain simple and interpretable models with a maximum of 4 concurrent descriptors. The beam search returned one model without misclassification (accuracy = 1.00) using four descriptors. To evaluate the robustness of this model a bootstrap validation (n = 100) was applied. The complete results are shown in Table 4.
Table 4.
Accuracies (raw and validated) of the best models and prediction systems with up to 4 descriptors constructed using a beam feature search in combination with random forest learners
| natts | Descriptor names | Accuracytrees=5 | Accuracybs, n=100 |
|---|---|---|---|
| 4 | TPSA, I3, QSUMN, QSUMN/Weight | 1.000 | 0.878 |
| 3 | TPSA, QSUMN, QSUMN/Weight | 0.985 | 0.868 |
| 2 | TPSA, QSUMN | 0.970 | 0.864 |
| 1 | TPSA | 0.926 | 0.834 |
The random forest prediction system based on four descriptors also achieves a high validated accuracy. The selected descriptors are similar to those obtained by multiple linear regressions and therefore highlight the importance of the following basic molecular properties:
TPSA was selected in all models.
QSUMN is the sum of charges on nitrogen atoms. This classifies all compounds by their charge on nitrogen atoms and subsequently also discriminates compounds having no nitrogen atom at all.
QSUMN/Weight represents a size-intensive descriptor calculated from QSUMN and the molecular weight. For large compounds the molecular weight is the dominating factor for this descriptor.
I3: Is −1 for acid compounds, +1 for basic compounds and 0 for the remaining compounds.
To analyse the dependence of the four descriptors we also calculated the intercorrelation matrix (Table 5). Although QSUMN and QSUMN/Weight are highly correlated, both seem be predictive for logBB. Especially compounds with higher molecular weight differ considerably for the two descriptors.
Table 5.
Intercorrelation matrix for the four descriptors used in the best random forest prediction model
| TPSA | I3 | QSUMN | QSUMN/Weight | |
|---|---|---|---|---|
| TPSA | 1.000 | |||
| I3 | 0.054 | 1.000 | ||
| QSUMN | −0.617 | −0.492 | 1.000 | |
| QSUMN/Weight | −0.374 | −0.536 | 0.790 | 1.000 |
Table 6 compares the results from our calculations with results from other publications. The results clearly show that random forest prediction systems are well-suited to classify BBB permeability. Altogether, we outperformed many other models trained on datasets with similar sizes in terms of validated accuracy, even using fewer molecular properties.
Table 6.
Results of classification systems for BBB permeability taken from the literature
| Method | Validation | Number of descriptors | Dataset size | Overall accuracy | Reference |
|---|---|---|---|---|---|
| SVM | 10-fold CV | <100 | 351 | 83.0 | [28] |
| DT | – | 2 | 43 | 86.0 | [56] |
| SVM | 10-fold CV | 8 | 351 | 80.0 | [28] |
| SVM | 5-fold CV | 5 | 415 | 79.1 | [38] |
| RF | 100 BS | 4 | 202 | 88.2 | This work |
SVM support vector machine, DT decision tree, RF random forest, CV cross-validation, BS bootstrapping
In contrast to results from quantitative models, qualitative classification models are able to predict BBB permeability with high accuracy, especially when aiming for simple models based on a small number of descriptors. To quantify BBB permeability a more sophisticated and complex model is needed. However, we have shown that the number of descriptors that can be used is limited when looking at validated performances. Using a high number of descriptors for small datasets bears the risk of overfitting and arbitrary correlations.
In contrast, we focused on a simple prediction system that links BBB permeability to easily understandable molecular properties. Focusing on a small number of descriptors it might be easier to construct a binary classifier than to quantitatively predict BBB permeability.
Strengths and limitations
In the present study there are several novel findings:
In addition to well-known descriptors, we added a significant number of descriptors that have never been evaluated and validated in the context of BBB prediction, for example size intensive descriptors (explained in [75]), and other novel descriptors listed in Table 1. Furthermore, we addressed the CSA which has been proposed as being predictive for BBB permeability. The qualitative models as shown in Table 4 include, in addition to TPSA, two of these novel descriptors.
All prediction systems are limited by the experimental error of the data they are based on. Therefore, our set consists of compounds with experimental logBB values only, compiled from various publications.
We developed an unparalleled compact and highly-predictive qualitative model validated by bootstrapping, that might act as general guideline for estimating BBB permeability.
Conclusion
In this work, we applied qualitative and quantitative in silico techniques to predict BBB permeability. For this purpose we created a reasonably large dataset (n = 362) of experimental logBB values. For each compound of the training set we calculated a broad set of descriptors ranging from simple atom count descriptors to computational more expensive descriptors like the CSA perdendicular to the amphiphilic axis. For this special descriptor were also able to validate calculated CSA with a set of experimentally measured values (n = 32).
The best quantitative prediction system based on multiple linear regression without overfitting yielded a bootstrapped squared correlation coefficient of 0.521. Qualitative models based on a random forest performed remarkably better. The best prediction system based on only four descriptors achieved a bootstrap validated accuracy of 88% (unvalidated 100%). Remarkably, the CSA was not chosen by the feature selection algorithm used to select the most predictive descriptors. In contrast, a combination of simple and well-known descriptors was found to be most useful to predict logBB.
Finally, we also showed that large and carefully comprised datasets, like the one presented here, reduce the risk of arbitrary correlations and result in more reproducible and robust models.
Support information
The SVL script to calculate and visualize the CSA perpendicular to the amphiphilic axis is provided as well as a spreadsheet file containing the whole set of compounds together with their corresponding logBB as well as a complete list of the descriptors calculated by ACD/Labs 10.0 and MOE 2010.10 for free download.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the German Ministry of Education and Research (BMBF Grant No. 01EX1015B).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- BBB
Blood-brain barrier
- CNS
Central nervous system
- CSA
Cross-sectional area
- LogBB
Logarithmic ratio between concentration of a compound in brain and blood
- P-Gp
P-glycoprotein transporter
- TPSA
Topological polar surface area
References
- 1.Bradbury MWB. The blood-brain barrier. Exp Physiol. 1993;78(4):453–472. doi: 10.1113/expphysiol.1993.sp003698. [DOI] [PubMed] [Google Scholar]
- 2.Wager TT, Chandrasekaran RY, Hou X, Troutman MD, Verhoest PR, Villalobos A, Will Y. Defining desirable central nervous system drug space through the alignment of molecular properties, in vitro ADME, and safety attributes. ACS Chem Neurosci. 2010;1(6):420–434. doi: 10.1021/cn100007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen PA, Andersson O, Hansen SH, Simonsen KB, Andersson G. Models for predicting blood-brain barrier permeation. Drug Discov Today. 2011;16(11–12):472–475. doi: 10.1016/j.drudis.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Lacombe O, Guyol AC, Videau O, Pruvost A, Bolze S, Prevost C, Mabondzo A. Brain penetration predictivity using in vitro primary rat and human cell-based blood-brain barrier models for drug discovery and development. Fundam Clin Pharmacol. 2010;24:8. [Google Scholar]
- 5.Muster W, Breidenbach A, Fischer H, Kirchner S, Müller L, Pähler A. Computational toxicology in drug development. Drug Discov Today. 2008;13(7–8):303–310. doi: 10.1016/j.drudis.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Pardridge WM. Blood-brain barrier delivery. Drug Discov Today. 2007;12(1–2):54–61. doi: 10.1016/j.drudis.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Stenehjem DD, Hartz AM, Bauer B, Anderson GW. Novel and emerging strategies in drug delivery for overcoming the blood brain barrier. Futur Med Chem. 2009;1(9):1623–1641. doi: 10.4155/fmc.09.137. [DOI] [PubMed] [Google Scholar]
- 8.Popescu BO. Pathological changes of the blood-brain barrier in dementias. J Neurol Sci. 2009;283(1–2):250. doi: 10.1016/j.jns.2009.02.045. [DOI] [PubMed] [Google Scholar]
- 9.Banks WA. Developing drugs that can cross the blood-brain barrier: applications to Alzheimer’s disease. BMC Neurosci. 2008;9(Suppl 3):S2. doi: 10.1186/1471-2202-9-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.HEd V, Kuiper J, AGd B, Berkel TJCV, Breimer DD. The blood-brain barrier in neuroinflammatory diseases. Pharmacol Rev. 1997;49(2):143–156. [PubMed] [Google Scholar]
- 11.Jong A, Huang S-H. Blood-brain barrier drug discovery for central nervous system infections. Curr Drug Targ Infect Disord. 2005;5:65–72. doi: 10.2174/1568005053174672. [DOI] [PubMed] [Google Scholar]
- 12.Bickel U. How to measure drug transport across the blood-brain barrier. NeuroRX. 2005;2(1):15–26. doi: 10.1602/neurorx.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehouck M-P, Méresse S, Delorme P, Fruchart J-C, Cecchelli R. An easier, reproducible, and mass-production method to study the blood-brain barrier in vitro. J Neurochem. 1990;54(5):1798–1801. doi: 10.1111/j.1471-4159.1990.tb01236.x. [DOI] [PubMed] [Google Scholar]
- 14.Fricker G (2008) In vitro models to study blood-brain barrier function. In: Ehrhardt C, Kim KJ (ed) Drug absorption studies: in situ, in vitro and in silico models, vol VII. Biotechnology: pharmaceutical aspects. Springer, US, pp 397–417. doi:10.1007/978-0-387-74901-3_17
- 15.Lacombe O, Videau O, Chevillon D, Guyot A-C, Contreras C, Blondel S, Nicolas L, Ghettas A, Benech H, Thevenot E, Pruvost A, Bolze S, Krzaczkowski L, Prevost C, As M. In vitro primary human and animal cell-based blood-brain barrier models as a screening tool in drug discovery. Mol Pharm. 2011;8(3):651–663. doi: 10.1021/mp1004614. [DOI] [PubMed] [Google Scholar]
- 16.Terasaki T, Hosoya K-I. Conditionally immortalized cell lines as a new in vitro model for the study of barrier functions. Biol Pharm Bull. 2001;24(2):111–118. doi: 10.1248/bpb.24.111. [DOI] [PubMed] [Google Scholar]
- 17.Boje KMK (2001) In vivo measurement of blood-brain barrier permeability. In: Current protocols in neuroscience. Wiley. doi:10.1002/0471142301.ns0719s15 [DOI] [PubMed]
- 18.Di L, Kerns EH, Bezar IF, Petusky SL, Huang YP. Comparison of blood-brain barrier permeability assays: in situ brain perfusion, MDR1-MDCKII and PAMPA-BBB. J Pharm Sci. 2009;98(6):1980–1991. doi: 10.1002/jps.21580. [DOI] [PubMed] [Google Scholar]
- 19.Mensch J, LJ L, Sanderson W, Melis A, Mackie C, Verreck G, Brewster ME, Augustijns P. Application of PAMPA-models to predict BBB permeability including efflux ratio, plasma protein binding and physicochemical parameters. Int J Pharm. 2010;395(1–2):182–197. doi: 10.1016/j.ijpharm.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 20.Reichel A, Begley DJ. Potential of immobilized artificial membranes for predicting drug penetration across the blood-brain barrier. Pharm Res. 1998;15(8):1270–1274. doi: 10.1023/A:1011904311149. [DOI] [PubMed] [Google Scholar]
- 21.Adenot M, Lahana R. Blood-brain barrier permeation models: discriminating between potential CNS and non-CNS drugs including P-glycoprotein substrates. J Chem Inf Comput Sci. 2004;44(1):239–248. doi: 10.1021/ci034205d. [DOI] [PubMed] [Google Scholar]
- 22.de Boer AG, van der Sandt ICJ, Gaillard PJ. The role of drug transporters at the blood-brain barrier. Annu Rev Pharmacol Toxicol. 2003;43:629–656. doi: 10.1146/annurev.pharmtox.43.100901.140204. [DOI] [PubMed] [Google Scholar]
- 23.de Lange ECM. Potential role of ABC transporters as a detoxification system at the blood-CSF barrier. Adv Drug Deliv Rev. 2004;56(12):1793–1809. doi: 10.1016/j.addr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Miller DS. Regulation of P-glycoprotein and other ABC drug transporters at the blood-brain barrier. Trends Pharmacol Sci. 2010;31(6):246–254. doi: 10.1016/j.tips.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer B, Hartz AMS, Fricker G, Miller DS. Modulation of p-glycoprotein transport function at the blood-brain barrier. Exp Biol Med. 2005;230(2):118–127. doi: 10.1177/153537020523000206. [DOI] [PubMed] [Google Scholar]
- 26.Schinkel AH. P-Glycoprotein, a gatekeeper in the blood-brain barrier. Adv Drug Deliv Rev. 1999;36(2–3):179–194. doi: 10.1016/S0169-409X(98)00085-4. [DOI] [PubMed] [Google Scholar]
- 27.Gratton JA, Abraham MH, Bradbury MW, Chadha HS. Molecular factors influencing drug transfer across the blood-brain barrier. J Pharm Pharmacol. 1997;49(12):1211–1216. doi: 10.1111/j.2042-7158.1997.tb06072.x. [DOI] [PubMed] [Google Scholar]
- 28.Kortagere S, Chekmarev D, Welsh WJ, Ekins S. New predictive models for blood-brain barrier permeability of drug-like molecules. Pharm Res. 2008;25(8):1836–1845. doi: 10.1007/s11095-008-9584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutter MC. Prediction of blood-brain barrier permeation using quantum chemically derived information. J Comput Aided Mol Des. 2003;17(7):415–433. doi: 10.1023/A:1027359714663. [DOI] [PubMed] [Google Scholar]
- 30.Feher M, Sourial E, Schmidt JM. A simple model for the prediction of blood-brain partitioning. Int J Pharm. 2000;201(2):239–247. doi: 10.1016/S0378-5173(00)00422-1. [DOI] [PubMed] [Google Scholar]
- 31.Kelder J, Grootenhuis PDJ, Bayada DM, Delbressine LPC, Ploemen J-P. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res. 1999;16(10):1514–1519. doi: 10.1023/A:1015040217741. [DOI] [PubMed] [Google Scholar]
- 32.Abraham MH, Chadha HS, Mitchell RC. Hydrogen bonding. 33. Factors that influence the distribution of solutes between blood and brain. J Pharm Sci. 1994;83(9):1257–1268. doi: 10.1002/jps.2600830915. [DOI] [PubMed] [Google Scholar]
- 33.Mente SR, Lombardo F. A recursive-partitioning model for blood–brain barrier permeation. J Comput Aided Mol Des. 2005;19(7):465–481. doi: 10.1007/s10822-005-9001-7. [DOI] [PubMed] [Google Scholar]
- 34.Bendels S, Kansy M, Wagner B, Huwyler J. In silico prediction of brain and CSF permeation of small molecules using PLS regression models. Eur J Med Chem. 2008;43(8):1581–1592. doi: 10.1016/j.ejmech.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Norinder U, Haeberlein M. Computational approaches to the prediction of the blood-brain distribution. Adv Drug Deliv Rev. 2002;54(3):291–313. doi: 10.1016/S0169-409X(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 36.Luco JM. Prediction of the brain − blood distribution of a large set of drugs from structurally derived descriptors using partial least-squares (PLS) modeling. J Chem Inf Comput Sci. 1999;39(2):396–404. doi: 10.1021/ci980411n. [DOI] [PubMed] [Google Scholar]
- 37.Crivori P, Cruciani G, Carrupt PA, Testa B. Predicting blood-brain barrier permeation from three-dimensional molecular structure. J Med Chem. 2000;43(11):2204–2216. doi: 10.1021/jm990968+. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Yap CW, Ung CY, Xue Y, Cao ZW, Chen YZ. Effect of selection of molecular descriptors on the prediction of blood-brain barrier penetrating and nonpenetrating agents by statistical learning methods. J Chem Inf Model. 2005;45(5):1376–1384. doi: 10.1021/ci050135u. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Yan A, Yuan Q. Classification of blood-brain barrier permeation by Kohonen’s self-organizing neural network (KohNN) and support vector machine (SVM) QSAR Comb Sci. 2009;28(9):989–994. doi: 10.1002/qsar.200960008. [DOI] [Google Scholar]
- 40.Zhang L, Zhu H, Oprea TI, Golbraikh A, Tropsha A. QSAR modeling of the blood-brain barrier permeability for diverse organic compounds. Pharm Res. 2008;25(8):1902–1914. doi: 10.1007/s11095-008-9609-0. [DOI] [PubMed] [Google Scholar]
- 41.Hemmateenejad B, Miri R, Safarpour MA, Mehdipour AR. Accurate prediction of the blood-brain partitioning of a large set of solutes using ab initio calculations and genetic neural network modeling. J Comput Chem. 2006;27(11):1125–1135. doi: 10.1002/jcc.20437. [DOI] [PubMed] [Google Scholar]
- 42.Teixido M, Belda I, Rosello X, Gonzalez S, Fabre M, Llora X, Bacardit J, Garrell JM, Vilaro S, Albericio F, Giralt E. Development of a genetic algorithm to design and identify peptides that can cross the blood-brain barrier 1. Design and validation in silico. QSAR Comb Sci. 2003;22(7):745–753. doi: 10.1002/qsar.200320004. [DOI] [Google Scholar]
- 43.Narayanan R, Gunturi SB. In silico ADME modelling: prediction models for blood-brain barrier permeation using a systematic variable selection method. Bioorgan Med Chem. 2005;13(8):3017–3028. doi: 10.1016/j.bmc.2005.01.061. [DOI] [PubMed] [Google Scholar]
- 44.Vastag M, Keserü GM. Current in vitro and in silico models of blood-brain barrier penetration: a practical view. Curr Opin Drug Discov Dev. 2009;12(1):115–124. [PubMed] [Google Scholar]
- 45.Gleeson MP. Generation of a set of simple, interpretable ADMET rules of thumb. J Med Chem. 2008;51(4):817–834. doi: 10.1021/jm701122q. [DOI] [PubMed] [Google Scholar]
- 46.Pardridge WM. Blood-brain barrier drug targeting: the future of brain drug developement. Mol Interv. 2003;3(2):90–105. doi: 10.1124/mi.3.2.90. [DOI] [PubMed] [Google Scholar]
- 47.Liu R, Sun H, So S–S. Development of quantitative structure − property relationship models for early ADME evaluation in drug discovery. 2. Blood-brain barrier penetration. J Chem Inf Comput Sci. 2001;41(6):1623–1632. doi: 10.1021/ci010290i. [DOI] [PubMed] [Google Scholar]
- 48.Waring MJ. Defining optimum lipophilicity and molecular weight ranges for drug candidates–Molecular weight dependent lower logD limits based on permeability. Bioorg Med Chem Lett. 2009;19(10):2844–2851. doi: 10.1016/j.bmcl.2009.03.109. [DOI] [PubMed] [Google Scholar]
- 49.Liu X, Testa B, Fahr A. Lipophilicity and its relationship with passive drug permeation. Pharm Res. 2011;28(5):962–977. doi: 10.1007/s11095-010-0303-7. [DOI] [PubMed] [Google Scholar]
- 50.Kramer C, Beck B, Clark T. A surface-integral model for log POW. J Chem Inf Model. 2010;50(3):429–436. doi: 10.1021/ci900431f. [DOI] [PubMed] [Google Scholar]
- 51.Muehlbacher M, Kerdawy AE, Kramer C, Hudson B, Clark T. Conformation-dependent QSPR models: logPOW. J Chem Inf Model. 2011;51(9):2408–2416. doi: 10.1021/ci200276v. [DOI] [PubMed] [Google Scholar]
- 52.Abraham MH. The permeation of neutral molecules, ions, and ionic species through membranes: brain permeation as an example. J Pharm Sci. 2010;100(5):1690–1701. doi: 10.1002/jps.22404. [DOI] [PubMed] [Google Scholar]
- 53.Fischer H, Gottschlich R, Seelig A. Blood-brain barrier permeation: molecular parameters governing passive diffusion. J Membr Biol. 1998;165(3):201–211. doi: 10.1007/s002329900434. [DOI] [PubMed] [Google Scholar]
- 54.Keseru GM, Molnar L. High-throughput prediction of blood − brain partitioning: a thermodynamic approach. J Chem Inf Comput Sci. 2000;41(1):120–128. doi: 10.1021/ci000043z. [DOI] [PubMed] [Google Scholar]
- 55.Seelig A. The role of size and charge for blood-brain barrier permeation of drugs and fatty acids. J Mol Neurosci. 2007;33(1):32–41. doi: 10.1007/s12031-007-0055-y. [DOI] [PubMed] [Google Scholar]
- 56.Gerebtzoff G, Seelig A. In silico prediction of blood-brain barrier permeation using the calculated molecular cross-sectional area as main parameter. J Chem Inf Model. 2006;46(6):2638–2650. doi: 10.1021/ci0600814. [DOI] [PubMed] [Google Scholar]
- 57.Hughes JD, Blagg J, Price DA, Bailey S, DeCrescenzo GA, Devraj RV, Ellsworth E, Fobian YM, Gibbs ME, Gilles RW, Greene N, Huang E, Krieger-Burke T, Loesel J, Wager T, Whiteley L, Zhang Y. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg Med Chem Lett. 2008;18(17):4872–4875. doi: 10.1016/j.bmcl.2008.07.071. [DOI] [PubMed] [Google Scholar]
- 58.Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem. 2000;43(20):3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- 59.Labute P. A widely applicable set of descriptors. J Mol Graph Model. 2000;18(4–5):464–477. doi: 10.1016/S1093-3263(00)00068-1. [DOI] [PubMed] [Google Scholar]
- 60.Molecular Operating Environment MOE 2010.10 (2010)
- 61.Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov Today. 2010;15(23–24):1052–1057. doi: 10.1016/j.drudis.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 63.Vilar S, Chakrabarti M, Costanzi S. Prediction of passive blood-brain partitioning: straightforward and effective classification models based on in silico derived physicochemical descriptors. J Mol Graph Model. 2010;28(8):899–903. doi: 10.1016/j.jmgm.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Platts JA, Abraham MH, Zhao YH, Hersey A, Ijaz L, Butina D. Correlation and prediction of a large blood-brain distribution data set—an LFER study. Eur J Med Chem. 2001;36(9):719–730. doi: 10.1016/S0223-5234(01)01269-7. [DOI] [PubMed] [Google Scholar]
- 65.Abraham MH, Ibrahim A, Zhao Y, Acree WE. A data base for partition of volatile organic compounds and drugs from blood/plasma/serum to brain, and an LFER analysis of the data. J Pharm Sci. 2006;95(10):2091–2100. doi: 10.1002/jps.20595. [DOI] [PubMed] [Google Scholar]
- 66.Garg P, Verma J. In silico prediction of blood brain barrier permeability: an artificial neural network model. J Chem Inf Model. 2006;46(1):289–297. doi: 10.1021/ci050303i. [DOI] [PubMed] [Google Scholar]
- 67.Guerra A, Páez JA, Campillo NE. Artificial neural networks in ADMET modeling: prediction of blood-brain barrier permeation. QSAR Comb Sci. 2008;27(5):586–594. doi: 10.1002/qsar.200710019. [DOI] [Google Scholar]
- 68.Rose K, Hall LH, Kier LB. Modeling blood-brain barrier partitioning using the electrotopological state. J Chem Inf Comput Sci. 2002;42(3):651–666. doi: 10.1021/ci010127n. [DOI] [PubMed] [Google Scholar]
- 69.Konovalov DA, Coomans D, Deconinck E, Vander Heyden Y. Benchmarking of QSAR models for blood-brain barrier permeation. J Chem Inf Model. 2007;47(4):1648–1656. doi: 10.1021/ci700100f. [DOI] [PubMed] [Google Scholar]
- 70.Zerara M, Brickmann J, Kretschmer R, Exner TE. Parameterization of an empirical model for the prediction of n-octanol, alkane and cyclohexane/water as well as brain/blood partition coefficients. J Comput Aided Mol Des. 2009;23(2):105–111. doi: 10.1007/s10822-008-9243-2. [DOI] [PubMed] [Google Scholar]
- 71.Xue Y, Yap CW, Sun LZ, Cao ZW, Wang JF, Chen YZ. Prediction of P-glycoprotein substrates by a support vector machine approach. J Chem Inf Comput Sci. 2004;44(4):1497–1505. doi: 10.1021/ci049971e. [DOI] [PubMed] [Google Scholar]
- 72.Wang Y-H, Li Y, Yang S-L, Yang L. Classification of substrates and inhibitors of P-glycoprotein using unsupervised machine learning approach. J Chem Inf Model. 2005;45(3):750–757. doi: 10.1021/ci050041k. [DOI] [PubMed] [Google Scholar]
- 73.Varma MVS, Sateesh K, Panchagnula R. Functional role of P-glycoprotein in limiting intestinal absorption of drugs: contribution of passive permeability to P-glycoprotein mediated efflux transport. Mol Pharm. 2004;2(1):12–21. doi: 10.1021/mp0499196. [DOI] [PubMed] [Google Scholar]
- 74.Advanced Chemistry Development Inc (2010) ACD/PhysChem. version 10.0 edn, Toronto
- 75.Purvis G. Size-intensive descriptors. J Comput Aided Mol Des. 2008;22(6):461–468. doi: 10.1007/s10822-008-9209-4. [DOI] [PubMed] [Google Scholar]
- 76.Andres C, Hutter MC. CNS permeability of drugs predicted by a decision tree. QSAR Comb Sci. 2006;25(4):305–309. doi: 10.1002/qsar.200510200. [DOI] [Google Scholar]
- 77.Fu X-C, Wang G-P, Shan H-L, Liang W-Q, Gao J-Q. Predicting blood-brain barrier penetration from molecular weight and number of polar atoms. Eur J Pharm Biopharm. 2008;70(2):462–466. doi: 10.1016/j.ejpb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 78.Furcy D, Koenig S (2005) Limited discrepancy beam search. Paper presented at the Proceedings of the 19th international joint conference on Artificial intelligence, Edinburgh, Scotland
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.