Abstract
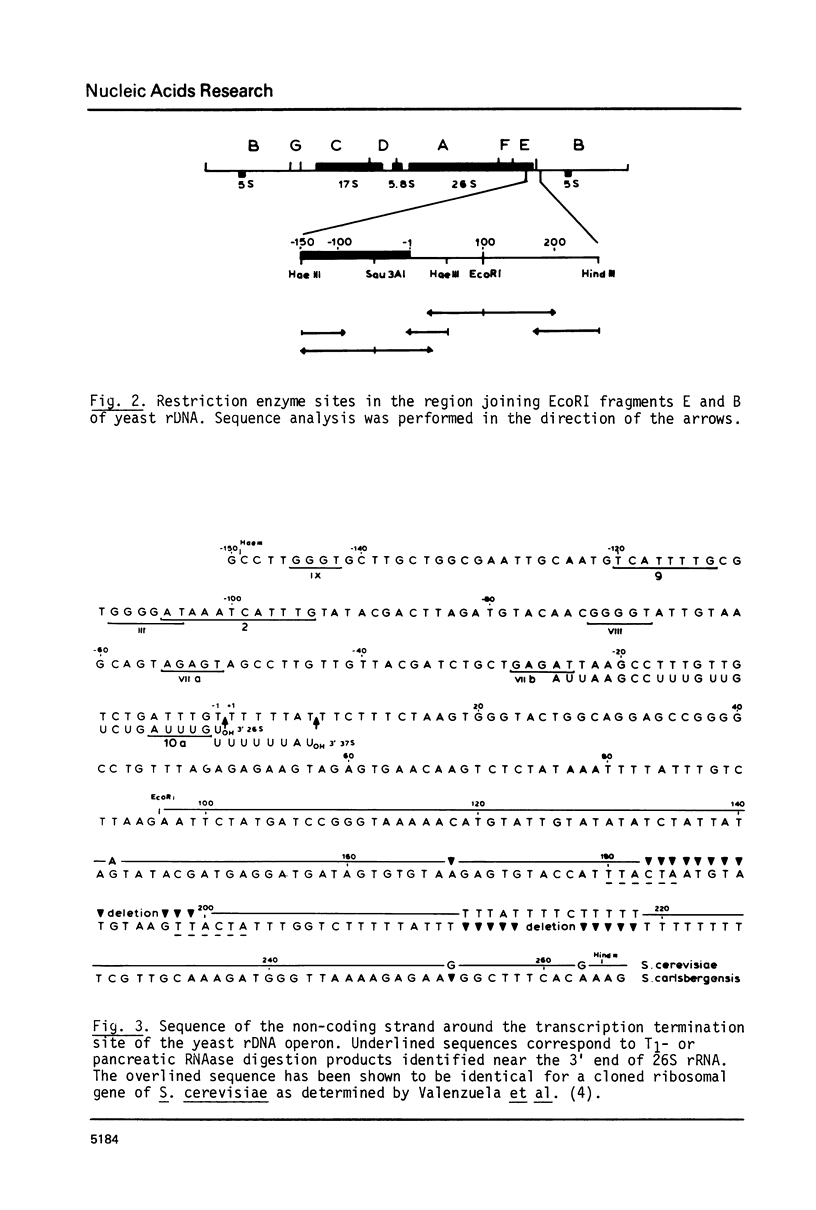
The site at which transcription of the ribosomal RNA operon in yeast is terminated was precisely localized. First, the exact position of the 3' end of the 26S rRNA gene was mapped on the rDNA on the basis of RNA- and DNA sequence data. Next, the 3' terminus of the primary transcript, 37S precursor rRNA, was established by hybridization experiments and sequence analysis. 37S pre-rRNA appears to be just 7 nucleotides longer at its 3' end than 26S rRNA. The non-coding strand around the termination site is extremely T-rich: 15 out of 18 nucleotides are T-residues. An extensive dyad symmetry is present in the sequence downstream from the termination site; a possible role of this structure in the regulation of transcription termination is discussed. The 3'-terminal 110 nucleotides of yeast 26S rRNA have approx. 50% and 60% homology with the corresponding regions of E. coli 23S rRNA and Xenopus laevis 28S rRNA, respectively.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Sequence of the 3'-terminal 21 nucleotides of yeast 17S ribosomal RNA. Nucleic Acids Res. 1977 Oct;4(10):3655–3663. doi: 10.1093/nar/4.10.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W., Bonewald R. Class of small multicopy plasmids originating from the mutant antibiotic resistance factor R1 drd-19B2. J Bacteriol. 1975 Aug;123(2):658–665. doi: 10.1128/jb.123.2.658-665.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Klootwijk J., de Jonge P., Planta R. J. The primary transcript of the ribosomal repeating unit in yeast. Nucleic Acids Res. 1979 Jan;6(1):27–39. doi: 10.1093/nar/6.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L. J., Brown D. D. Nucleotide sequence of Xenopus borealis oocyte 5S DNA: comparison of sequences that flank several related eucaryotic genes. Cell. 1978 Dec;15(4):1145–1156. doi: 10.1016/0092-8674(78)90042-9. [DOI] [PubMed] [Google Scholar]
- Leer J. C., Tiryaki D., Westergaard O. Termination of transcription in nucleoli isolated from Tetrahymena. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5563–5566. doi: 10.1073/pnas.76.11.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim V. I., Mazanov A. L. Tertiary structure for palindromic regions of DNA. FEBS Lett. 1978 Apr 1;88(1):118–123. doi: 10.1016/0014-5793(78)80621-8. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Tinoco I., Jr DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980 May 24;8(10):2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Rosenberg M. Isolation and sequence determination of the 3'-terminal regions of isotopically labelled RNA molecules. Nucleic Acids Res. 1974 May;1(5):653–671. doi: 10.1093/nar/1.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler D. G., Davies J. E. Specific cleavage of ribosomal RNA caused by alpha sarcin. Nucleic Acids Res. 1977 Apr;4(4):1097–1110. doi: 10.1093/nar/4.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Hunt J. A., Dalgarno L. Studies on the 3'-terminal sequences of the large ribosomal ribonucleic acid of different eukaryotes and those associated with "hidden" breaks in heart-dissociable insects 26S ribonucleic acid. Biochem J. 1974 Sep;141(3):617–625. doi: 10.1042/bj1410617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Trapman J., Planta R. J. Maturation of ribosomes in yeast. I Kinetic analysis by labelling of high molecular weight rRNA species. Biochim Biophys Acta. 1976 Sep 6;442(3):265–274. doi: 10.1016/0005-2787(76)90301-4. [DOI] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Venegas A., Sewell E. T., Masiarz F. R., DeGennaro L. J., Weinberg F., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. II. Physical map and nucleotide sequence of the 5 S ribosomal RNA gene and adjacent intergenic regions. J Biol Chem. 1977 Nov 25;252(22):8126–8135. [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Chapman A. B., Platt T., Guarente L. P., Beckwith J. Deletions of distal sequence after termination of transcription at the end of the tryptophan operon in E. coli. Cell. 1980 Apr;19(4):829–836. doi: 10.1016/0092-8674(80)90073-2. [DOI] [PubMed] [Google Scholar]
- van den Bos R. C., Retèl J., Planta R. J. The size and the location of the ribosomal RNA segments in ribosomal precursor RNA of yeast. Biochim Biophys Acta. 1971 Mar 25;232(3):494–508. doi: 10.1016/0005-2787(71)90603-4. [DOI] [PubMed] [Google Scholar]