Abstract
Reprogramming energy metabolism from oxidative phosphorylation to aerobic glycolysis, a common feature of human cancer, is associated with a relative acidic tumor microenvironment which can sometimes be further accentuated by hypoxia operating within most solid tumors. We found that alteration of extracellular pH induces marked and rapid changes of autophagic activity. Interestingly, acidic and basic conditions induced completely opposite effect on autophagy, with its activity suppressed at lower pH whereas stimulated at higher pH. Gene knockdown experiments indicated that pH induced-autophagy requires Beclin 1, Vps34 and Atg5, key components of the autophagy pathway. Of note, an acidic condition not only inhibits the basal but also blocks the starvation-induced autophagy activity. Significantly, examination of different areas of tumor mass revealed a lower autophagic activity within the inner region than the outer region. These findings have important implications on the connections between autophagy and cancer as well as a wide range of other physiological and pathological processes.
Keywords: autophagy, extracellular pH, cancer, microenvironment, Beclin 1, VPS34, Atg5
Introduction
Autophagy, a self-catabolic process which maintains intracellular homeostasis by degrading redundant or faulty cell components, occurs both as a cell's constitutive activities and as a response to stressful stimuli, such as starvation.1,2 Although accumulating evidence has indicated a close connection between autophagy and cancer, the role of autophagy in cancer is still complex, with associations with both tumor suppression and cancer cell survival.3 For instance, reduced expression of autophagy-specific genes, such as beclin 1, results in increased tumorigenesis in mice, and enforced expression of such genes (becn1, atg5) suppresses the formation of tumors in vivo.4,5 In contrast, autophagy has also been reported to be important for the maintenance of oxidative metabolism and tumorigenesis in models of aggressive cancers with active Ras oncogene.6
Published data suggest that the tumor microenvironment can significantly affect the activity of autophagy. For example, low oxygen tension or hypoxia, a common condition in solid tumors, activates autophagy in cancer cells through both HIF-1 dependent and independent pathway.7,8 Indeed, Degenhardt et al. showed that autophagic activity was induced predominantly in the tumor prior to acquisition of a blood supply during epithelial tumorigenesis.9 These data indicate a close interaction between tumor microenvironment and autophagy.
An important change in the tumor microenvironment is pH. Extracellular pH in a wide array of cancers has been determined to be significantly more acidic than that in normal tissues, which may further decrease as tumor size increases.10–14 For instance, electrode measurements show that pH in human brain tumors can be as low as 5.9, whereas normal brain tissue has a pH of about 7.1.11 Similar observations have also been made by magnetic resonance spectroscopy.13 Together, these data demonstrate that acidic stress is a common feature of solid tumors.
A change in the environmental pH may play an important pathological role in the context of tumorigenesis. On the one hand, cancer cells must evolve a mechanism to survive in the acidic microenvironment; on the other hand, acidic extracellular pH has a wide impact on various biological processes, including cell proliferation, migration and angiogenesis.11,13–17 For instance, acidic extracellular pH activates Ras and the ERK1/2 MAPK pathway to enhance VEGF production in human cancer cells.18 Recently, Hjelmeland et al. showed that culturing glioma cells under acidic conditions promoted the expression of HIF-2a as well as its downstream targets, and induced a cancer stem cell phenotype.19 Interestingly, recent reports have demonstrated that tumoral acidosis can be reduced by increasing systemic concentrations of pH buffers, inhibition of carbonic anhydrase IX and proton pump, which leads to suppression of malignant growth, suggesting the potential exploitation of the pH environments for the treatment of cancer.12,20–23 Moreover, changes in extracellular pH also play important roles in immune function, bone function and the nervous system.24–26
Here we sought to determine the effect of pH environment on autophagy and to show that higher pH induces autophagy, whereas lower pH suppresses both basal and starvation-induced autophagy. Interestingly, examination of different areas of tumor mass revealed a lower autophagic activity within the inner region than the outer region of tumor. Our data implicated the potential interaction between changes in tissue microenvironment and autophagy regulation.
Results and Discussion
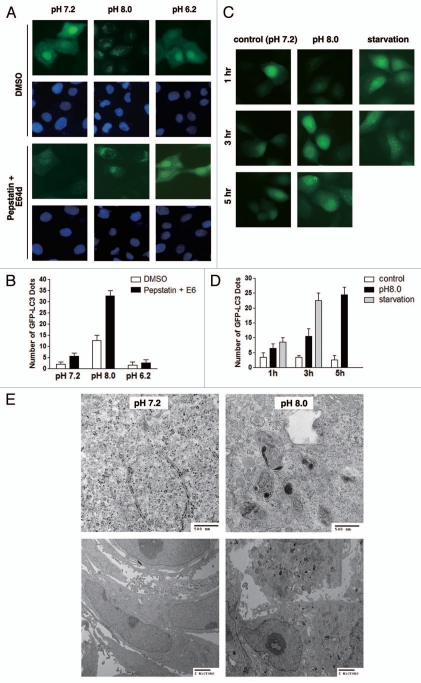
To investigate whether autophagic activity is modulated by extracellular pH, CO2-independent medium was used to avoid the interference of CO2 in the medium pH. We first studied GFP-LC3 punctate formation in MCF-10A cells that were cultured in media of different pHs. As shown in Figure 1A and B, after 6 h of incubation, whereas cells in basic pH (8.0) exhibited significantly increased punctate formation, culturing cells in lower pH (6.2) resulted in a marked reduction in the number of GFP punctates, suggesting the regulation of autophagy by extracellular pH environment. Furthermore, we determined the time dependency of punctate formation by performing live cell imaging. We found that GFP-LC3 punctate formation in MCF-10A cells started as early as 3 h after switching to basic pH media, which was further enhanced over time (Fig. 1C and D). To further confirm this finding, MCF-10A cells were cultured under condition of basic pH (pH = 8.0) for 6 h, fixed, embedded and examined by transmission electron microscope. Indeed, we found cells exposed to basic pH displayed clear induction of autophagy as evidenced by the formation of numerous autophagic vacuoles (Fig. 1E).
Figure 1.
Assessment of pH-regulated autophagy by microscopy. (A and B) Regulation of autophagy by extracellular pH. MCF-10A cells stably expressing GFP-LC3 minigene were incubated in CO2-independent media of different pHs for 6 h. Cells were fixed with methanol and DAPI-staining was employed to visualize the nuclei. Fluorescent images were taken at 600X magnification (A) and quantification was performed in at least 50 cells (B). (C and D) Rapid induction of GFP-LC3 punctate formation by basic pH. MCF-10A cells stably expressing GFP-LC3 minigene were incubated in control or pH 8.0 CO2-independent medium for the indicated period of time. Cells starved with EBSS served as a positive control of GFP-LC3 punctate formation. Fluorescent images were taken at 600x magnification (C) and quantification was performed in at least 50 cells (D). (E) Ultrastructural examination of MCF-10A cells after incubation in control or basic medium for 6 h by transmission electron microscopy.
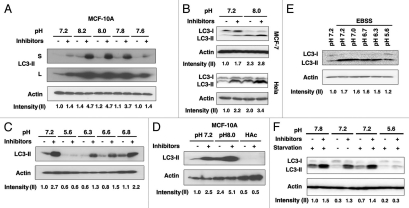
To eliminate the possibility that basic pH environment blocks lysosomal degradation rather than induces autophagy, LC3 was used as a molecular marker to investigate the autophagic flux.27 Consistent with the microscopic data, we found a clear increase of LC3-II after 6 h of incubation in basic pH (Fig. 2A). Importantly, LC3-II levels further accumulated even in the presence of lysosomal protease inhibitors comprising Pepstatin and E64d (Fig. 2A), suggesting that basic pH induces the autophagic flux in MCF-10A cells. Since the expression of LC3-I in MCF-10A cells was not detectable under our experimental conditions, we performed this LC3 conversion assay in additional cell lines including HeLa, MCF-7 and PANC-1 and observed a similar effect in all the cell lines tested (Fig. 2B and data not shown). These data suggested that induction of autophagy is a general cellular response upon basic pH environment. Interestingly, lower pH (<6.8) suppressed the basal autophagic activity as shown by the smaller accumulation of LC3-II upon lysosomal blockage (Fig. 2C). The suppressive effect of acidic pH on basal autophagic activity was further confirmed by incubating the cells in medium acidified with acetic acid (0.1%, Fig. 2D). To examine the effect of acidic pH on induced autophagic activity, cells were starved under the condition of either normal or acidic pH for 3 h and subjected to protein gel blot analysis for LC3 conversion. As shown in Figure 2E and F, LC3 I-II conversion was increased upon starvation. However, this induction was remarkably suppressed by acidic pH, suggesting that acidic extracellular pH inhibits not only basal but also starvation-induced autophagy. Interestingly, we did not see a significant additive effect when the cells were starved under basic pH condition, suggesting that the same regulatory pathway may be involved (Fig. 2F). These data implicated the potential effect of acidic tumor microenvironment on autophagy regulation.
Figure 2.
Assessment of pH-regulated autophagy by LC3 protein gel blotting in various cell lines. (A) Dose-dependency of autophagy induction by basic pH in MCF-10A cells. Cells were incubated in medium of indicated pH for 6 h in the presence or absence of lysosome inhibitors. Cell lysates were subjected to protein gel blot for LC3. Both longer (L) and shorter (S) exposures of the LC3 blot were shown. Quantification was done based on the shorter exposure. (B) Basic pH induces autophagy in MCF-7 and HeLa cells. Cells were treated with medium of indicated pH and LC3 conversion was evaluated by protein gel blotting in the presence or absence of lysosomal inhibitors. (C) Inhibition of basal autophagy by acidic pH in MCF-10A cells. Cells were incubated in medium of indicated pH for 6 h in the presence or absence of lysosome inhibitors. Cell lysates were subjected to protein gel blot for LC3. (D) Acetic acid (HAc) inhibits basal autophagy. MCF-10A cells were incubated in medium of indicated pH or acidified with 0.1% HAc, plus or minus lysosomal inhibitors. (E) Acidic pH suppresses starvation-induced autophagy. HeLa cells were starved with EBSS at indicated pH for 3 h. After that, cells were lysed and lysates were subjected to protein gel blotting. (F) HeLa cells were starved with EBSS at indicated pH for 3 h in the presence or absence of lysosomal inhibitors. After that, cells were lysed and lysates were subjected to protein gel blotting. Band densitometry quantification and the calculation of relative LC3-II intensity were performed as described in Materials and Methods.
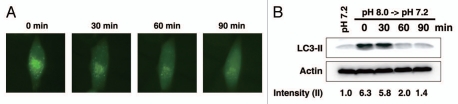
We further investigated the reversibility of autophagic induction in response to basic pH by performing live cell imaging. Cells were incubated in basic medium (pH 8.0) for 6 h, following which the medium was changed to control pH (pH 7.2) and time-lapse images were taken every 30 min (Fig. 3). Interestingly, we found that GFP-LC3 punctates began vanishing as soon as 30 min after cells were incubated in control medium, which completely disappeared after 2 h of incubation, indicating that autophagy regulation is a rapid cellular stress response to pH changes.
Figure 3.
Reversibility of basic pH-induced autophagy. MCF-10A cells were incubated in pH 8.0 CO2-independent medium for 6 h. After that (0 min), medium was replaced with control (pH 7.2) CO2-independent medium. Live cell imaging (A) and protein gel blot for LC3 (B) were employed to examine the reversibility of basic pH-induced autophagy. Green fluorescent images were taken every 30 min. Images were taken at 600x magnification. Band densitometry quantification and the calculation of relative LC3-II intensity were performed as described in Materials and Methods.
To gain further understanding of environmental pH-modulated autophagy, we measured the intracellular pHs when cells were exposed to media of different pHs with a pH-sensitive fluorophore SNARF-1 by flow cytometry.28 We found that the intracellular pH changed along with the extracellular pH, albeit to a lesser extent. Specifically, cells were incubated in media of different pHs including pH 8.7, 8.0, 7.8, 7.2, 6.8 and 6.3. The intracellular pHs measured after 6 h were 7.9, 7.5, 7.4, 7.1, 6.9 and 6.4, respectively. These data suggested that changes of extracellular pH can induce corresponding pH changes inside the cells, affecting autophagic activity.
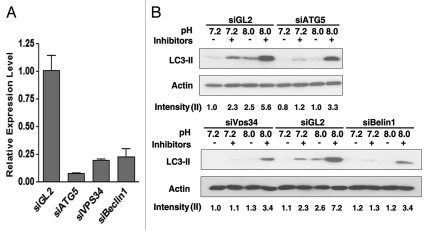
To explore mechanisms for the pH-dependent autophagic response, we examined the canonical components of the autophagy pathway including Vps34, Beclin1 and Atg5.29 To this end, we transfected MCF-10A cells with siRNA targeting each gene. After 48 h, cells were incubated in basic pH medium (pH = 8.0). Autophagy induction was measured by protein gel blot for LC3. As shown in Figure 4A, transfection of siRNAs efficiently reduced the mRNA levels of target genes. Furthermore, silence of these genes suppressed the induction of autophagy upon basic pH treatment (Fig. 4B). The incomplete suppression as observed in this assay is likely due to incomplete knockdown of the target genes. These data indicated that basic pH induces autophagy at least partially mediated by the canonical autophagy pathway.
Figure 4.
Basic pH-induced autophagy is mediated by Atg5, Vps34 and Beclin 1. MCF-10A cells were reverse-transfected with indicated siRNAs and cultured for 48 h. (A) Knockdown efficiency was measured by real-time PCR assay. siGL2, which targeted the luciferase gene in the pGL2 construct, was used as the negative control. (B) Cells were incubated in medium of indicated pH in the presence or absence of lysosomal inhibitors Pepstatin and E64d. After 6 h, LC3 level was measured by protein gel blotting followed by densitometry quantification using the Image J program. The calculation of the relative LC3-II intensity was performed as described in Materials and Methods.
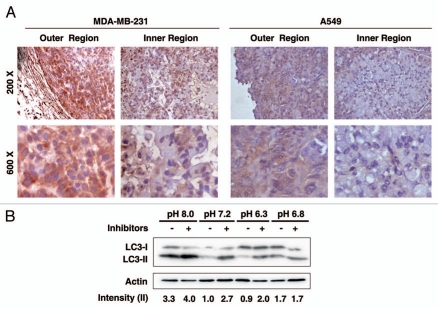
Previous studies have shown that, in a wide array of cancers, the extracellular pH in solid tumor is significantly more acidic than that in normal tissues. The pH may further decrease as tumor size increases.10–13 Interestingly, Hjelmeland et al. showed that the intratumoral extracellular pH at the center of the glioma xenograft was even further decreased when compared with the tumor edge.19 Therefore, we tested the potential relevance of the pH-regulated autophagy in the context of solid xenograft tumors. MDA-MB-231 cells were implanted subcutaneously into the posterior flank of BALB/c athymic nude mice at 4–6 weeks of age. Two weeks after injection, two independent tumors were dissected and examined. Immunohistochemical staining of LC3 was used as a surrogate marker for autophagy in the tumor specimens as described previously in reference 30. Interestingly, we found that LC3 accumulated at a noticeably higher level in the outer region as compared with the inner region within the tumor mass (Fig. 5A, left part), suggesting the potential correlation between autophagic activity and the pH environment. This finding was also validated in xenograft tumor derived from A549 cells (Fig. 5, right part). The autophagy response to different extracellular pHs in MDA-MB-231 cells was also confirmed by an in vitro pH treatment followed by the LC3 conversion assay (Fig. 5B), indicating that the regulation of autophagy by extracellular pH is a common mechanism in a variety of cell types.
Figure 5.
Differential autophagic activity in the outer and inner regions of solid tumors. (A) MDA-MB-231 or A549 cells were injected subcutaneously into the posterior flank of BALB/c athymic nude mice. Two weeks after implantation, tumors were dissected and stained with LC3 antibody. Pictures were taken in both the outer and inner region within the same tumor slide. Two independent tumors were examined for each cancer cell line and representative pictures were shown at magnifications of 200x (upper part) and 600x (lower part). (B) MDA-MB-231 cells were incubated in medium of indicated pH for 6 h in the presence or absence of lysosome inhibitors. Cell lysates were subjected to protein gel blot for LC3. Band densitometry quantification and the calculation of relative LC3-II intensity were performed as described in Materials and Methods.
Taken together, our data indicated that autophagic activity is regulated by extracellular pH, which may have important implications for the connections between autophagy and cancer as well as for a wide range of other physiological and pathological processes such as neuronal activity and immune function.24–26 Further studies will be necessary to decipher how the extracellular pH is sensed by the cells and the subsequent regulation of autophagy.
Materials and Methods
Cell culture.
The following human cell lines were originally obtained from the American Type Culture Collection (ATCC): MCF-10A (nontransformed mammary epithelial cells, CRL-10317), MCF-7, MDA-MB-231 (mammary carcinoma, HTB-22 and HTB-26, respectively), HeLa (cervical carcinoma, CCL2), PANC-1 (pancreatic carcinoma, CRL-1469), A549 (lung carcinoma, CCL-185). All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM, 10-013-CV Cellgro) containing 10% heat-inactivated FBS (Invitrogen, 10082-147), except for MCF-10A cells which were cultured as described previously in reference 31. For starvation, cells were incubated in Earle's Balanced Salt Solution (EBSS, Invitrogen, 14155063). As the pH of EBSS was 7.2, which was comparable to that of the control culture medium, starvation with EBSS did not have significant impact on the extracellular pH.
pH manipulation of cell culture medium.
To achieve a more stable extracellular pH environment, cells were cultured in 37°C under atmospheric CO2 in CO2-independent medium (Invitrogen, 18045088) supplemented with 4 mM L-glutamine (Invitrogen, 25030) and 10% FBS during the experiment. Cells cultured in medium of pH 7.2 were used as control to demonstrate the basal autophagic activity. The CO2-independent medium of pH 7.2 by itself did not have any significant impact on the basal autophagic activity as assessed by protein gel blotting for LC3 and GFP-LC3 punctate formation (Fig. S1). Acidic and basic pHs of cell culture medium were adjusted with 50 mM HEPES buffer (Fisher Scientific, BP310) containing either hydrogen chloride or sodium hydroxide, and measured by a Corning pH meter 340 (Corning Inc.). No detectable effect on cell viability was observed after 6 h of treatment at neither basic nor acidic pH (Fig. S2). In addition, cell culture medium acidified with 0.1% acetic acid, which resulted in an acidic pH of 5.9, was used to further validate the effect of acidic pH on autophagic activity. To monitor the medium pH during the experiment, we measured the medium pH after 6 h of incubation. Although we didn't find any significant change within 6 h, we did notice a trend of pH neutralization. This effect might be due to cellular metabolism, as the pH remained stable in the cell-free setting (Fig. S3).
Autophagy assays.
The activity of autophagy was determined by quantification of punctate GFP-LC3. Cells stably expressing GFP-LC3 minigene were fixed with methanol (when nuclear staining was performed) and GFP fluorescence was visualized using a Nikon microscope (Nikon Inc.,), and images were taken using a 40x objective. DAPI (1 ng/ml, Invitrogen, D1306) staining was used to visualize the nuclei. The numbers of GFP-LC3 punctates were quantified from at least 50 cells. For analysis of autophagy by electron microscopy, cells were prepared as described previously and images were taken under the microscope using either 6,000x or 30,000x magnifications.32 For analysis of endogenous LC-3 conversion by protein gel blot, cells were incubated in CO2-independent medium of indicated pH for 6 h unless otherwise indicated, and lysed with RIPA buffer. Proteins were then separated in 15% PAGE gels and blotted with anti-LC3 antibody (Medical and Biological Laboratories, PM036) and anti-β-actin antibody (Santa Cruz Biotechnology, sc-47778). Band densitometry quantification was performed using the Image J program (Version 1.43u, National Institute of Health). β-actin was used as the internal control to adjust sample loading in all the quantifications performed. In each protein gel blot, the LC3-II intensity of the control sample (pH 7.2) without lysosomal inhibitors were set as 1 and used as the standard. The relative LC3-II intensities in other samples were calculated and shown after comparing to the control sample. All the quantitative data of LC3 protein gel blots were presented as bar graphs in the Table S1. For inhibition of lysosomal degradation, 1 uM Pepstatin (Sigma-Aldrich, P4265) and 10 ug/ml E64d (Enzo Life Sciences, BML-PI107) were added to the culture medium at the time when the pH treatment was started. Immunohistochemistry staining was performed as described previously using anti-LC3 antibody.30
Measurement of intracellular pH.
Intracellular pH was measured using a standard ratiometric method with a pH-sensitive fluorophore SNARF-1 by flow cytometry.28 Briefly, cells were incubated in cell culture media of different pHs for 6 h. After that, cells were trypsinized and resuspended back into their respective medium. SNARF (10 uM, Invitrogen, C1272) and Pluronic F-127 (0.02%, Invitrogen, P3000MP) were then loaded into the cells for 15 min before the flow cytometry measurement. pH calibration was performed using high potassium buffer with nigericin (5 ug/ml, Invitrogen, N1495).
siRNA-mediated gene knockdown.
All the siRNAs were purchased from Sigma-Aldrich. siGL2, which targets the luciferase gene in pGL2 construct, was used as the negative control. The target sequences of siRNAs were as follows: GL2, 5′-UCG AAG UAU UCC GCG UAC G-3′; Atg5, 5′-GCA ACU CUG GAU GGG AUU G-3′; Vps34, 5′-GUG AUA AGU CUG UCA GAG U-3′; Beclin1, 5′-CAC UUG UUC CUU ACG GAA A-3′. Cells were reverse-transfected with 50 nM of indicated siRNA using Lipofectamine RNAiMAX (Invitrogen, 13778). Forty-eight hours after transfection, total RNA was purified from the transfected cells with Trizol (Invitrogen, 15596). Reverse transcription was conducted using the iScript™ cDNA Synthesis Kit (BioRad, 1708890). Real-time PCR analysis was performed using the following primers: ATG5, 5′-TGG GAT TGC AAA ATG ACA GA-3′ and 5′-TCC GGG TAG CTC AGA TGT TC-3′; Vps34, 5′-AAG CAG TGC CTG TAG GAG GA-3′, 5′-GCT TGT GGT GTT TGC TCT CA-3′; Beclin1, 5′-AAC CTC AGC CGA AGA CTG AA-3′, 5′-GGA TCA GCC TCT CCT CCT CT-3′.
Statistical analysis.
Data are expressed as the mean ± SEM from at least three separate experiments. Unless otherwise noted, the differences between groups were analyzed using the Student's t-test. All tests performed were two sided.
Acknowledgements
This work is supported by the Cancer Center Support Grant (P30CA54174) and an NIH/NCI grant (RO1 CA125144) for Z.Y.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 5.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 6.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3 and BNIP3L. Cell Death Differ. 2008;15:1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 9.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallinowski F, Vaupel P. pH distributions in spontaneous and isotransplanted rat tumours. Br J Cancer. 1988;58:314–321. doi: 10.1038/bjc.1988.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 12.Gerweck LE, Seetharaman K. Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer Res. 1996;56:1194–1198. [PubMed] [Google Scholar]
- 13.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 14.Chiche J, Brahimi-Horn MC, Pouyssegur J. Tumour hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. J Cell Mol Med. 2010;14:771–794. doi: 10.1111/j.1582-4934.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber V, De Milito A, Harguindey S, Reshkin SJ, Wahl ML, Rauch C, et al. Proton dynamics in cancer. J Transl Med. 2010;8:57. doi: 10.1186/1479-5876-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 17.Rofstad EK, Mathiesen B, Kindem K, Galappathi K. Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 2006;66:6699–6707. doi: 10.1158/0008-5472.CAN-06-0983. [DOI] [PubMed] [Google Scholar]
- 18.Xu L, Fukumura D, Jain RK. Acidic extracellular pH induces vascular endothelial growth factor (VEGF) in human glioblastoma cells via ERK1/2 MAPK signaling pathway: mechanism of low pH-induced VEGF. J Biol Chem. 2002;277:11368–11374. doi: 10.1074/jbc.M108347200. [DOI] [PubMed] [Google Scholar]
- 19.Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, Macswords J, Lathia JD, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69:4894–4903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva AS, Yunes JA, Gillies RJ, Gatenby RA. The potential role of systemic buffers in reducing intratumoral extracellular pH and acid-mediated invasion. Cancer Res. 2009;69:2677–2684. doi: 10.1158/0008-5472.CAN-08-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lou Y, McDonald PC, Oloumi A, Chia S, Ostlund C, Ahmadi A, et al. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res. 2011;71:3364–3376. doi: 10.1158/0008-5472.CAN-10-4261. [DOI] [PubMed] [Google Scholar]
- 23.De Milito A, Canese R, Marino ML, Borghi M, Iero M, Villa A, et al. pH-dependent antitumor activity of proton pump inhibitors against human melanoma is mediated by inhibition of tumor acidity. Int J Cancer. 2010;127:207–219. doi: 10.1002/ijc.25009. [DOI] [PubMed] [Google Scholar]
- 24.Chesler M. The regulation and modulation of pH in the nervous system. Prog Neurobiol. 1990;34:401–427. doi: 10.1016/0301-0082(90)90034-E. “ http://dx.doi.org/10.1016/0301-0082(90)90034-E”. [DOI] [PubMed] [Google Scholar]
- 25.Lardner A. The effects of extracellular pH on immune function. J Leukoc Biol. 2001;69:522–530. [PubMed] [Google Scholar]
- 26.Arnett TR. Extracellular pH regulates bone cell function. J Nutr. 2008;138:415–418. doi: 10.1093/jn/138.2.415S. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 28.Chow S, Hedley D, Tannock I. Flow cytometric calibration of intracellular pH measurements in viable cells using mixtures of weak acids and bases. Cytometry. 1996;24:360–367. doi: 10.1002/(SICI)1097-0320(19960801)24:4<360::AIDCYTO7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 29.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 31.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.