Summary of recent advances
Multiphoton imaging is a promising approach for addressing current issues in systems biology and high-content investigation of embryonic development. Recent advances in multiphoton microscopy, including light-sheet illumination, optimized laser scanning, adaptive and label-free strategies, open new and promising opportunities for embryo imaging. However, the literature is often unclear about which microscopy technique is most adapted for achieving specific experimental goals. In this review, we describe and discuss the key concepts of imaging speed, imaging depth, photodamage, and nonlinear contrast mechanisms in the context of recent advances in live embryo imaging. We illustrate the potentials of these new imaging approaches with a selection of recent applications in developmental biology.
Keywords: Nonlinear microscopy; Light-sheet microscopy; 2-photon excited fluorescence; Second-harmonic generation (SHG); Third-harmonic generation (THG); Coherent raman scattering (CARS, SRS)
Introduction
From a microscopy perspective, live embryos present uniquely challenging characteristics compared to other biological samples. Embryos are smaller than 1 millimeter, at least during early developmental stages, making them accessible for three-dimensional (3D) imaging with light microscopy. However, they typically have an ellipsoidal shape and their inner structure is inhomogeneous and constantly changing. In addition, embryos are sensitive to manipulation and photodamage, and their labeling can be difficult. These properties challenge the performance of microscopy techniques in terms of imaging depth, imaging speed, photodamage and contrast. Since its introduction in 1990 [1], 2-photon excited fluorescence (2PEF) microscopy has proven to be the most effective approach for deep tissue fluorescence microscopy. It has found many applications in neuroscience [2–3] and more recently in other fields, such as in immunology [4]. Multiphoton (or nonlinear) imaging is attractive also for embryo imaging and in recent years has been applied to an increasing number of published studies in developmental biology using various model systems, such as fruit fly [5–8], quail [9], zebrafish [10], or mouse embryos [11–12]. Multiphoton imaging is also promising for addressing current issues in systems biology and high-content experimental investigation of embryonic development [13] requiring novel methods for faster and deeper imaging of embryos with better contrast and resolution. In this review we analyze the parameters limiting imaging speed and depth in the currently available imaging modalities, and we discuss promising recent advances in multiphoton microscopy of live embryos, including light-sheet excitation and label-free imaging.
Fast imaging of live embryos with multiphoton light-sheet microscopy
Imaging developmental processes often requires time-lapse 3D-image acquisitions (4D imaging). The imaging speed of a microscope can be defined by its pixel (or voxel) rate, i.e. the number of pixels per unit time that can be obtained with sufficient signal and contrast. A high pixel rate permits capturing with adequate time resolution fast processes such as heart development (50–130 frames per second (fps) in [14–16]), cilia beating (900 fps in [17]) or fluid flow in developing embryos (44 fps in [18]). A high pixel rate is also required to study slower large-scale processes such as collective cell migration or cell division patterns with a large number of pixels per image to reach the appropriate spatial resolution: for instance, in toto imaging of early development [16,19–20] typically requires acquiring ~100 million voxels per 3D-image stack in less than a minute.
In this context, point-scanning confocal or multiphoton approaches are usually too slow, as the image is recoreded one pixel at a time (Fig. 1). Indeed, in these approaches signal level prescribes pixel accumulation times of typically 1–10 μs, corresponding to pixel rates of only 105 to 106 pixels.s−1.
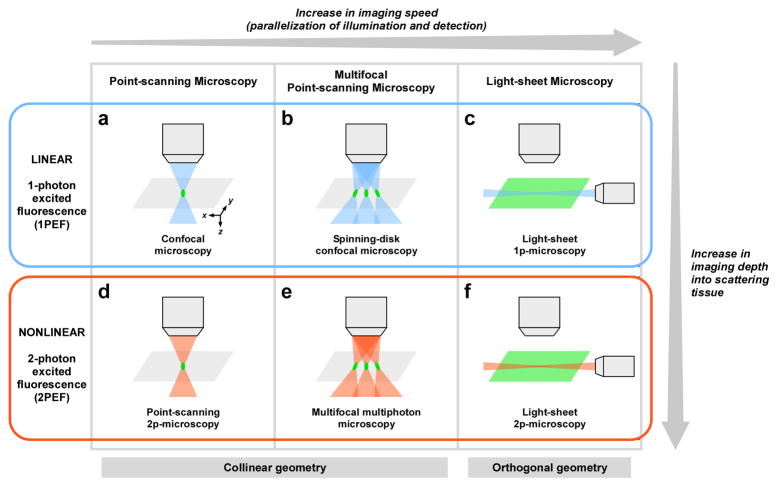
Figure 1. Strategies for improving acquisition speed in current fluorescence microscopy techniques.
Similar strategies have been developed in linear (a-c) and nonlinear (d-f) microscopy. Both point-scanning (a and d) and multifocal (b and e) approaches use a collinear geometry: the illumination and the detection paths are collinear. Light-sheet microscopy (c and f) uses an orthogonal geometry: the illumination path is orthogonal to the detection path. Pixel rate range typically from 105–106 pixels.s−1 in point-scanning microscopy to 107–108 pixels.s−1 in light-sheet microscopy. While nonlinear microscopy generally provides deeper imaging than linear microscopy, differences exist in imaging depth performance between the different implementations of nonlinear microscopy, as discussed in the text.
Several approaches have been explored during the last 15 years to improve the imaging speed of multiphoton microscopy up to ~107 pixels.s−1, including fast point-scanning and multifocal approaches (Fig. 1, Tables 1 and 2, and [21] for a review). However, besides hardware limitations (i.e. scanning speed, readout time, data transfer or storage) the pixel rate of any microscope is fundamentally limited by the signal level that can be obtained within the pixel accumulation time without causing fluorophore saturation or photodamage (including phototoxicity to the biological sample and photobleaching of the fluorophores). Hence, even though fast point-scanning can be implemented using resonant scanners, polygonal mirrors or acousto-optic deflectors [21], the useful pixel rate is still limited by fluorophore photophysics of the single-point excitation approach (third column in Table 1). The main strategy to circumvent this limitation is to parallelize the sample illumination and the signal detection. Using multifocal excitation (Fig. 1), overall pixel rate can be increased while maintaining the same illumination time per pixel (Table 1). However with this approach, an increase in imaging speed requires a proportional increase in laser average power (Table 1 and Table 2), similar to linear microscopy (Supp. Table 1). Available laser power therefore limits the achievable speed gain. Moreover, increasing the laser average power may eventually lead to linear absorption and photodamage, as it is the case in linear microscopy.
Table 1. Strategies for fast acquisition speed in multiphoton microscopy: theoretical scaling of illumination parameters.
This table summarizes how illumination parameters scale in different implementations of fast multiphoton microscopy. Using point-scanning 2p-microscopy as a reference, we compare the parameters of fast point-scanning, multifocal, and light-sheet 2p-microscopy (with Gaussian beam scanning) approaches. Assuming the pixel rate is increased N times, the illumination laser power is scaled to obtain the same level of emitted fluorescence for the entire image. Other conditions are kept unchanged (Gaussian beams, same axial resolution, same pulse width, repetition rate). In the case of fast point-scanning 2p-microscopy, the laser is scanned N times faster, resulting in an illumination time N times shorter, which is compensated by times more illumination laser average power. The most limiting factor is the short illumination time: a limited signal can be generated before reaching photo-physical limits and saturation of the fluorophore. In the case of multifocal multiphoton microscopy, the use of N foci increase pixel rate N times but requires increasing the laser average power N times to reach similar signal levels. While other 2p-techniques requires only more power due to the quadratic dependence of excitation on illumination power, the multifocal approach loses this advantage and requires N times more power (similar to linear techniques, see Supp. Table 1). Finally, the light-sheet 2p-microscopy strategy uses lower illumination NA without changing axial resolution. α is chosen to keep the same axial resolution between point-scanning and light-sheet 2p-microscopies (typically α~5–10): for NA<0.7 [59]. In this case, illumination time and intensity are significantly longer and weaker, respectively, compared to other techniques (in practice, with N up to several 100s). This reduces photodamage and avoids fluorophore saturation. Interestingly, since α scales as 1/NA, it appears that the advantages of light-sheet compared to point-scanning are more pronounced at low NA and low spatial resolution. The color codes indicate a comparison with point-scanning 2p-microscopy: blue is unchanged, green is advantageous, light red is moderately disadvantageous, and red is severely disadvantageous.
| Point-scanning 2p-microscopy | Fast point- scanning 2p-microscopy (N-time faster scan) | Multifocal multiphoton microscope (N foci) | Light-sheet 2p-microscopy | |||
|---|---|---|---|---|---|---|
| Scanning scheme |
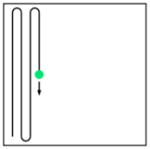 Point scanning |
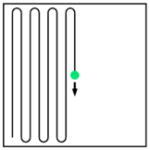 Fast point scanning |
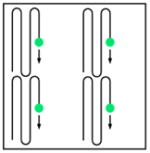 Multiple point scanning |
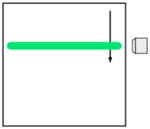 Fast line scanning |
||
| Acquisition speed (pixel rate) | r | Nr | Nr | Nr | ||
| Numerical aperture of illumination objective | NA | NA | NA |
|
||
| Illumination time per 2p-excited volume* | t |
|
t |
|
||
| Illumination laser intensity | I |
|
I |
|
||
| Illumination laser average power | P |
|
NP |
|
If the spatial sampling (or pixel size) is the same for every technique, this value is related to the illumination time per pixel. See Supp. Table 1 for the comparison with linear techniques.
Table 2. Strategies for fast acquisition speed in multiphoton microscopy: example of experimental illumination parameters.
Experimental values from published work confirm the parameter scaling presented in Table 1. Importantly, they demonstrate the advantageous use of long illumination time and low illumination intensity in light-sheet 2p-microscopy with limited increase in laser power compared to point-scanning 2p-microscopy. In addition, they show that the main limitation of multifocal multiphoton microscopy is the requirements for high laser average power. Note that these experimental values have been used for imaging different biological sample with different labeling: therefore, the comparison is only indicative.
| Reference | Mc Mahon et al. [6] | Bahnmann et al. [59] | Truong et al. [16] |
|---|---|---|---|
| Microscopy | Point-scanning 2p-microscopy | Multifocal multiphoton microscopy | Light-sheet 2p-microscopy |
| Sample | Live Drosophila embryos | Dissociated adult rat cells (cardiac myocytes) | Live zebrafish embryonic heart |
| Fluorophore | GFP | Fluo3 calcium dye | GFP |
| Image (or frame) size | 400×400 pixels 200×200 μm2 |
128×128 pixels 64×64 μm2 |
400×400 pixels 160×160 μm2 |
| Frame rate | 2.2 fps | 640 fps | 70 fps |
| Acquisition speed (pixel rate) | 0.36 106 pix/s | 10.5 106 pix/s | 11.2 106 pix/s |
| Illumination time per pixel | 2.8 μs | 3.4 μs | ~ 100 μs |
| Illumination laser intensity per focus | ~ 10 MW.cm−2 | ~ 10 MW.cm−2 | ~ 0.1 MW.cm−2 |
| Illumination laser average power | 30 mW | 360 mW | 50 mW |
| Excitation wavelength | 940 nm | 780 nm | 920 nm |
Among the strategies for improving the imaging speed of multiphoton microscopy, the recent implementation of scanned light-sheet microscopy using two-photon excitation (2p-SPIM in [16] and light-sheet 2p-microscopy in this review) introduces a new paradigm. In this technique, a sheet of light is generated by scanning a weakly focused Gaussian beam faster than the image acquisition time to illuminate an entire plane of the sample, which is then imaged with a camera oriented orthogonally to the sheet. Compared to a static light-sheet generated with a cylindrical lens [22], this scanned light-sheet approach generates typically 100 times stronger 2PEF signal [16], which is critical for live imaging. Light-sheet 2p-microscopy is the only technique improving overall pixel rate over point-scanning 2p-microscopy with longer pixel accumulation time and lower peak intensity (Table 1 and Table 2). This fundamental property results from the orthogonal geometry of the illumination and detection pathways (which are collinear in conventional microscopy), allowing the use of a low numerical aperture (NA) illumination focusing (resulting in a large illumination volume) without degrading the axial resolution and the overall signal rate [16]. The use of low-NA illumination has three important advantages for multiphoton live imaging. First, it results in lower peak intensity, and therefore less higher-order nonlinear photodamage to the tissue [23]. Second, parallelization of the illumination is done along the light propagation direction, reusing the same excitation energy, thus requiring less laser power than in multifocal approaches, in turn limiting linear absorption and photodamage. Finally, the weakly focused excitation beam is less sensitive to sample-induced optical aberrations and resolution loss with depth than in the case of high-NA focusing [16]. In addition, in the conditions presented in Table 3 [16], the laser is scanned ~15 times during the image acquisition with 1 ms between two passes. This temporal excitation pattern potentially results in lower photobleaching, as time is given for fluorophore dark state relaxation [24].
Overall, compared to other fast multiphoton techniques, light-sheet 2p-microscopy provides fast acquisition while reducing photodamage and requiring minimal increase in laser power as demonstrated in live embryos [16] (Fig. 2a). To date, it is the fastest implementation of multiphoton microscopy with up to ~1.1 107 pix/s (Table 2). We note however that light-sheet microscopy relies on widefield (camera-based) detection, which leads to compromises in terms of imaging depth, as discussed in the next section.
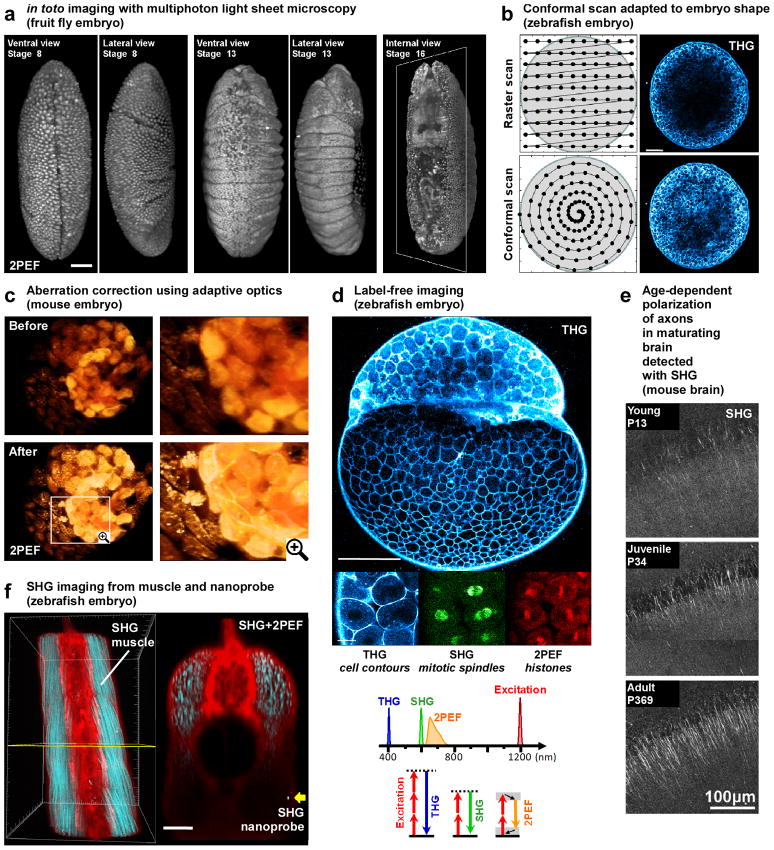
Figure 2. Selected applications of advanced multiphoton microscopy in developmental biology.
This figure illustrates various recent applications of multiphoton microscopy in developmental biology. (a) Long term imaging of Drosophila embryos using light-sheet 2p-microscopy (reprinted from [16], scale bar is 50 μm). (b) Standard raster scanning in multiphoton microscopy produces inhomogeneous signal levels across the embryo. Conformal scanning adapted to the embryo shape provide homogeneous signal for the entire image (image adapted from [30] and reprinted with permission from AAAS, scale bar is 100 μm). (c) 3D rendering of point-scanning 2p-microscopy imaging of a mouse embryo before and after correction of sample-induced aberrations showing improvement in both signal intensity and spatial resolution (adapted from [33] and reprinted with permission from OSA). (d) Simultaneous 2PEF, SHG and THG imaging of zebrafish embryos allows detection of histones, mitotic spindles and cell contours, respectively. Such label-free imaging with SHG and THG signals has been used for 3D cell segmentation and tracking and for reconstructing the cell lineage of early zebrafish development (image adapted from [30] and reprinted with permission from AAAS, scale bars are 200 μm (top) and 20 μm (bottom)). (e) Maturation of axons and dendrites is probed with SHG in mouse hippocampal slices (adapted from [49], copyright (2008) National Academy of Sciences, USA). (f) 3D reconstruction of combined 2PEF (from Bodipy TR dye in red) and SHG (from muscle endogenous signal and BaTiO3 nanoparticle in blue) signals recorded in zebrafish embryos: the strong SHG signal from the nanoprobe (yellow arrow) is detected twice deeper than the endogeneous SHG from muscles (image adapted from [51] and reprinted with permission from authors, scale bar is 50 μm).
Parameters governing imaging depth in tissue microscopy
The imaging depth corresponds to how deep into the tissue images can be recorded with sufficient quality (resolution, signal intensity, contrast). Imaging depth is limited by light scattering [25] and by sample-induced optical aberrations. Point-scanning 2p-microscopy is usually considered as the gold-standard in imaging depth for in vivo fluorescence imaging of tissues and embryos [26] (Fig. 3). For instance, the large-scale dynamic analysis of the deepest mesoderm cells during Drosophila gastrulation has been made possible only using point-scanning 2p-microscopy [6].
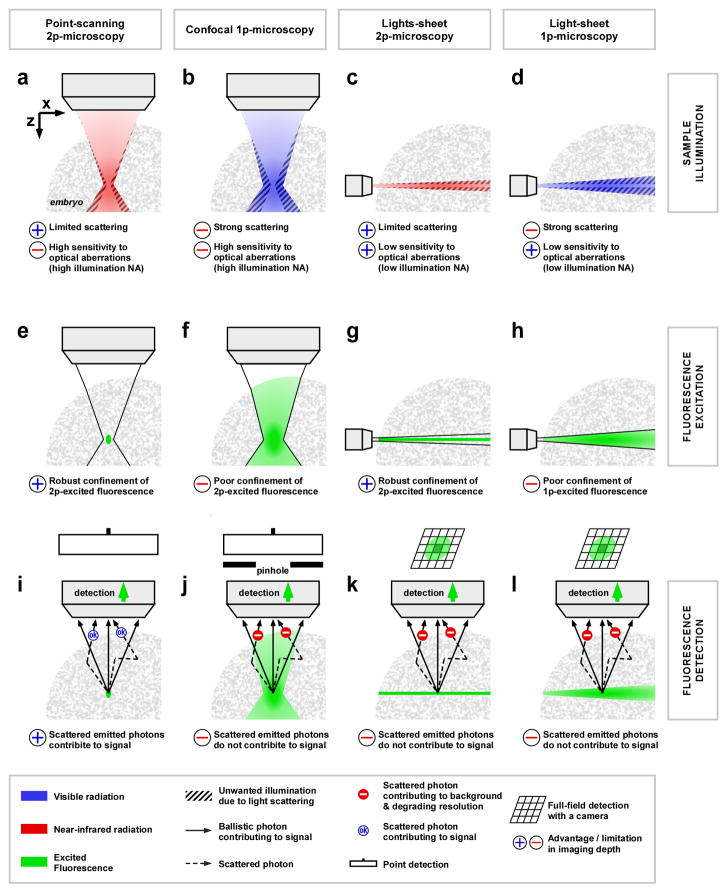
Figure 3. Imaging depth in point-scanning and light-sheet microscopy: comparison of 1-photon and 2-photon excited fluorescence.
The principles governing imaging depth are analyzed in three steps: sample illumination (a-d), fluorescence excitation (e-h) and fluorescent detection (i-l). 2p-microscopy uses longer excitation wavelengths (red in a and c) than 1p-microscopy (blue in b and d), resulting in reduced scattering inside biological tissues, preserving focus quality in depth with less out-of-focus sample illumination (dashed area in a-d). Compared to confocal and point-scanning 2p-microscopy, the light-sheet illumination has the advantage of using low-NA illumination focusing, which is less sensitive to sample-induced optical aberrations (c-d). The linear fluorescence excitation in 1p-microscopy, results in a direct equivalence between illumination (blue in b and d) and fluorescence excitation volumes (green in f and h). It results in a high sensitivity to illumination light scattering and a poor confinement of fluorescence excitation. In light-sheet 1p-microscopy, this poor confinement results in lower axial sectioning and degraded axial resolution due to the scattering-induced thickening of the light-sheet at high sample depth (h). The nonlinearity of fluorescence excitation in 2p-microscopy confines the excitation to the region with highest illumination intensity, resulting in an excitation volume (green in e and g) smaller than the illumination volume (red in a and c). This robust confinement of the fluorescence excitation makes 2p-microscopy less sensitive to scattering of the illumination light and allows preserving spatial resolution deeper into biological tissues (e and g). Finally, concerning fluorescence signal detection, point-scanning 2p-microscopy has a key advantage compared to other techniques. Namely, the 3D-confinement of the fluorescence excitation (green in i) guarantees the spatial origin of emitted photons, which allows collecting both scattered and non-scattered photons to build the fluorescence signal. Hence, point-scanning 2p-microscopy has the most efficient signal collection using scattered emitted photons as part of the signal. In any other techniques of microscopy, these scattered photons are either rejected (using a pinhole such as in confocal microscopy, (j)) or degrade the image quality. For instance, in light-sheet microscopy, scattering of the fluorescence on its way to the camera results in cross talk between adjacent pixels and image blurring (k and l): only ballistic photons contribute to the signal while scattered photons cause contrast and resolution degradation.
The depth performance of point-scanning 2p-microscopy relies on three phenomena: (i) superior penetration of illumination light, (ii) robust confinement of excitation volume, and (iii) efficient collection of the fluorescence. Let us compare this technique with confocal microscopy, light-sheet 2p-microscopy and light-sheet 1p-microscopy with respect to these three points (Fig. 3).
Scattering of near infrared light is reduced compared to that of visible light in biological tissues. For this reason, both point-scanning and light-sheet 2p-microscopies benefit from greater penetration of the illumination light, and reduced degradation of the illumination volume with depth compared to linear techniques (Fig. 3a-d).
In multiphoton microscopy, the nonlinear dependence of fluorescence generation on illumination confines excitation to the regions with highest intensity. As a result, in both point-scanning and light-sheet 2p-microscopy fluorescence excitation is robustly confined in space and is less sensitive to scattering of illumination light. In contrast, in light-sheet 1p-microscopy, the excitation volume is identical to the illumination volume, resulting in a direct loss of axial resolution at high sample depth due to scattering-induced thickening of the light-sheet (Fig. 3e-h).
Finally, in point-scanning 2p-microscopy, all fluorescence is emitted from a confined volume corresponding to a single voxel in the 3D-image, meaning that both scattered and ballistic (non-scattered) photons can be collected and attributed to the signal (Fig. 3i). This collection efficiency is a unique feature of point-scanning 2p-microscopy: in all other techniques, only ballistic photons contribute to the signal while scattered photons need to be rejected with a pinhole or otherwise would cause contrast degradation (Fig. 3j–l). In light-sheet microscopy, the scattering of the fluorescence on its way to the camera results in cross talk between adjacent pixels and image blurring.
However, light-sheet microscopy does has one advantage other collinear techniques (confocal and point-scanning 2p-microscopy): the use of lower illumination NA leads to less sensitivity to optical aberrations and thus contributes to maintain better axial resolution in light-sheet 2p-microscopy imaging of inhomogeneous embryos [16].
In summary, light-sheet microscopy with 2p-excitation provides deeper imaging than with 1p-excitation for two fundamental reasons: deeper penetration of illumination light and robust confinement of fluorescence excitation. At large depths, light-sheet 2p-microscopy lacks the background-free collection advantage of point-scanning 2p-microscopy, but is less sensitive to aberration-induced degradation in axial resolution.
Adaptive advantages of point-scanning for multiphoton imaging of embryos
A developing embryo is a dynamic and inhomogeneous biological system. Optical properties vary between species and tissues [26], and they also constantly evolve in time and space during embryonic development [27]. As a consequence, embryo imaging would strongly benefit from the ability of microscope illumination and acquisition schemes to adapt to the changing properties of the developing tissue. In this context, point-scanning multiphoton techniques have a fundamental advantage compared to parallelized illumination strategies, which is the ability to readily tailor the imaging parameters for each individual point of the embryo.
Recent advances in active and adapted control of microscope illumination and acquisition hold great potential for embryo imaging. The active adjustment of illumination power depending on local signal levels in confocal microscopy [28] and point-scanning 2p-microscopy [29] have been shown to reduce photodamage and avoid fluorophore and detection saturation. Recently, the novel concept of conformal scanning was demonstrated for multiphoton imaging of zebrafish embryos [30]: spiral scanning was used to match the embryo spherical shape, allowing the constant adjustment of scanning speed to the imaging depth. Using slow scanning in deep regions and fast scanning in peripheral regions, the illumination is optimized to obtain homogeneous signal and reduced phototoxicity while minimizing the acquisition time (Fig. 2b), providing effective in toto imaging [30]. More sophisticated techniques using adaptive optics to correct for optical aberrations within embryos are promising directions for improved imaging of embryos [31–33]. Adaptive multiphoton microscopy has been shown to correct for resolution losses during deep imaging (Fig. 2c), and can be performed dynamically in evolving embryos [34].
Beyond fluorescence: other nonlinear contrast mechanisms (SHG, THG, CARS, and SRS)
Another advantage of nonlinear microscopy is that in addition to fluorescence, other multiphoton processes can be used as contrast mechanisms to provide complementary information. These include second-harmonic generation (SHG), third-harmonic generation (THG), coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS). These imaging modalities [35] share the benefits of point-scanning 2p-microscopy in terms of 3D resolution and penetration depth. However they rely on coherent optical processes, and therefore have more complex contrast mechanisms than fluorescence microscopy. For example, signal strength is generally sensitive to the spatial distribution of molecules within the excitation volume, and signal radiation usually occurs in the direction of the excitation beam.
In many cases such signals can be obtained from unstained tissues, with the additional benefit of not suffering from photobleaching. SHG is exclusively observed from dense organized non-centrosymmetric electronic structures. Some natural sources of SHG are fibrillar collagen, myofilaments, astroglial fibers, starch, and polarized tubulin assemblies such as mitotic spindles that can be observed in embryos (Fig. 2d-f). THG does not require molecular asymmetry but is observed only near optical heterogeneities. In practice, THG signals are obtained from dense non-aqueous objects such as lipid droplets [36], mineralized or absorbing structures, and generally from interfaces between media of different refractive indices. Coherent Raman processes such as CARS and SRS derive their contrast from molecular vibrational modes and can be used for micro-spectroscopy and chemically selective imaging. SRS provides increased contrast compared with CARS at the cost of increased experimental complexity. The most widespread use of coherent Raman microscopy for biological studies is currently the selective imaging of lipid distribution in tissues based on contrast from CH-bond vibration [37–40].
THG and SHG are efficiently produced using femtosecond excitation pulses and require a single laser. For that reason, combination with fluorescence-based point-scanning 2p-microscopy is straightforward. Several studies have reported harmonic and multimodal harmonic/fluorescence imaging of embryos in various models: fruit fly [5,41], zebrafish [30,42–43], mouse [44], or worm [45]. SHG imaging carried out in light-sheet mode and combined with light-sheet 2p-microscopy, has also been recently demonstrated [16]. CARS/SRS microscopy requires two synchronized and overlapped excitation beams, and contrast is optimized with picosecond rather than femtosecond pulses. For these reasons combination of coherent Raman scattering with 2PEF is more complex, but multimodal fluorescence/harmonic/Raman imaging is becoming a reality.
One particularly attractive aspect of these imaging approaches for embryo imaging is that they provide label-free structural or molecular vibrational imaging. In some experimental situations, it is challenging to obtain long-term, strong, specific, and non-invasive fluorescent labeling. Label-free nonlinear imaging has proven to be a particularly effective addition to fluorescence for studying early division patterns in the zebrafish embryo [30] (Fig. 2d), and lipid storage in worms [37–38,46]. Harmonic-generation contrast usually provides additional structural information on molecular or supra-molecular order that would not be easily detectable with fluorescent labeling strategies: for instance, collagen macromolecular organization into fibrils [47], myosin structural conformation in sarcomere [48], microtubule array polarization during brain maturation [49] (Fig. 2e), or detection of sub-micron-scale anisotropy in the cornea [50]. In addition, we note that artificial nanostructures usually produce strong nonlinear optical signals that, unlike fluorescence, do not bleach or saturate with high laser intensity. Thus, harmonic signals from nanoprobes may be detected with high sensitivity, as illustrated by SHG imaging of nanocrystals inside zebrafish embryos [51] (Fig. 2f).
To summarize, non-fluorescent nonlinear signals should generally not be viewed only as a label-free substitute to fluorescence, but rather as providing additional information, the potential of which we think has not been fully explored yet for developmental biology.
Conclusion and perspectives
Recent advances in multiphoton microscopy, including light-sheet, adaptive and label-free strategies, open promising avenues for embryo imaging. However, the literature is often unclear about the comparative performances of microscopy modalities. Therefore, it is important to understand the principles, advantages and limitations of each microscope implementation. In this review, we provide keys to understand recent methodological developments in the perspective of their application to developmental biology. We dicsuss why light-sheet illumination provides faster imaging with lower photodamage than other multiphoton microscopy geometries. We review how multiphoton microscopy achieves high imaging depth into tissues and clarify why the standard point-scanning approach, though lacking in imaging speed, hold fundamental advantages compared to parallelized illumination strategies for imaging dynamic and inhomogeneous embryos. We discuss the mechanisms and advantages of non-fluorescent techniques of multiphoton microscopy and illustrate how label-free imaging can be applied to developmental biology. A number of additional experimental developments are still under investigation with potential benefits for application to developmental biology. These include the use of laser pulse shaping [52] and of Bessel beam illumination [53–55]. Finally, we note that many recent developments in linear microscopy can be also be applied in a straightforward manner to multiphoton microscopy: for instance light-sheet 2p-microscopy would benefit from techniques developed for light-sheet 1p-microscopy, such as deconvolution [56], background rejection using structured illumination [57] or HiLo [58].
Supplementary Material
Box 1. Definition of microscopy terms and acronyms used in this review.
| Linear microscopy | Microscopy using on a linear contrast mechanism (the signal scales linearly with the laser illumination intensity). |
| Nonlinear microscopy | Microscopy using on a nonlinear contrast mechanism (the signal scales nonlinearly with the laser illumination intensity), including 2PEF, SHG, THG, CARS, and SRS signals. |
| Multiphoton microscopy | Synonym to nonlinear microscopy. |
| 1p | 1-photon (corresponding to a linear excitation) |
| 2p | 2-photon (corresponding to a nonlinear excitation) |
| Contrast mechanisms: | |
| 1PEF | 1-Photon Excited Fluorescence |
| 2PEF | 2-Photon Excited Fluorescence |
| SHG | Second Harmonic Generation |
| THG | Third Harmonic Generation |
| CARS | Coherent Anti-stokes Raman Scattering |
| SRS | Stimulated Raman Scattering |
| Microscope implementation: | |
| Point-scanning 2p-microscopy | Microscopy based on 2PEF signal using raster scanning of single point (also called TPLSM, 2p-LSM, 2p-microscopy, 2PEF microscopy, two-photon microscopy in the literature). |
| Light-sheet illumination | Microscopy using light-sheet illumination in orthogonal geometry (also called SPIM, DSLM, Plane illumination microscopy,… in the litterature) |
| Light-sheet 2p-microscopy | Microscopy based on 2PEF signal using light-sheet illumination (also called 2p-SPIM in [16]) |
| Light-sheet 1p-microscopy | Microscopy based on 1PEF signal using light-sheet illumination (also called 1p-SPIM in [16], DSLM in [20]) |
| Multifocal Multiphoton Microscopy | Microscopy based on 2PEF signal using raster scanning of multiple points |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 2.Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Wilt BA, Burns LD, Wei Ho ET, Ghosh KK, Mukamel EA, Schnitzer MJ. Advances in Light Microscopy for Neuroscience. Annual Review of Neuroscience. 2009;32:435–506. doi: 10.1146/annurev.neuro.051508.135540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahalan MD, Parker I. Choreography of cell motility and interaction dynamics imaged by two-photon microscopy in lymphoid organs. Annual Review of Immunology. 2008;26:585–626. doi: 10.1146/annurev.immunol.24.021605.090620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Supatto W, Debarre D, Moulia B, Brouzes E, Martin J-L, Farge E, Beaurepaire E. In vivo modulation of morphogenetic movements in Drosophila embryos with femtosecond laser pulses. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1047–1052. doi: 10.1073/pnas.0405316102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon A, Supatto W, Fraser SE, Stathopoulos A. Dynamic Analyses of Drosophila Gastrulation Provide Insights into Collective Cell Migration. Science. 2008;322:1546–1550. doi: 10.1126/science.1167094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebollo E, Roldán M, Gonzalez C. Spindle alignment is achieved without rotation after the first cell cycle in Drosophila embryonic neuroblasts. Development. 2009;136:3393–3397. doi: 10.1242/dev.041822. [DOI] [PubMed] [Google Scholar]
- 8.Brodland GW, Conte V, Cranston PG, Veldhuis J, Narasimhan S, Hutson MS, Jacinto A, Ulrich F, Baum B, Miodownik M. Video force microscopy reveals the mechanics of ventral furrow invagination in Drosophila. Proceedings of the National Academy of Sciences. 2010;107:22111–22116. doi: 10.1073/pnas.1006591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato Y, Poynter G, Huss D, Filla MB, Czirok A, Rongish BJ, Little CD, Fraser SE, Lansford R. Dynamic Analysis of Vascular Morphogenesis Using Transgenic Quail Embryos. Plos One. 2010;5:e12674. doi: 10.1371/journal.pone.0012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien GS, Rieger S, Martin SM, Cavanaugh AM, Portera-Cailliau C, Sagasti A. Two-photon axotomy and time-lapse confocal imaging in live zebrafish embryos. J Vis Exp. 2009 doi: 10.3791/1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol. 1999;17:763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDole K, Xiong Y, Iglesias PA, Zheng Y. Lineage mapping the pre-implantation mouse embryo by two-photon microscopy, new insights into the segregation of cell fates. Developmental Biology. 2011;355:239–249. doi: 10.1016/j.ydbio.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truong TV, Supatto W. Toward high-content/high-throughput imaging and analysis of embryonic morphogenesis. Genesis. 2011;49:555–569. doi: 10.1002/dvg.20760. [DOI] [PubMed] [Google Scholar]
- 14.Liebling M, Forouhar AS, Wolleschensky R, Zimmermann B, Ankerhold R, Fraser SE, Gharib M, Dickinson ME. Rapid three-dimensional imaging and analysis of the beating embryonic heart reveals functional changes during development. Developmental Dynamics. 2006;235:2940–2948. doi: 10.1002/dvdy.20926. [DOI] [PubMed] [Google Scholar]
- 15.Arrenberg AB, Stainier DYR, Baier H, Huisken J. Optogenetic Control of Cardiac Function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 16**.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nature Methods. 2011 doi: 10.1038/nmeth.1652. advance online publication. Implemetation of multiphoton scanned light-sheet microscopy for live imaging of embryos. Experimental comparison with point-scanning 2p-microscopy and light-sheet 1p-microscopy. [DOI] [PubMed] [Google Scholar]
- 17.Hirota Y, Meunier A, Huang S, Shimozawa T, Yamada O, Kida YS, Inoue M, Ito T, Kato H, Sakaguchi M, et al. Planar polarity of multiciliated ependymal cells involves the anterior migration of basal bodies regulated by non-muscle myosin II. Development. 2010 doi: 10.1242/dev.050120. [DOI] [PubMed] [Google Scholar]
- 18.Supatto W, Fraser SE, Vermot J. An all-optical approach for probing microscopic flows in living embryos. Biophysical Journal. 2008;95:L29–L31. doi: 10.1529/biophysj.108.137786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Megason SG, Fraser SE. Digitizing life at the level of the cell: high-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mechanisms of Development. 2003;120:1407–1420. doi: 10.1016/j.mod.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of Zebrafish Early Embryonic Development by Scanned Light Sheet Microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 21.Carriles R, Schafer DN, Sheetz KE, Field JJ, Cisek R, Barzda V, Sylvester AW, Squier JA. Invited Review Article: Imaging techniques for harmonic and multiphoton absorption fluorescence microscopy. Rev Sci Instrum. 2009:80. doi: 10.1063/1.3184828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palero J, Santos S, Artigas D, Loza-Alvarez P. A simple scanless two-photon fluorescence microscope using selective plane illumination. Opt Express. 2010;18:8491–8498. doi: 10.1364/OE.18.008491. [DOI] [PubMed] [Google Scholar]
- 23.Ji N, Magee JC, Betzig E. High-speed, low-photodamage nonlinear imaging using passive pulse splitters. Nature Methods. 2008;5:197–202. doi: 10.1038/nmeth.1175. [DOI] [PubMed] [Google Scholar]
- 24.Donnert G, Eggeling C, Hell SW. Major signal increase in fluorescence microscopy through dark-state relaxation. Nat Meth. 2007;4:81–86. doi: 10.1038/nmeth986. [DOI] [PubMed] [Google Scholar]
- 25.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nature Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 26.Ntziachritos V. Going deeper than microscopy: the optical imaging frontier in biology. Nature Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 27*.Supatto W, McMahon A, Fraser SE, Stathopoulos A. Quantitative imaging of collective cell migration during Drosophila gastrulation: multiphoton microscopy and computational analysis. Nature Protocols. 2009;4:1397–1412. doi: 10.1038/nprot.2009.130. Practical considerations for multiphoton imaging of embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoebe RA, Van Oven CH, Gadella TWJ, Dhonukshe PB, Van Noorden CJF, Manders EMM. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat Biotech. 2007;25:249–253. doi: 10.1038/nbt1278. [DOI] [PubMed] [Google Scholar]
- 29.Chu KK, Lim D, Mertz J. Practical implementation of log-scale active illumination microscopy. Biomed Opt Express. 2010;1:236–245. doi: 10.1364/BOE.1.000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Olivier N, Luengo-Oroz MA, Duloquin L, Faure E, Savy T, Veilleux I, Solinas X, Debarre D, Bourgine P, Santos A, et al. Cell Lineage Reconstruction of Early Zebrafish Embryos Using Label-Free Nonlinear Microscopy. Science. 2010;329:967–971. doi: 10.1126/science.1189428. Demonstration of conformal scan strategy for improved illumination and in toto imaging of zebrafish embryos. Reconstruction of cell lineage in early zebrafish embryos using label-free multiphoton microscopy. [DOI] [PubMed] [Google Scholar]
- 31.Rueckel M, Mack-Bucher JA, Denk W. Adaptive wavefront correction in two-photon microscopy using coherence-gated wavefront sensing. PNAS. 2006;103:17137–17142. doi: 10.1073/pnas.0604791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jesacher A, Thayil A, Grieve K, Debarre D, Watanabe T, Wilson T, Srinivas S, Booth M. Adaptive harmonic generation microscopy of mammalian embryos. Optics Letters. 2009;34:3154–3156. doi: 10.1364/OL.34.003154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Débarre D, Botcherby EJ, Watanabe T, Srinivas S, Booth MJ, Wilson T. Image-based adaptive optics for two-photon microscopy. Opt Lett. 2009;34:2495–2497. doi: 10.1364/ol.34.002495. Adaptive optics for improving multiphoton imaging of fixed mouse embryo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivier N, Debarre D, Beaurepaire E. Dynamic aberration correction for multiharmonic microscopy. Optics Letters. 2009;34:3145–3147. doi: 10.1364/OL.34.003145. [DOI] [PubMed] [Google Scholar]
- 35.Masters BR, So PT. Handbook of biomedical nonlinear optical microscopy. New York: Oxford University Press; 2008. [Google Scholar]
- 36.Debarre D, Supatto W, Pena AM, Fabre A, Tordjmann T, Combettes L, Schanne-Klein MC, Beaurepaire E. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nature Methods. 2006;3:47–53. doi: 10.1038/nmeth813. [DOI] [PubMed] [Google Scholar]
- 37.Hellerer T, Axäng C, Brackmann C, Hillertz P, Pilon M, Enejder A. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proceedings of the National Academy of Sciences. 2007;104:14658–14663. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. RNAi screening for fat regulatory genes with SRS microscopy. Nature Methods. 2011;8:135–U152. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Video-Rate Molecular Imaging in Vivo with Stimulated Raman Scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huff TB, Shi Y, Sun W, Wu W, Shi R, Cheng J-X. Real-Time CARS Imaging Reveals a Calpain-Dependent Pathway for Paranodal Myelin Retraction during High-Frequency Stimulation. Plos One. 2011;6:e17176. doi: 10.1371/journal.pone.0017176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Debarre D, Supatto W, Farge E, Moulia B, Schanne-Klein MC, Beaurepaire E. Velocimetric third-harmonic generation microscopy: micrometer-scale quantification of morphogenetic movements in unstained embryos. Optics Letters. 2004;29:2881–2883. doi: 10.1364/ol.29.002881. [DOI] [PubMed] [Google Scholar]
- 42.Chu S-W, Chen S-Y, Tsai T-H, Liu T-M, Lin C-Y, Tsai H-J, Sun C-K. In vivo developmental biology study using noninvasive multi-harmonic generation microscopy. Optics Express. 2003;11:3093–3099. doi: 10.1364/oe.11.003093. [DOI] [PubMed] [Google Scholar]
- 43.Sun CK, Chu SW, Chen SY, Tsai TH, Liu TM, Lin CY, Tsai HJ. Higher harmonic generation microscopy for developmental biology. Journal of Structural Biology. 2004;147:19–30. doi: 10.1016/j.jsb.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 44.Thayil A, Watanabe T, Jesacher A, Wilson T, Srinivas S, Booth M. Long-term imaging of mouse embryos using adaptive harmonic generation microscopy. Journal of Biomedical Optics. 2011:16. doi: 10.1117/1.3569614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tserevelakis GJ, Filippidis G, Krmpot AJ, Vlachos M, Fotakis C, Tavernarakis N. Imaging Caenorhabditis elegans embryogenesis by third-harmonic generation microscopy. Micron. 2010;41:444–447. doi: 10.1016/j.micron.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Yen K, Le TT, Bansal A, Narasimhan D, Cheng JX, Tissenbaum HA. A Comparative Study of Fat Storage Quantitation in Nematode Caenorhabditis elegans Using Label and Label-Free Methods. Plos One. 2010:5. doi: 10.1371/journal.pone.0012810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strupler M, Pena AM, Hernest M, Tharaux PL, Martin JL, Beaurepaire E, Schanne-Klein MC. Second harmonic imaging and scoring of collagen in fibrotic tissues. Optics Express. 2007;15:4054–4065. doi: 10.1364/oe.15.004054. [DOI] [PubMed] [Google Scholar]
- 48.Nucciotti V, Stringari C, Sacconi L, Vanzi F, Fusi L, Linari M, Piazzesi G, Lombardi V, Pavone FS. Probing myosin structural conformation in vivo by second-harmonic generation microscopy. Proceedings of the National Academy of Sciences. 2010;107:7763–7768. doi: 10.1073/pnas.0914782107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwan AC, Dombeck DA, Webb WW. Polarized microtubule arrays in apical dendrites and axons. Proceedings of the National Academy of Sciences. 2008;105:11370–11375. doi: 10.1073/pnas.0805199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olivier N, Aptel F, Plamann K, Schanne-Klein M-C, Beaurepaire E. Harmonic microscopy of isotropic and anisotropic microstructure of the human cornea. Opt Express. 2010;18:5028–5040. doi: 10.1364/OE.18.005028. [DOI] [PubMed] [Google Scholar]
- 51*.Pantazis P, Maloney J, Wu D, Fraser SE. Second harmonic generating (SHG) nanoprobes for in vivo imaging. Proceedings of the National Academy of Sciences. 2010;107:14535–14540. doi: 10.1073/pnas.1004748107. Imaging SHG nanoprobes in live zebrafish embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labroille G, Pillai RS, Solinas X, Boudoux C, Olivier N, Beaurepaire E, Joffre M. Dispersion-based pulse shaping for multiplexed two-photon fluorescence microscopy. Optics Letters. 2010;35:3444–3446. doi: 10.1364/OL.35.003444. [DOI] [PubMed] [Google Scholar]
- 53.Olivier N, Mermillod-Blondin A, Arnold CB, Beaurepaire E. Two-photon microscopy with simultaneous standard and extended depth of field using a tunable acoustic gradient-index lens. Optics Letters. 2009;34:1684–1686. doi: 10.1364/ol.34.001684. [DOI] [PubMed] [Google Scholar]
- 54.Fahrbach FO, Simon P, Rohrbach A. Microscopy with self-reconstructing beams. Nature Photonics. 2010;4:780–785. [Google Scholar]
- 55.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nature Methods. 2011;8:417–U468. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verveer PJ, Swoger J, Pampaloni F, Greger K, Marcello M, Stelzer EHK. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nature Methods. 2007;4:311–313. doi: 10.1038/nmeth1017. [DOI] [PubMed] [Google Scholar]
- 57.Keller PJ, Schmidt AD, Santella A, Khairy K, Bao ZR, Wittbrodt J, Stelzer EHK. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nature Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mertz J, Kim J. Scanning light-sheet microscopy in the whole mouse brain with HiLo background rejection. Journal of Biomedical Optics. 2010:15. doi: 10.1117/1.3324890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nature Biotechnology. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.