Abstract
Two main mechanisms of protein turnover exist in eukaryotic cells: the ubiquitin-proteasome system and the autophagy-lysosomal pathway. Autophagy is an emerging important constituent of many physiological and pathological processes, such as response to nutrient deficiency, programmed cell death and innate immune response. In mammalian cells the selectivity of autophagy is ensured by the presence of cargo receptors, such as p62/SQSTM1 and NBR1, responsible for sequestration of the ubiquitinated proteins. In plants no selective cargo receptors have been identified yet. The present report indicates that structural and functional homologs of p62 and NBR1 proteins exist in plants. The tobacco protein, named Joka2, has been identified in yeast two-hybrid search as a binding partner of a small coiled-coil protein, a member of UP9/LSU family of unknown function, encoded by the UP9C gene strongly and specifically induced during sulfur deficiency. The typical domains of p62 and NBR1 are conserved in Joka2. Similarly to p62, Joka2-YFP has dual localization (cytosolic speckles and the nucleus); it forms homodimers and interacts with a member of the ATG8 family. Increased expression of Joka2 and ATG8f was observed in roots of tobacco plants grown for two days in nutrient-deficient conditions. Constitutive ectopic expression of Joka2-YFP in tobacco resulted in attenuated response (manifested by lesser yellowing of the leaves) to nutrient deficiency. In conclusion, Joka2, and presumably the process of selective autophagy, might constitute an important part of plant response to environmental stresses.
Keywords: ATG8, autophagy, cargo receptor, NBR1, nutrient deficiency, p62/SQSTM1, selective autophagy, sulfur, nitrogen, tobacco
Introduction
Autophagy, or “self-eating” is a ubiquitous catabolic process in eukaryotic cells. Although it was first described about 40 years ago, our molecular understanding of this process started only about a decade ago.1,2 The best characterized type of autophagy, macroautophagy, occurs in a wide range of eukaryotes including mammals, plants and fungi, and leads to the degradation of portions of the cytoplasm, which may include cell organelles. During this process a double-membrane structure, called the autophagosome, sequesters the cargo (e.g., cell material such as organelles, soluble cytosolic proteins and protein aggregates) for degradation. Subsequently, the outer membrane of the autophagosome fuses with the vacuole membrane resulting successively in uptake of the cargo enclosed by the inner autophagosomal membrane (the autophagic body) in the vacuole, the degradation of the cargo and the release of the products for reuse. The membrane origins of autophagosomes are unclear and may involve multiple sources, including the endoplasmic reticulum, Golgi apparatus, mitochondria and plasma membrane.3–6 At least 34 various proteins, which transiently associate and act in a hierarchical order during autophagosome assembly, have been identified so far. Genes encoding most of these proteins—autophagy-related (ATG) genes—have been found in screenings of yeast mutants defective in autophagy.7 The process and core molecular machinery components are evolutionarily conserved,8 however the higher eukaryote autophagy pathway might require more elaborate molecular machinery, including factors that are absent in yeast. The human autophagy system has a tremendous influence on protein homeostasis and involves multiple protein-protein interactions.9 It is needed for appropriate response to nutrient stress, innate and adaptive immunity and autophagic cell death. Malfunction of autophagy has been linked to a wide range of human pathologies, including cancer, different neurodegenerative diseases, immunological disorders and pathogen infection.10–12 Autophagy is also important during development of mammals, flies and worms.13
The autophagy specific ubiquitin-like (UBL) proteins of the ATG8 family (known also in mammals as LC3 or GABARAP) are central regulators of autophagosome assembly, maturation and lysosomal fusion. In addition, interaction of the conserved surface of ATG8 with a conserved hydrophobic W/YXXL/I motif (referred as LIR region) in cargo receptors is necessary for the selective cargo recruitment to the autophagosomes.14,15 In mammals, at least two proteins, p62/SQSTM1/Sequestosome-1 and NBR1 (neighbor of BRCA1 gene 1), can function as cargo receptors (or cargo binding proteins) in autophagic clearance of protein aggregates.16–19 The published data are mostly available for human p62, which itself is degraded by autophagy.20,21 The p62 is found in cellular inclusion bodies together with polyubiquitinated proteins, in protein aggregates that accumulate in various chronic, toxic and degenerative diseases.21–24 The p62 protein mainly facilitates the autophagic clearance of the aggregates of the ubiquitinated proteins, however, it is also capable of binding nonubiquitinated proteins such as, TRAF6,25,26 ALFY27 or Keap1.21 The human NBR1 protein is larger than p62 and in addition to the three domains common for both proteins, such as N-terminal phox/Bem1p (PB1), ZZ-type zinc finger and C-terminal ubiquitin association domain (UBA), it also contains two coiled-coil regions (CC1 and CC2), an amphipathic helical domain (JUBA) capable of binding phosphatidylinositol-phosphates28,29 and the functionally uncharacterized NBR1 domain conserved in all NBR1-like proteins.30 The NBR1 protein interacts with p62 in a PB1-PB1 fashion,31 but homodimerizes via its N-terminal CC region.32 Like p62, NBR1 is degraded by autophagy and interacts with ATG8-like proteins.28
Autophagy is a well-known process in yeasts and animals but it has only been recently established in plants. Studies of autophagy in plants are greatly facilitated by the functional and structural conservation of ATG proteins.33–35 Similarly to the yeast and metazoan systems the plant ATG8 proteins are critical components of the autophagy pathway, therefore in many studies the GFP-ATG8 fusions have been used as markers of autophagosomes in plants.36–41 The lack of obvious phenotypes of atg mutants grown under nutrient-sufficient conditions suggested that autophagy was not essential for plants. However, more detailed studies reveal that the atg mutants senescent earlier and were hypersensitive to nitrogen starvation and carbon limitation.38,39,42–44 Moreover, it was demonstrated that autophagy could be induced by treating plants with hydrogen peroxide or methyl viologen.45 An interesting link between nutrient availability, leaf senescence and autophagy was first revealed by Hanaoka at al.46 The autophagy-deficient mutants of Arabidopsis thaliana were capable of completing the normal life cycle but displayed early leaf senescence, which was exaggerated under nutrient-deficient conditions. Recently, it was established that under nutrient deficiency, RUBISCO and whole chloroplasts were delivered to the vacuole by autophagy and degraded.47,48 In general, autophagy in plants seems to be involved in nutrient recycling. It provides substrates during nutrient deprivation and acts as a cell survival mechanism through recycling cell waste. On the other hand, other evidence indicates that a constitutive basal autophagy occurs also under normal growth conditions.49,50 Moreover, it was recently found that autophagy operates a negative feedback loop modulating NPR1-dependent salicylic acid signaling and that this negative feedback is necessary to limit excessive senescence and the programmed cell death in response to pathogen infection.51
It was commonly believed that in plants no selective autophagy receptors exist and only the core molecular autophagy machinery operates.30,35 However, we demonstrate that the Joka2 protein from Nicotiana tabacum actually is a structural and possibly a functional homolog of p62 and NBR1 proteins. Expression of Joka2 gene was increased in tobacco roots but not in the shoots during nitrogen (N) or sulfur (S) deficiency. The overproduction of Joka2 reduced plant bleaching in both normal growth conditions and during nutrient shortage. We propose that Joka2, by analogy to p62 and NBR1—the selective cargo receptors from the mammalian system—participates in the process of selective autophagy in plants. Our data associate plant response to nutrient deprivation with Joka2, a member of p62/NBR1 family, and indicate for the first time a link between nutrient-deficiency response and the process of selective autophagy.
Results
Identification of tobacco Joka2 as a partner of UP9C.
The UP9C protein belongs to the family of UP9/LSU like proteins present in many plant species.52–54 At least six isoforms of these small (about 100 residues) proteins, containing coiled-coil domain exist in tobacco, while A. thaliana has four of them (LSU1-LSU4). Analysis of transgenic tobacco plants with silenced expression of UP9-like genes (due to expression of UP9C in the antisense orientation) strongly argues for the significant role of some of the UP9/LSU proteins in regulation of plant response to S-deficit.53 This function is possibly mediated by protein-protein interactions. We previously identified 17 clones encoding putative partners of UP9C originated from the cDNA library prepared from N. tabacum plants grown for 2 d in S-deficient conditions (GenBank Accession No: GU066878– GU066894). Surprisingly, the similar yeast two-hybrid (Y2H) experiment with the cDNA library prepared from N. plumbaginifolia seedlings grown in normal (nutrient sufficient) conditions resulted in identification of only three clones denoted pJoka2, pJoka8 and pJoka20 (Fig. 1A) encoding different proteins than those identified from N. tabacum library. Database searches and location of characteristic domains within the predicted open reading frames of Joka2, Joka8 and Joka20 allowed either for identification of a corresponding protein previously known in tobacco (the case of Joka20) or for identification of homologs in other plant species (the case of Joka2 and Joka8). The Joka20 protein was identified as L7/L12, a nuclear-encoded component of chloroplast ribosomes.55 The Joka8 protein appeared to contain a basic helix-loop-helix (bHLH) motif and was classified as a member of a huge family of bHLH transcription factors.56 Initially, no function to Joka2 could be assigned but it appeared to contain two well-characterized domains: Phox/Bem 1p (PB1; pfam:00564) and ZZ-type zinc finger (ZZ; pfam:00569). The Joka2 open reading frame was incomplete since no translation initiation and no translation stop codons were present in the cloned cDNA fragment.
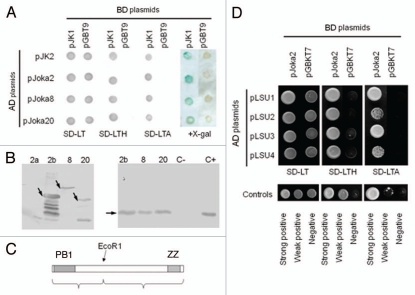
Figure 1.
Interactions of UP9C and LSU proteins. (A) The pJK1 and pJK2 plasmids contain UP9C in the BD- and AD-vectors, respectively. The yeast strains co-transformed with indicated BD- and AD-plasmids were grown on selective media without leucine and tryptophan (SD-LT), leucine, tryptophan and adenine (SD-LTA), leucine, tryptophane and histidine (SD-LTH) or were screened for β-galactosidase expression (+X-gal). (B) The protein gel blot shown on the left-hand side verifies expression of the GST-fusion proteins in the extracts from bacteria producing GST-ZZ (2b), GsT-Joka8 (8) and GST-Joka20 (20); the GST-PB1 protein was not detected. The protein gel blot shown on the righthand side shows results of the “pull-down” assay performed as described in Materials and Methods. The results confirm interaction of UP9c with Joka8, Joka20 and the ZZ domain of Joka2; the extract containing His-tagged UP9C protein (C+) was loaded as a positive control and the arrows indicate the positions of the proteins corresponding to the expected sizes of the recombinant proteins: GST-ZZ (54.5 kDa), GST-Joka8 (66.5 kDa), GST-Joka20 (42 kDa) and His-UP9C (17.2 kDa). (C) The scheme of the protein (truncated NpJoka2) encoded by the insert present in pJoka2. The position of EcoRI used for the subcloning is shown and the domains PB1 and ZZ are indicated. (D) The pJK11 plasmid contains the insert as pJoka2 but in pGAD424. Plasmids pLUs1–4 contain the corresponding orfs (LSU1–4 from Arabidopsis) cloned into pGBT9. The remaining explanations as in (A).
Interactions between UP9C and the identified partners were confirmed by the “pull-down” assay (Fig. 1B). In order to check which of the two well-characterized protein domains, PB1 and ZZ, present in Joka2 (Fig. 1C) is involved in the interaction with UP9C, the DNA fragments containing each of the domains were cloned separately into the bacterial expression vectors. From the results of the pull-down experiment it can be concluded that either the ZZ domain or the PB1-ZZ interdomain region is responsible for interaction with UP9C. Subsequent Y2H experiments allowed us to exclude that the PB1-ZZ interdomain region is capable of UP9C binding (Fig. S1) and thus, the region of interaction can be limited to the ZZ domain. Additionally, we demonstrated that the LSU1–LSU4 proteins of A. thaliana are able to interact with Joka2 (Fig. 1D). This finding indicates that despite the relatively limited sequence similarity, the protein features responsible for interactions with Joka2 are conserved in the UP9/LSU family. It is worth to emphasize a clear difference in the strength of interaction of different LSU proteins with Joka2; LSU1 and LSU3 interact stronger than LSU2 and LSU4.
It was reasonable to anticipate that the Joka2 protein is longer than the one deduced from the cloned sequence, therefore we focused on cloning of the full-length cDNA from N. tabacum. Using the RT-PCR, 5′-RACE and 3′-RACE methods the cDNA encoding the full-length protein have been cloned. In addition to PB1 and ZZ domains, the deduced protein contained also a duplication of the ubiquitin associated domain (UBA/TS-N; pfam00627). All subsequent experiments were performed only using plasmids containing the N. tabacum cDNA.
Joka2 contains domains conserved in p62/SQSTM1 and NBR1 and forms homodimers.
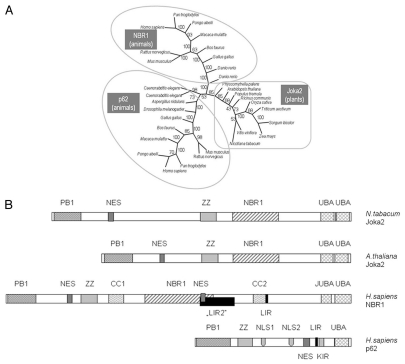
Proteins with a similar to Joka2 layout of domains exist in other eukaryotic organisms, both animals and plants (Fig. 2). The animal homologs can be divided into two families, NBR1-like and p62-like. The best characterized protein with a comparable arrangement of domains is the human protein p62/SQSTM1 also called Sequestosome-1, A170 or ZIP, which is a multifunctional protein implicated in several signal transduction pathways and is required for autophagic clearance of protein aggregates. It acts as a selective autophagy receptor by interacting with both ubiquitin conjugated to the target proteins and the ATG8 proteins present on the autophagosome.14,19,57 As shown in Figure 2B, p62 comprises a N-terminal region that includes PB1 domain (residues 20–102) and zinc finger (ZZ, residues 122–167), a central region containing LIR (LC3-interacting; residues 337–343) and KIR (Keap 1-interacting; residues 346–359), and C-terminal region encompassing a ubiquitin-associated domain (UBA, residues 391–436).21 A nuclear export signal (NES) and two basic monopartite nuclear localization signals (NLS1 and NLS2) are located between residues 303–320, 186–189 and 264–277, respectively.58 The three domains (PB1, ZZ and UBA) characterized in p62 can be also found in its plant homologs, though the plant proteins are generally larger (Fig. 2B and Table S1). The human NBR1 is not as well characterized as p62, however there is growing evidence about its role as a selective autophagy cargo receptor. Although the plant proteins are generally smaller than animal NBR1, a unique future of NBR1-like proteins (NBR1 domain) can be found between their ZZ and UBA domains. In addition, Joka2 proteins from tobacco and arabidopsis contain the potential nuclear export signal (NES) between PB1 and ZZ, and two adjacent UBA domains (UBA1 and UBA2) at their C-termini. The UBA1 domains from either NtJoka2 or AtJoka2 and the UBA2 domain from NtJoka2 are capable of forming amphipathic helix (not shown). Therefore, it is tempting to speculate that this region might have a similar role as the membrane-interacting JUBA domain of human NBR1, which is essential for NBR1 localization to the late endosomes.29
Figure 2.
The family of p62/NBR1/Joka2 proteins. (A) The phylogenic tree was constructed using full-length protein sequences by the parsimony methods and 100 bootstrap replicates using SEQBOOT, PROTPARS and CONSENS of the Phylip v.3.69 program package. The bootstrap values are given at the respective branches. The three subfamilies are marked and identified by the name of a typical member (p62, NBR1 or Joka2). The accession numbers of the proteins used in the analysis are shown in Table S1. (B) characteristic domains present in the p62, NBR1 and Joka2 proteins from Nicotiana tabacum, Arabidopsis thaliana and Homo sapiens. Proteins and domains are drawn to scale. See text for details and domains description.
We observed formation of Joka2-Joka2 dimers independently for NpJoka2 (Fig. 1) and for NtJoka2 (Fig. 3), in two separate Y2H experiments. The region necessary for such interaction appeared to contain PB1 domain and was mapped to the first 228 residues of NpJoka2 (see: pJK6, Table S2) and the first 422 residues (see: pDEST22/PB1 and pDEST32/PB1, Table S2) of NtJoka2. These results are consistent with the data for p62 showing that PB1 domain is crucial for the formation of multimers31 and seems to be in contrast with the general information available for NBR1 that this protein homodimerizes via its CC1 domain.59
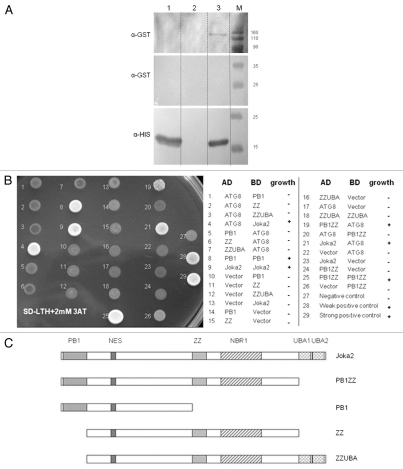
Figure 3.
Interactions of Joka2 and ATG8f. (A) The protein gel blot with anti-GST (α-GST) and anti-His (α-HIS) antibodies showing the results of the “pull-down” experiment using the mixture of crude exctracts prepared from the bacteria producing His-ATG8f and GST-Joka2. After affinity purification through the Ni column, both proteins are detected (lane 3) due to ATG8f-Joka2 interaction. The controls, without GST-Joka2 and without His-ATG8f, are shown in lanes 1 and 2, respectively. The negative controls with the GST-tag (produced from the empty vector pGEX4T-1) and his-ATG8f protein and with the GST-Joka2 protein and the His-tag (produced from the empty vector pET28a) are shown in lanes 1 and 2, respectively. For α-GST the high and the low range proteins are shown in the upper part and the middle part, respectively. The molecular marker (M) with the size (kDa) of the proteins is indicated. (B) The results of Y2H experiment and (C) the scheme of the Joka2 fragments present in the used plasmids. AD means fusions of the activating domain of GAL4 with the indicated protein or domain. BD means fusions of the DNA binding domain of GAL4 with the indicated protein or domain. Pluses and minuses indicate the growth and lack of growth, respectively, on the plate shown on the lefthand side. The growth is an indicator of protein-protein interaction. Proteins and domains shown in (B and C) are drawn to scale.
Joka2 interacts with ATG8.
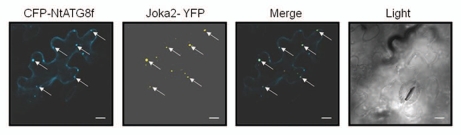
Despite identification of many autophagy-related (ATG) genes in plants, no reports on the presence or a potential function of any of the Joka2 homologs could be found in the literature. Therefore, it was important to confirm that Joka2 indeed resembles p62 concerning its most crucial and well-established feature, namely binding to ATG8/UBL proteins. In higher eukaryotes, both mammals and plants, there is a large family of ATG8/UBL-like proteins. For example, in the human genome there are six genes encoding ATG8 family members,9 while in A. thaliana nine genes encoding ATG8 family (AtATG8a–AtATG8i) are present and expressed.41,60 Function of AtATG8f under both favorable growth conditions and starvation stresses was previously investigated in reference 36 and 37, thus this isoform was our candidate of choice for testing if the members of ATG8 family would interact with Joka2. There are 15 known tobacco EST sequences encoding full-length proteins corresponding to ATG8 (Table S1). Based on phylogenic analysis (not shown) we estimated that TC107227 encodes the closest homolog of AtATG8f. This cDNA, referred to below as NtATG8f, has been cloned and used in pull-down and Y2H experiments to investigate its potential interaction with Joka2. In fact, the results shown in Figure 3 clearly indicated that such interaction takes place in both cases when the full-length NtATG8f and NtJoka2 proteins were used. Additionally, it was also possible to limit the region of NtJoka2 necessary for the interaction with NtATG8f to the residues 1–751 (out of 843 in total) in Y2H experiment. However, we found interaction of the trimmed down NtJoka2 with NtATG8f could only be observed if the former was present in the AD-plasmid but not in the BD-plasmid (compare colonies 19 and 20 in Fig. 3B). The LIR motif of mammalian p62, responsible for binding to ATG8/UBL proteins, is located between ZZ and UBA domains. The similar LIR motif as well as the atypical one have been located in NBR1. Several potential LIR motifs might be predicted in Joka2 and our results do not yet enable us to distinguish which of them function as binding sites for ATG8/UBL proteins. In addition to the interactions shown in Figure 3, the co-localization of Joka2-YFP and CFPATG8f was observed after transient expression of both proteins in N. benthamiana leaves (Fig. 4). This experiment indicated that Joka2-YFP and CFP-ATG8f might be present in common bodies, presumably autophagosomes. Taking into consideration the results shown in Figures 3 and 4 and the presence of potential LIR motifs in Joka2, it can be implied that both proteins interact in plant cells.
Figure 4.
Co-localization of Joka2-YFP and CFP-NtATG8f in Nicotiana benthamiana grown in sulfur-deficient medium for 2 d before agroinfiltration. The parts show accumulation of CFP-NtATG8f fluorescence signal (CFP-NtATG8f), accumulation of NtJoka2-YFP fluorescence signal (Joka2-YFP), overlay of both (Merge) and the corresponding tissues under bright field (Light). The white arrow indicates the site of signals co-localization. Scale bars (10 µm) are shown.
Dual localization of Joka2 in plant cells.
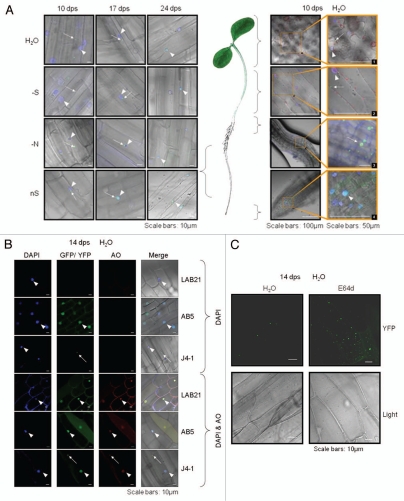
The J4 and J5 lines of tobacco (containing Joka2-YFP and Joka2-CFP expression cassettes, respectively) were obtained by agrobacteriummediated transformation of tobacco line LA Burley 21 with the binary pJ4 and pJ5 plasmids, respectively (Table S2). Expression of the transgenes in the selected transformants was confirmed (not shown) and the seeds from several transformants (J4-1, J4-2, J4-10, J5-1, J5-2, J5-3 and J5-6) were obtained after self-pollination. To monitor Joka2 localization in planta the seeds were germinated and seedlings were maintained in the following liquid media: H2O, S-deficient (S-), N-deficient (N-) and nutrient-sufficient (nS). Observations were performed on the 10th, 17th and 24th day-post-sowing (dps) in several lines. Since no apparent differences between J4 and J5 lines were noticed, localization of Joka2-YFP in J4-1 seedlings is shown as a typical example (Fig. 5). Distribution of Joka2-YFP was different depending on plant part, growth medium and the number of dps (Fig. 5A and Figs. S2–4). The largest differences were observed in the root elongation zone (Fig. 5A, the lefthand side parts). On the 10th dps the small cytosolic granules were present in all seedlings, however they were most numerous in seedlings grown in H2O. On the 17th dps the granules were larger, less frequent (1–2 per cell), located close to the nucleus and, in seedlings grown in nS, the fluorescence was observed also in the nucleus. On the 24th dps, regardless from the medium, the signal was present only in the nucleus. In contrast to the root elongation zone, distribution of Joka2-YFP in leaflets, hypocotyls and the root tips was not much affected by nutrients availability nor the number of dps, therefore for these parts only the results for the 10th dps and H2O are shown (Fig. 5A, the righthand side parts). Interestingly, in the root tips only the nuclear location of Joka2-YFP was observed. In leaflets and hypocotyl Joka2-YFP formed small cytosolic spots (a few per cell). It is worthwhile to emphasize a clear difference, which was observed in signal distribution between the hypocotyl (less intense signal) and roots (more intense spots).
Figure 5.
Localization of Joka2-YFP in tobacco seedlings. (A) The overlays of the signal for Joka2-YFP, DNA staining with DAPI, chlorophyll signal (observed as a red fluorescence of the chloroplasts; seen only in green parts) and brightfield image are shown for the indicated numbers of days post sawing (dps) in the indicated parts of the J4-1 seedlings grown in the indicated conditions (-S, sulfur starvation; -N, nitrogen starvation; nS, nutrient sufficient medium and water). Images 1–4 are magnifications of the indicated parts. Notice, a difference in the number of green spots between the roots (Joka2-YFP abundant) and the shoots (Joka2-YFP hardly detected), which is clearly observed in the shoot-root transition zone (magnified picture #3). Arrows point the cytosolic spots, while arrowheads the nuclei. (B) The signals of DAPI-stained nuclei, fluorescent protein (Joka2YFP for J4-1 or eGFP for AB5), red signal after acridine orange staining (AO) and the overlay of all is shown for the indicated plants: LA Burley 21 (LAB21) and transgenic AB5 (producing eGFP) used as a control and J4-1 grown in water for 14 d. extranuclear Joka2-YFP speckles and red spots stained by AO (if present) are shown by arrows. Arrowheads point the nuclei. (C) The treatment with protease inhibitor E64d increases the number of spots including Joka2-YFP. Both upper parts show the Joka2-YFP signal, while both lower parts show the corresponding tissues in the transparent view (Light). The scale bars are indicated. The separate signals used for the overlay are shown in Figures S2–5.
The punctated location of Joka2-YFP in the roots of seedlings grown in H2O was further investigated by acridine orange (AO) staining (Fig. 5B). The AO dye has been previously used to stain the autophagosomes in mammals,61 giving a red signal for the acidic compartments (autophagosomes but also RNA-containing structures) and a green signal when bound with DNA (nucleus). In addition to the J4-1 line producing Joka2-YFP, two tobacco lines served as controls: the parental line, LA Burley 21 and its derivative, the transgenic AB5 line producing EGFP. All seedlings were either stained with DAPI to visualize nuclei or double-stained with DAPI and AO to visualize also intracellular acidic vesicles. The single DAPI-staining allowed nuclei visualization in all three lines. Furthermore, as expected, the signal of Joka2-YFP in J4-1 was found only in cytosolic spots and no green signal was observed in LA Burley 21, while in AB5 seedlings it was abundantly observed in the nuclei and the cytosol due to a free distribution of GFP between these two compartments. Moreover, all seedlings double stained with DAPI and AO displayed blue, green and red fluorescence in the nucleus but only J4-1 displayed considerable fluorescence in intracellular spots outside the nucleus. The observed co-localization of the Joka2-YFP signal with the red AO signal in J4-1 seedlings indicates that these vesicles have acidic character, which in turn, is a characteristic feature of autophagic lysosomes.
An additional proof for location of Joka2-YFP in autophagosomes was obtained after application of cysteine protease inhibitor, E64d, to the J4-1 seedlings. This procedure resulted in an increased number of Joka2-YFP granules (Fig. 5C). It is well documented that E64d is an inhibitor of autophagic degradation of the proteins, what results in accumulation of undegraded cytoplasmic components in newly formed autolysosomes.49 This result suggests that Joka2, as its mammalian homologs, might be degraded during autophagy process.
Sulfur and nitrogen deficiency affects expression of Joka2 and ATG8.
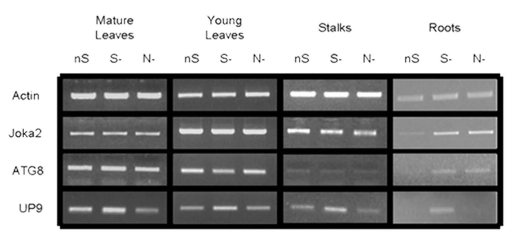
Expression of NtJoka2 and NtATG8f was monitored in different parts (young leaves, mature leaves, stalks and roots) of 2-mo-old LA Burley 21 plants grown for 2 d without sulfur (S-), without nitrogen (N-) or in the control (nutrient-sufficient; nS) conditions. The semiquantitative RT-PCR (sqRT-PCR) indicated that the expression of both genes, ATG8f and Joka2 was upregulated in roots but not in any shoot parts after 2 d of growth in either S- or N-conditions (Fig. 6). The expression of UP9C and UP9A genes (the primers used do not allow to distinguish between both genes) was used as a control and, as previously reported in reference 53, these genes were induced in all parts of the plants in S- but not in N-conditions. Interestingly, a previously unreported slight reduction of UP9C expression was observed in N- in mature leaves as compared with nS. It is an intriguing observation, however, the significance of such regulation is unclear.
Figure 6.
Expression of Joka2, ATG8f and UP9 in various parts of LA Burley 21 plants. The 8-week-old plants grown in nutrient-sufficient (nS) medium were transferred for 2 d into S- or N-deficient medium and, as a control, to nS again. Gene expression was monitored by sqRT-PCR. expression of actin (Tac9) served as a control. The UP9 contains the mixture of UP9C and UP9A (both induced by sulfur starvation) because the used primers amplify both products.
Overexpression of Joka2 decreases nutrient deficiency-induced yellowing of leaves.
Microscopy studies clearly suggested that transgenic tobacco lines generated in this study contained a high level of Joka2 fusion proteins. Preliminary examination of the transgenic J4 and J5 lines in the optimal growth conditions did not reveal any obvious phenotypes. Therefore, it was of interest to check if an excess of Joka2 protein would result in any noticeable phenotypes in suboptimal conditions.
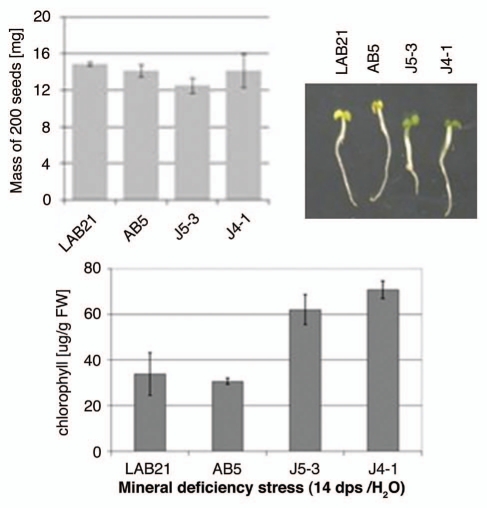
To check how Joka2 overproduction affects plant response to mineral deficiency stress, two transgenic lines, J4-1 and J5-3, were germinated in water in parallel with two control lines, the parental line, LA Burley 21 and the previously mentioned AB5 line, overexpressing EGFP. The yellowing of leaf tissue and chlorophyll content were monitored at the 14th dps. The J4-1 and J5-3 seedlings were greener and had higher chlorophyll contents than the control seedlings when maintained in H2O (Fig. 7), while no apparent difference was observed in seedlings maintained in the nutrient-sufficient medium (not shown). To exclude the possibility that the performance of the seedlings is linked to the seeds' mass, the weight of 200 seeds from each line was determined in triplicates. The calculated average values (shown in Fig. 7) revealed a lack of association of the response to mineral deficiency with the seed mass. Therefore, the observed difference in seedlings performance must depend on other factors, for example on efficiency of usage of the limiting nutrients.
Figure 7.
Effect of mineral deficiency stress on plants overproducing Joka2 fusion proteins. The mass of 200 seeds determined before sowing as well as the phenotype and chlorophyll content of tobacco seedlings germinated and maintained for 14 d in water are shown. LAB21, LA Burley 21; AB5, the line producing EGFP; J4-1, the line producing Joka2-YFP, J5-3, the line producing Joka2-CFP.
Discussion
Autophagy might be a highly selective process implicating cargo receptors in removing protein aggregates and damaged or excess organelles. A number of ubiquitin-binding proteins containing LIR motifs, responsible for their attachment to autophagosome-specific ATG8-like proteins, exist in mammals. Among them the best characterized are p62/SQSTM1 and NBR1. In plants, proteins sharing the characteristic motifs of p62 and NBR1 are present. The structural similarity suggests that they may possess a similar function. In this study we focused on the tobacco protein Joka2. Several lines of evidence certify that this protein is a structural and functional homolog of the mammalian autophagy cargo receptors: (i) conservation of characteristic domains (Fig. 2), (ii) formation of Joka2-Joka2 homodimers via PB1 domain (Fig. 3B), (iii) interaction and co-localization with ATG8 (Figs. 3 and 4), (iv) subcellular localization pattern and increased number of Joka2-YFP speckles in response to E64d treatment (Fig. 5). Accordingly, we propose that the process of selective autophagy functions in plants similarly to mammalian cells and involves both the core molecular machinery and the selective cargo receptor(s).
Only one isoform of Joka2-like protein was identified in N. tabacum. In addition, we were able to identify only one copy of Joka2-like gene in A. thaliana, At4g24690. This result suggests that Joka2 might be the only autophagy cargo receptor for protein aggregates in plants, in contrast to mammals, where at least two of them (p62 and NBR1) are present.17 Intriguingly, none of the several available T-DNA insertional mutants (SALK_053992, SALK_135513, SALK_144852, GK536D03.01) within At4g24690 gene appeared to be homozygous. Despite our intensive efforts we failed in obtaining homozygous T-DNA insertional mutants in the next generations and observed strongly reduced seed germination efficiency in these lines (Zientara-Rytter K, Sirko A, unpublished). It is tempting to speculate that At4g24690 might encode an essential protein, needed at the stage of seed germination, however the experimental evidence supporting this notion is still limited.
In our experiments (Fig. 5) Joka2-YFP was present in both the cytosolic granules (co-stained with AO, co-localized with ATG8) and in the nucleus (co-stained with DAPI). Mammalian p62 can also shuttle continuously between cytoplasm and the nucleus and it has the nuclear localization (NLS) and the nuclear export signals (NES). Shuttling of p62 between both compartments is modulated by its phosphorylation and aggregation.27,58 Unlike the cytosol, where two complementary proteolytic systems, lysosomes and proteasomes, are responsible for protein degradation, the nucleus is believed to have only the proteasomal system. Therefore, it was suggested that in the nucleus p62 could facilitate the recruitment of protein aggregates to the proteasome.58 Other researchers suggested that p62 can serve as an adaptor for proteasomal degradation of certain ubiquitinated proteins and that it has a ubiquitin-independent role in degradation of some autophagy substrates.62,63 On the other hand, it has been postulated that p62 can be implicated in two different, but not mutually exclusive, mechanisms of the crosstalk between the ubiquitin-proteasome and autophagy-lysosome systems. At time when autophagy operates at normal rate, p62 serves to deliver the ubiquitinated proteins for autophagosomal destruction. However, in the situation when autophagy is impaired, p62 prevents the proteasomal degradation of some proteins by competing with other ubiquitin binding proteins, which facilitate proteasomal degradation.64,65 In any case, the ability to reside in cytoplasmic speckles and in the nucleus suggests that p62 serves a complex role. Moreover, at least two distinct p62 subpopulations exist in cytoplasm, one within membrane-free aggregates and one with membrane-confined autophagosomal and lysosomal structures. A novel regulatory mechanism was recently proposed, where a membrane-free aggregate containing p62 would form in a rapidly reversible manner facilitating sequestration of specific cargo away from their normal, functionally important sites within the cell.66 Similar functions of the plant Joka2 protein can be considered, however at present they are too speculative.
Our results, including organ-specific regulation of NtJoka2 and NtATG8f transcription by N- and S-deficiency, interaction between NtJoka2 and NtUP9C, and increased tolerance of J4 and J5 plants to such environmental stresses as nutrient deficiency, strongly suggest the involvement of Joka2 (and indirectly, of selective autophagy) in plant response to these stresses. As shown in Figure 6 transcription of Joka2 and ATG8f is apparently induced in tobacco plants grown in either N or S deficiency. Interestingly, in the conditions of our experiments (2 d of nutrients deficit) expression of both genes was upregulated only in the roots but not in any shoot parts. This observation might be important in identification of the regulatory elements of the long-distance signaling of nutrients availability and of the overall plant nutritional status. The autophagy process in plants occurs as a general response to different abiotic stresses, including nutrient starvation and oxidative stress. It is also a mechanism for regulation of programmed cell death in response to pathogens and of developmentally regulated programmed cell death.67 Regulation of autophagy in mammals is better characterized. The process is controlled by pathways that interpret the status of cellular energy (AMP-dependent protein kinase) and nutrients (target of rapamycin), and by growth factors such as insulin.9 The previously considered candidates for regulators of ATG genes in plants include hormones such as cytokinins37 and ABA.68 The physiological role of autophagy in plants grown under starvation has been studied before and it has been reported that autophagy-defective mutants are impaired in autophagosome formation and cannot survive certain starvation conditions.46 The better performance of our transgenic tobacco lines containing increased amounts of Joka2YFP and Joka2-CFP resemble effects observed in A. thaliana overproducing GFP-ATG8f-HA fusion.37 One could speculate that the decreased leaf yellowing of J4-1 and J5-3 lines during stress might be an indication of better recycling of the waste due to the enhanced autophagy, however more studies are needed to understand this process and the observed phenotypes.
To our knowledge no reports about any role of autophagy in plants during sulfur starvation existed previously despite work on autophagy function in response to sugar, carbon and nitrogen starvation.67 Therefore, identification of Joka2 as a partner of UP9C protein was surprising. The function of UP9/LSU-like proteins is unknown but UP9C has been recently recognized as required for adequate plant response to sulfur deficit.53 It is possible to speculate that some UP9/LSU-like proteins, especially those encoded by genes strongly induced during sulfur deprivation, might link sulfur deficiency response with selective protein degradation through autophagy or in proteasomes. As demonstrated in our laboratory, UP9C is able to interact with a variety of protein partners, including proteins involved in synthesis and signaling of important phytohormones, such as jasmonic acid and ethylene. Concerning the possible mechanisms and consequences of Joka2-UP9/LSU interactions several hypotheses can be considered: (i) UP9/LSU can modulate certain properties of Joka2, e.g., subcellular localization, cargo recruitment and/or ability to form multimers or aggregates—to our knowledge this would be a novel mechanism not found for p62; (ii) UP9/LSU could act as adaptors between different cargo proteins and the cargo receptor, Joka2—to our knowledge no such adaptors were proposed for p62; (iii) during sulfur starvation (or possibly other conditions) Joka2 could sequester UP9/LSU away from their functionally relevant sites or targets in cells. This is an interesting hypothesis, because at least two examples of such effects exist for p62. The first concerns cyclic AMP phosphodiesterase66 and the second—Keap1 (kelch-like ECH-associated protein 1).21 Unfortunately, since the molecular function of UP9/LSU is not yet characterized, it is rather difficult to verify the consequences of Joka2 binding. In any case, Joka2-UP9/LSU interaction might form a novel regulatory mechanism responsible for fast functional inactivation or protection of the target proteins. Consequently, the process would ensure the swift plant adjustment to nutrient availability. The above hypotheses are currently under study in our laboratory.
Although a complete understanding of the mechanisms explaining the role of Joka2 in tobacco plants is missing, our results strongly suggest that this protein is a structural and functional homolog of mammalian selective autophagy receptors, p62 and NBR1.
Finally, it is worthwhile to mention that the transgenic tobacco plants described in this work, which overproduce Joka2-YFP and Joka2-CFP fusion proteins, might be a useful tool for studying the process of autophagy in plants. Recently, a reporter system to monitor autophagy in mammalian cells based on p62 has been proposed.69 According to the authors, the GFP-p62 performed the best among the three tested reporter fusions (GFP-LC3, GFP-NBR1 and GFP-p62) and was the most useful in screening for compounds or conditions that affected the rate of autophagy.
Materials and Methods
DNA methods and plasmids construction.
Plasmids used in this work are described in Table S2. Details on their construction are available upon request. Gateway BP recombination by using attB-tailed gene-specific primers or cDNA cloning into pENTR™/D-TOPO vector and Gateway LR recombination reactions were done as described in the Gateway® Technology—a universal technology to clone DNA sequence for functional analysis and expression in multiple systems (Invitrogen, 12535-019 and 12535-027). Oligonucleotides for PCR, RT-PCR, RACE and DNA sequencing are listed in Table S3. All plasmids were checked by restriction digestion and/or by DNA sequencing. Conventional techniques were used for DNA manipulation and Escherichia coli transformation.70 The cDNA synthesis and semiquantitative RT-PCR (sqRT-PCR) were conducted as previously described in reference 71. RNA isolation and mRNA purification for the 5′- and the 3′-RACE were performed using the SMART™ RACE cDNA Amplification Kit (BD Biosciences-Clontech, 634914) according to the procedure recommended by the manufacturer.
Yeast two-hybrid experiments.
The N. plumbaginifolia cDNA library in pGAD10 was a kind gift of Dr. Witold Filipowicz, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland. Manipulation of yeast cells and library screening were performed according to standard protocols (Clontech Yeast Protocol Handbook, PT3024-1). The PJ69-4 72 strain of Saccharomyces cerevisiae was used for transformation and approximately 6.5 x 106 transformants were placed on selective medium lacking leucine, tryptophan and histidine (SD-LTH). True positive prey clones, after retransformation, were confirmed for their ability to activate the three reporter genes, HIS3, ADE2 and lacZ, when cotransforming yeasts with pJK1 as a bait. For simultaneous transformation with the defined “bait” and “prey” plasmids, since just several transformants were sufficient for further applications, the “Quick and Easy TRAFO Protocol” available at www.umanitoba.ca/faculties/medicine/biochem/gietz/ and the S. cerevisiae AH109 strain with four reporter genes (HIS3, ADE2, MEL1 and lacZ) were used.
Immunoblots and “pull-down” assay.
Prior to the “pulldown” assay the presence of recombinant proteins fused to GST or his-tag in the respective bacterial extracts was verified by immunoblots using mouse monoclonal or rabbit polyclonal anti-GST IgG and rabbit polyclonal anti-his IgG (Sigma-Aldrich, G1160 or Santa Cruz Biotechnology Inc., SC-33613 and H-15 SC-803) as primary antibody and anti-rabbit IgG or anti-mouse IgG conjugated to alkaline phosphatase (Sigma-Aldrich, A3687 or A3562) as secondary antibody. The “GST pull-down” assay consisted of the following steps: (1) the protein extract from bacteria producing recombinant His-tagged UP9C was mixed with the extracts from bacteria producing GST-ZZ (the part of Joka2 with ZZ domain), GST-Joka8, GST-Joka20 or GST; (2) extracts were incubated on ice for 6 h with gentle rocking; (3) proteins were purified in the native condition on Glutathione Sepharose column (GE Healthcare Bio-Sciences AB, 17.0756-01); (4) protein gel blots were performed with appropriate antibodies. The “HIS pull-down” assay consisted of the following steps: (1) the protein extract from bacteria producing recombinant His-tagged ATG8f was purified in the native condition on HIS-select HF Nickel Affinity Gel (Sigma-Aldrich, H0537), (2) the protein extract from bacteria producing GST-tagged Joka2 protein was incubated with gel in 4°C overnight with gentle rocking; (3) proteins were purified and eluated in the native condition; (4) protein gel blots were performed with apropriate antibodies.
Plant transformation and transient expression.
The pJ4 and pJ5 plasmids were used for stable agrobacterium-mediated transformation of N. tabacum cv LA Burley 21 as previously described in reference 73. Kanamycin- or hygromycin-resistant plants were selected and self-pollinated to obtain T2 generation. For transient co-expression of CFP-NtATG8f and Joka2-YFP proteins in N. benthamiana leaves a fresh overnight cultures of A. tumefaciens, containing the appropriate binary plasmids, were spun down, washed twice and re-suspended in sterile water to obtain a cell density of 2 × 108 cfu/ml (OD600 ∼ 0.2), then cultures were mixed and diluted to a final OD600 ∼ 0.4. Young N. benthamiana plants with fully expanded leaves of about 5 cm in diameter were transfer to S-conditions 2 d before infiltration of the leaves with the prepared bacterial suspension using needleless syringe. Leaves were harvested and analyzed under confocal microscope 3 d after agroinfiltration.
Plant growth conditions and chlorophyll measurement.
Tobacco seeds were surface-sterilized in microcentrifuge tubes using a vapor-phase seed sterilization method. Shortly, tubes with seeds were placed into a desiccator jar along with two beakers, each containing 50 ml of bleach and 1.5 ml of concentrated HCl. Sterilization by chlorine fumes was continued for 3 h. Then, centrifuge tubes containing seeds were placed in a sterile laminar flow hood and left open for 1 h. Depending on the experiment, the seeds were spread into water or on plates with modified 0.5 x Hoagland medium either full (nS) or lacking nitrogen (N-) or sulfur (S-). In N-medium the equimolar amounts of KOH, KH2PO4 and CaCl2 were used instead of KNO3, NH4H2PO4 and Ca(NO3)2, while in S-medium the equimolar amounts of MgCl2 replaced MgSO4. The medium was either liquid (in Erlenmayer flasks or hydroponic containers) or solidified with agar (0.8% w/v). The seedlings were incubated in a growth chamber under a long-day regime of 16 h light/8 h dark cycle at 24°C. For testing the effect of mineral deficiency, the sterilized seeds were germinated and seedlings were maintained for 14 d in H2O. Entire seedlings were collected and the chlorophyll content was measured as previously described in reference 74.
Microscopy methods.
The stock solution of E64d protease inhibitor (Alexis, BML PI107 0001; 10 mg/ml in ethanol) and fluorescent dyes [DAPI (Sigma-Aldrich, D9561; 1 mg/ml in DMSO), acridine orange (AO) (Invitrogen, A3568; 10 mg/ml in H2O)] were prepared. Tobacco seedlings were washed and incubated with DAPI (1 µg/ml) and/or AO (1 µg/ml) for 15 min in the darkness at room temperature or with E64d (1 µg/ml) for 1 d. After the treatment, seedlings were washed in water (3 times, 5 min each) and observed using a confocal microscope. The images were obtained at the Laboratory of Confocal and Fluorescence Microscopy at IBB PAS with a Nicon confocal microscope, Eclipse TE2000-E and processed using EZ-C1 3.60 FreeViewer software. For GFP/YFP or AO (green signal) imaging, the 488nm line from an Argon-Ion Laser (40 mW) was used for excitation, and a 500–530 nm or a 565–640 nm band pass filter for detection of emission. The 543 nm line of a Green He-Ne Laser (1.0 mW) and a 650 nm long pass filter were used for chlorophyll emission and detection, respectively. The blue fluorescence of DAPI or CFP was imaged using 404 nm Violet-Diode Laser MOD (44.8 mW) for excitation and a 430–465 nm or 435–485 nm band pass filter for emission.
Computer analysis and accession numbers.
Similarity searches were performed with BLASTP at NCBI (www.ncbi.nlm. nih.gov/BLAST/) or with BLASTN using TGI Database of plant EST at DFCI (http://compbio.dfci.harvard.edu/tgi/plant.html). Translation of nucleotide sequences was generated at EMBL-EBI (www.ebi.ac.uk/services/index.html). Multiple sequence alignment was generated by MAFFT ver.5.667 using the E-INS-i strategy (http://timpani.genome.ad.jp/∼mafft/server/).75 Phylogeny was inferred by programs from the Phylip v3.36 package.76 Sequences were aligned in Phylip-format using T-coffee (www. ch.embnet.org/cgi-bin/tcoffee_parser). The following programs were accessed through the ExPASy Proteomic Server (www.expasy.ch): GOR477 used for secondary structure prediction, PSORT78 used for predicting the localization, SMART (http://smart.embl-heidelberg.de),79 MOTIFSCAN80 and EML Server (http://elm.eu.org) for identification of the protein domains and patterns.
The accession numbers of the sequences used in this study are listed in Table S1.
Acknowledgements
Marta Piecho is acknowledged for excellent technical assistance, Dali Gaganidze for help with pull-down and Anna Anielska-Mazur for help with microscope techniques. We are grateful to Prof. Teresa Zoładek for her comments on the manuscript. This work was supported by the Polish Ministry of Higher Education and Science (grant Nr N N302 119435).
Abbreviations
- AD
activation domain (of GAL4)
- AO
acridine orange
- BD
DNA binding domain (of GAL4)
- CFP
cyan fluorescent protein
- dps
days post sowing
- EGFP
enhanced green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3
- LIR
LC3 interacting region
- LSU
low sulfur upregulated
- NBR1
neighbor of BRCA1 gene 1
- NES
nuclear export signal
- NLS
nuclear localization signal
- PB1 domain
Pho and Bem1 domain
- UBA domain
ubiquitin-binding domain
- UBL
ubiquitin-like
- Y2H
yeast two-hybrid
- YFP
yellow fluorescent protein
- ZZ domain
ZZ-type zinc finger domain
Supplementary Material
References
- 1.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. nrm2245. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6:438–448. doi: 10.4161/auto.6.4.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol-3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 7.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis and necrotic signals promotes T-cell homeostasis. Immunol Rev. 2010;236:95–109. doi: 10.1111/j.1600-65X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreau K, Luo S, Rubinsztein DC. Cytoprotective roles for autophagy. Curr Opin Cell Biol. 2010;22:206–211. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo EK. Innate immunity to mycobacteria: vitamin D and autophagy. Cell Microbiol. 2010;12:1026–1035. doi: 10.1111/j.1462-5822.2010.01491.x. [DOI] [PubMed] [Google Scholar]
- 13.Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 16.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Kirkin V, Lamark T, Johansen T, Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 19.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 22.Du Y, Wooten MC, Gearing M, Wooten MW. Ageassociated oxidative damage to the p62 promoter: implications for Alzheimer disease. Free Radic Biol Med. 2009;46:492–501. doi: 10.1016/j.freeradbiomed.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Y, Wooten MC, Wooten MW. Oxidative damage to the promoter region of SQSTM1/p62 is common to neurodegenerative disease. Neurobiol Dis. 2009;35:302–310. doi: 10.1016/j.nbd.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NFkappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NFkappaB activation. J Biol Chem. 2010;285:2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 28.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Mardakheh FK, Auciello G, Dafforn TR, Rappoport JZ, Heath JK. Nbr1 is a novel inhibitor of ligandmediated receptor tyrosine kinase degradation. Mol Cell Biol. 2010;30:5672–5685. doi: 10.1128/MCB.00878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 31.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 32.Waters S, Marchbank K, Solomon E, Whitehouse C, Gautel M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 2009;583:1846–1852. doi: 10.1016/j.febslet.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 33.Reumann S, Voitsekhovskaja O, Lillo C. From signal transduction to autophagy of plant cell organelles: lessons from yeast and mammals and plant-specific features. Protoplasma. 2010 doi: 10.1007/s00709-010-0190-0. [DOI] [PubMed] [Google Scholar]
- 34.Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 35.Yoshimoto K, Takano Y, Sakai Y. Autophagy in plants and phytopathogens. FEBS Lett. 2010;584:1350–1358. doi: 10.1016/j.febslet.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Slavikova S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, et al. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot. 2005;56:2839–2849. doi: 10.1093/jxb/eri276. [DOI] [PubMed] [Google Scholar]
- 37.Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G. An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot. 2008;59:4029–4043. doi: 10.1093/jxb/ern244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitinlike proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
- 41.Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010;62:483–493. doi: 10.1111/j.1365313X.2010.04166.x. [DOI] [PubMed] [Google Scholar]
- 42.Phillips AR, Suttangkakul A, Vierstra RD. The ATG12conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178:1339–1353. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson AR, Vierstra RD. Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol. 2005;8:165–173. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277:33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- 45.Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, et al. Mobilization of rubisco and stromalocalized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 2008;148:142–155. doi: 10.1104/pp.108.122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006;47:1641–1652. doi: 10.1093/pcp/pcl031. [DOI] [PubMed] [Google Scholar]
- 50.Moriyasu Y, Hattori M, Jauh GY, Rogers JC. Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process. Plant Cell Physiol. 2003;44:795–802. doi: 10.1093/pcp/pcg100. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewandowska M, Wawrzynska A, Kaminska J, Liszewska F, Sirko A. Identification of novel proteins of Nicotiana tabacum regulated by short term sulfur starvation. In: Saito K, De Kok LJ, Stuhlen I, Hawkesford MJ, Schnug E, Sirko A, editors. Sulfur transport and assimilation in plants in the postgenomic era. Leiden, The Netherlands: Backhuys Publishers; 2005. pp. 153–156. [Google Scholar]
- 53.Lewandowska M, Wawrzynska A, Moniuszko G, Lukomska J, Zientara K, Piecho M, et al. A Contribution to identification of novel regulators of plant response to sulfur deficiency: Characteristics of a tobacco gene UP9C, its protein product and the effects of UP9C silencing. Mol Plant. 2010;3:347–360. doi: 10.1093/mp/ssq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myakushina YA, Milyaeva EL, Romanov GA, Nikiforova VY. Mutation in LSU4 gene affects flower development in Arabidopsis thaliana. Doklady Biochem Biophys. 2009;428:257–260. doi: 10.1134/S16076729050093. [DOI] [PubMed] [Google Scholar]
- 55.Elhag GA, Thomas FJ, McCreery TP, Bourque DP. Nuclear-encoded chloroplast ribosomal protein L12 of Nicotiana tabacum: characterization of mature protein and isolation and sequence analysis of cDNA clones encoding its cytoplasmic precursor. Nucleic Acids Res. 1992;20:689–697. doi: 10.1093/nar/20.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cel. 2003;15:2497–2502. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 58.Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285:5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 60.Hayward AP, Tsao J, Dinesh-Kumar SP. Autophagy and plant innate immunity: Defense through degradation. Semin Cell Dev Biol. 2009;20:1041–1047. doi: 10.1016/j.semcdb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 61.Mitou G, Budak H, Gozuacik D. Techniques to study autophagy in plants. Int J Plant Genomics. 2009;2009:451357. doi: 10.1155/2009/451357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geetha T, Seibenhener ML, Chen L, Madura K, Wooten MW. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem Biophys Res Commun. 2008;374:33–37. doi: 10.1016/j.bbrc.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, et al. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 64.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 65.Lamark T, Johansen T. Autophagy: links with the proteasome. Curr Opin Cell Biol. 2010;22:192–198. doi: 10.1016/j.ceb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 66.Christian F, Anthony DF, Vadrevu S, Riddell T, Day JP, McLeod R, et al. p62 (SQSTM1) and cyclic AMP phosphodiesterase-4A4 (PDE4A4) locate to a novel, reversible protein aggregate with links to autophagy and proteasome degradation pathways. Cell Signal. 2010;22:1576–1596. doi: 10.1016/j.cellsig.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 67.Bassham DC. Function and regulation of macroautophagy in plants. Biochim Biophys Acta. 2009;1793:1397–1403. doi: 10.1016/j.bbamcr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 68.Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larsen KB, Lamark T, Overvatn A, Harneshaug I, Johansen T, Bjorkoy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy. 2010:6. doi: 10.4161/auto.6.6.12510. [DOI] [PubMed] [Google Scholar]
- 70.Sambrook J, Frisch EF, Maniattis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 71.Wawrzynska A, Lewandowska M, Hawkesford MJ, Sirko A. Using a suppression subtractive library-based approach to identify tobacco genes regulated in response to short-term sulphur deficit. J Exp Bot. 2005;56:1575–1590. doi: 10.1093/jxb/eri152. [DOI] [PubMed] [Google Scholar]
- 72.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wawrzynski A, Kopera E, Wawrzynska A, Kaminska J, Bal W, Sirko A. Effects of simultaneous expression of heterologous genes involved in phytochelatin biosynthesis on thiol content and cadmium accumulation in tobacco plants. J Exp Bot. 2006;57:2173–2182. doi: 10.1093/jxb/erj176. [DOI] [PubMed] [Google Scholar]
- 74.Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophyls a and b of leaf extracts in different solvents. Biochem Soc Trans. 1983;11:591–592. [Google Scholar]
- 75.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Felsenstein J. PHYLIP (Phylogeny Interference Package) version 3.6. Seattle: Department of Genome Sciences, University of Washington; 2004. Distributed by the author. [Google Scholar]
- 77.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 78.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 79.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, et al. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:142–144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360138502022513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.