Macrophage polarization is an integral part of many disease states, and macrophages are polarized through either the classical or alternative activation pathways. It has been shown that transcription factor networks controlling specialized gene expression programs regulate the functional polarization of macrophages. In this study, Lazar and colleagues investigate the role of histone acetylation in macrophage response to infection. By using a mouse model with macrophage-specific deletion of HDAC3, they demonstrate that HDAC3 functions in macrophages as a negative regulator of the alternative activation gene expression program. After inducing an immune response, the HDAC3 mutant mice show reduced inflammation and presence of alternatively activated macrophages. Thus, this study provides novel insights into a macrophage-autonomous mechanism regulating polarization.
Keywords: HDAC3, macrophage, epigenomic
Abstract
Macrophages, a key cellular component of inflammation, become functionally polarized in a signal- and context-specific manner. Th2 cytokines such as interleukin 4 (IL-4) polarize macrophages to a state of alternative activation that limits inflammation and promotes wound healing. Alternative activation is mediated by a transcriptional program that is influenced by epigenomic modifications, including histone acetylation. Here we report that macrophages lacking histone deacetylase 3 (HDAC3) display a polarization phenotype similar to IL-4-induced alternative activation and, furthermore, are hyperresponsive to IL-4 stimulation. Throughout the macrophage genome, HDAC3 deacetylates histone tails at regulatory regions, leading to repression of many IL-4-regulated genes characteristic of alternative activation. Following exposure to Schistosoma mansoni eggs, a model of Th2 cytokine-mediated disease that is limited by alternative activation, pulmonary inflammation was ameliorated in mice lacking HDAC3 in macrophages. Thus, HDAC3 functions in alternative activation as a brake whose release could be of benefit in the treatment of multiple inflammatory diseases.
Macrophages are a heterogeneous population of immune cells, functionally polarized in a signal- and context-specific manner. Polarization is often simplified into two broad opposing categories: classical activation (M1) induced by interferon γ in conjunction with TRL ligands such as lipopolysaccharide (LPS), and alternative activation (M2) as a result of exposure to Th2 cytokines such as interleukin 4 (IL-4) and IL-13 (Mosser and Edwards 2008; Geissmann et al. 2010; Gordon and Martinez 2010). Macrophage polarization is an important component of many disease states, including infection, insulin resistance, atherosclerosis, arthritis, and cancer. While classically activated macrophages contribute to the progression of inflammation, alternatively activated macrophages are considered anti-inflammatory and, due to the robust production of arginase 1 and other molecules that can impact the extracellular matrix, are proposed to promote wound healing. In particular, the tissue immune response to eggs laid by Schistosoma mansoni is characterized by a robust Th2 inflammatory response in which alternatively activated macrophages have a critical protective role, limiting inflammation likely through suppression of T-cell proliferation and response (Herbert et al. 2004, 2010; Anthony et al. 2007; Nair et al. 2009; Pesce et al. 2009).
At the molecular level, functional polarization of macrophages is dependent on complex networks of transcription factors, resulting in very distinct signal-specific gene expression programs (Barish et al. 2010; Ghisletti et al. 2010; Natoli et al. 2011). Recent reports have demonstrated that an efficient transcriptional response during polarization can be dependent on changes in the epigenome that are mediated by chromatin-modifying enzymes (De Santa et al. 2009; Ishii et al. 2009; Ghisletti et al. 2010; Satoh et al. 2010). In particular, Jmjd3, a histone demethylase, has been implicated in the removal of repressive chromatin modifications during the inflammatory response to LPS (De Santa et al. 2009). Furthermore, underscoring signal specificity, histone demethylation of the promoter of Irf4 by Jmjd3 was also shown to be necessary for the alternative activation-like response to chitin but not to IL-4 (Satoh et al. 2010).
Histone acetylation is a reversible chromatin modification that has also been implicated in macrophage response to environmental cues (Halili et al. 2009; Roger et al. 2011; Shakespear et al. 2011). Acetylation of lysine residues including K9 and K27 in the tail of histone H3 (H3K9 and H3K27, respectively) has been linked to gene activation (Roh et al. 2007; Rada-Iglesias et al. 2011). Histone acetyltransferases (HATs) including p300 (Marmorstein and Trievel 2009) place acetylation marks on histones, and these are removed by histone deacetylases (HDACs) (Clayton et al. 2006). HDACs can be grouped into several classes, and inhibitors of class I HDACs alter macrophage function in a variety of models of inflammation (Huang 2006; Haberland et al. 2009; Shakespear et al. 2011). However, the underlying mechanisms and the specific HDACs responsible for these effects remain unclear. Here we combine gene targeting with whole-genome expression analysis and chromatin immunoprecipitation (ChIP) coupled to genome-wide sequencing (ChIP-seq) to uncover a critical role for HDAC3 in macrophage polarization, specifically limiting alternative activation.
Results
Deletion of HDAC3 in macrophages leads to a gene expression program characteristic of IL-4-induced alternative activation
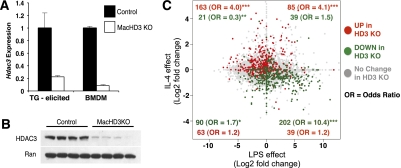
Germline deletion of HDAC3 in mice results in early embryonic lethality (Knutson et al. 2008). We generated mice with exons 4–7 of Hdac3 flanked by loxP sites (HDAC3f/f) (Supplemental Fig. 1) and bred them with mice expressing Cre recombinase under the control of the myeloid-specific Lysozyme M promoter (LysMCre+) (Clausen et al. 1999) to generate mice lacking HDAC3 in macrophages. LysMCre+, HDAC3f/f (MacHD3 knockout) mice were viable and displayed no overt abnormalities (data not shown). HDAC3 mRNA and protein were deleted in both thioglycollate-elicited peritoneal macrophages and bone marrow-derived macrophages (BMDMs) (Fig. 1A,B).
Figure 1.
Deletion of HDAC3 in macrophages leads to a gene expression program characteristic of IL-4-induced alternative activation. (A) Expression of Hdac3 measured by qPCR in thioglycollate (TG)-elicited macrophages and BMDMs. (B) Western blot for HDAC3 and Ran (loading control) in BMDMs (n = 4 of each genotype). (C) Scatter plot generated from overlaying the expression fold changes measured by whole-genome arrays in response to 6 h of LPS (X-axis) (Ghigo et al. 2010) and 24 h of IL-4 (Y-axis) in BMDMs, with the significant changes observed in unstimulated HDAC3-deficient macrophages. The numbers in the plot are the number of genes significantly changed in the HDAC3 knockout macrophages in each quadrant. Red dots represent genes that were increased, and green dots represent genes that were decreased in the HDAC3 knockout macrophages compared with controls. Odds ratios (OR), defined here as the fold enrichment within a quadrant relative to what would be expected if they were randomly dispersed, were calculated using the Fisher's exact test. (*) P < 1 × 10−5; (**) P < 1 × 10−10; (***) P < 1 × 10−15.
The gene expression changes in unstimulated BMDMs deleted of HDAC3 were examined by whole-genome microarray. In comparison with control macrophages, genes whose expression was significantly altered in HDAC3-deficient macrophages were enriched for immune and inflammatory response pathways (Supplemental Table 1). To determine whether these changes were related to classical versus alternative activation pathways, the HDAC3-regulated genes were overlaid on the set of genes classically regulated by LPS (Ghigo et al. 2010) as well as alternatively activated by IL-4 in wild-type macrophages (Fig. 1C). Remarkably, the majority of genes increased in the HDAC3 knockout macrophages were found to be positively regulated by IL-4 exposure of wild-type macrophages (Fig. 1C, top quadrants). Genes decreased upon HDAC3 deletion were genes that were negatively regulated by IL-4 treatment of wild-type macrophages, and (Fig. 1C, left quadrants), importantly, a large subset of these genes was positively regulated by LPS (Fig. 1C, bottom right quadrant). In contrast, very few genes positively regulated by LPS were increased upon HDAC3 deletion, and, of those, the majority were not signal-specific changes, since they increased with both IL-4 and LPS (Fig. 1C, top right quadrant). These data indicate that deletion of HDAC3 in macrophages results in a gene expression program that is strikingly similar to that associated with alternative activation.
Alternative activation is enhanced in HDAC3-deficient macrophages
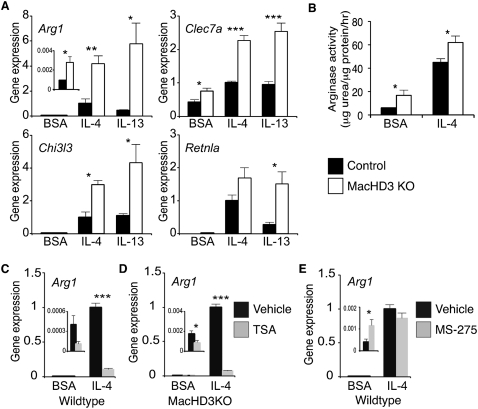
Using quantitative PCR (qPCR), alternative activation markers Arg1 and Clec7a were noted to be induced in HDAC3-deficient macrophages in the absence of cytokine treatment (Fig. 2A), confirming the skewing of these cells toward alternative activation. Moreover, treatment of HDAC3-deficient macrophages with Th2 cytokines IL-4 or IL-13 markedly potentiated the induction of several genes characteristic of alternative activation, including Arg1, Clec7a, Chi3l3, and Retnla (Fig. 2A). Enhancement of the alternative activation transcriptional program was functional, as shown by increased activity of arginase 1 protein in both the absence and presence of IL-4 (Fig. 2B). These results demonstrate that HDAC3 normally limits the expression of genes characteristic of IL-4-induced alternative activation in both the absence and presence of cytokine stimulation.
Figure 2.
Deletion of HDAC3 results in enhanced IL-4 macrophage alternative activation. (A) Expression of alternative activation signature genes as determined by qPCR (n = 4). Values shown are relative to the corresponding IL-4-treated control macrophages. (B) Arginase activity as determined by measurement of urea production (n = 3–5). (C,D) Gene expression determined by qPCR after pretreatment with TSA in wild-type macrophages (C) and HDAC3 knockout macrophages (D). (E) Gene expression determined by qPCR after pretreatment with MS-275 in wild-type macrophages. Values shown are relative to the corresponding IL-4-exposed macrophages pretreated with vehicle. (BSA) Bovine serum albumin; error bars, SEM; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Interestingly, rather than phenocopy the IL-4-potentiating effect of HDAC3 depletion, treatment of wild-type macrophages with the pan-HDAC inhibitor trichostatin A (TSA) actually decreased both basal and IL-4-stimulated expression of Arg1 in an HDAC3-independent manner (Fig. 2C,D). However, TSA had inconsistent effects on other IL-4-induced genes (Supplemental Fig. 2). Moreover, another pan-HDAC inhibitor, MS-275, had no significant effect on IL-4 induced Arg1 expression (Fig. 2E). These findings underscore the pleotropic effects of pharmacologic HDAC inhibitors that lack specificity toward individual HDACs and have idiosyncratic off-target effects (Shakespear et al. 2011).
HDAC3 localizes to a subset of Pu.1-defined macrophage-specific enhancers
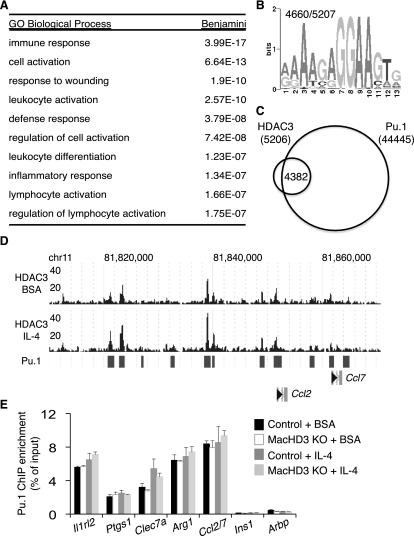
We next explored the mechanism of gene regulation by HDAC3 in BMDMs using ChIP-seq to identify genomic regions of HDAC3 enrichment. The distribution of these genomic sites of HDAC3 recruitment was similar to other reported transcription factor and coregulator cistromes (Farnham 2009; Visel et al. 2009), with most enrichment being observed in intergenic and intronic regions (Supplemental Fig. 3B). The nearest gene to each HDAC3-binding site was identified using gene-centered annotation (Shin et al. 2009), and gene ontology (GO) analysis was performed. Strikingly, all of the biological pathways that were significantly enriched in this unbiased analysis were related to immune or macrophage functions (Fig. 3A). This pattern is highly macrophage-specific, since, in liver, HDAC3 localizes most commonly near genes important in lipid and fatty acid metabolism (Feng et al. 2011). Indeed, the macrophage and liver HDAC3 cistromes show very little overlap, demonstrating substantial tissue specificity for HDAC3 binding (Supplemental Fig. 3C).
Figure 3.
HDAC3 localizes to a subset of Pu.1-defined macrophage-specific enhancers. (A) Summary of GO categories enriched among genes nearest to HDAC3 regions in basal conditions (BSA). (B) Top most-enriched motif found in HDAC3-enriched regions upon de novo motif search; the number of times this motif was observed in all regions considered is reported in the top left. (C) Venn diagram demonstrating overlap of HDAC3 regions with published Pu.1 sites (Ghisletti et al. 2010). (D) Genome browser image of HDAC3-enriched sequence for part of chromosome 11 in BMDMs and the overlap of reported Pu.1 sites (bars). (E) Validation of Pu.1 binding at regions of HDAC3 enrichment by ChIP-qPCR. Regions were identified by nearest transcriptional start site (TSS). Regions at Ins1 and Arbp TSS served as controls (n = 3–5). Error bars, SEM.
We next performed a de novo motif search within the HDAC3-enriched genomic regions in macrophages to predict transcription factors that colocalize at these sites. In this analysis, the top six most-enriched motifs (Fig. 3B; Supplemental Table 2) matched the binding specificity of Ets transcription family member Pu.1, the master regulator of macrophage identity that constitutively occupies macrophage-specific enhancers and prepares them for signal-specific transcription factor regulation (Ghisletti et al. 2010; Heinz et al. 2010). Of note, in contrast to similar analysis of the HDAC3 cistrome in liver (Feng et al. 2011), binding motifs for nuclear receptors were not among the top 10 enriched motifs. Comparison of the HDAC3 cistrome with the previously described BMDM Pu.1 cistrome (Ghisletti et al. 2010) revealed that the vast majority (84%) of HDAC3-enriched regions are indeed at sites of Pu.1 binding (Fig. 3C,D). The presence of Pu.1 at HDAC3-enriched regions was validated by ChIP-qPCR (Fig. 3E). From these analyses, we conclude that HDAC3 binds mainly at a subset of Pu.1-defined macrophage enhancers.
HDAC3 regulates genes characteristic of IL-4-induced alternative activation by deacetylation of nearby enhancer regions
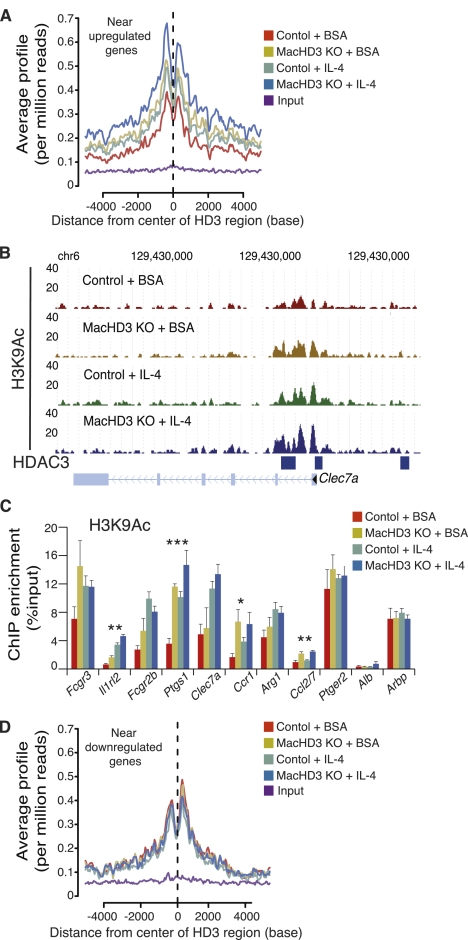
We next assessed genome-wide acetylation of histone H3 on Lys 9 (H3K9Ac), an epigenomic mark that is associated with gene activation (Roh et al. 2007), in control and HDAC3 knockout macrophages treated with IL-4 or vehicle. We focused on areas of HDAC3 enrichment near genes that increased upon IL4-induced alternative activation and HDAC3 deletion. In wild-type macrophages, H3K9Ac was increased at these regions upon IL-4 stimulation, consistent with transcriptional activation (Fig. 4A). The details of a representative IL-4-induced gene, Clec7a, are shown in Figure 4B. Moreover, a marked stimulatory effect of HDAC3 deletion on H3K9Ac was observed in regions bound by HDAC3 in BMDMs, consistent with the notion that genome-associated HDAC3 acts locally to deacetylate histone, and this effect of HDAC3 deletion was exaggerated in the IL-4-treated macrophages (Fig. 4A,B). The HDAC3 dependence of H3K9 deacetylation was confirmed at several HDAC3-binding sites by ChIP-qPCR (Fig. 4C). Similar trends were observed at histone H3 on Lys 27, another residue marked by acetylation at enhancers (Rada-Iglesias et al. 2011), demonstrating that the effect of HDAC3 was not residue-specific and likely impacting global histone acetylation in these regions (Supplemental Fig. 4). Changes in acetylation were not observed at HDAC3-enriched regions near genes whose expression decreases upon HDAC3 deletion or IL-4 treatment (Fig. 4D), suggesting that the effects of HDAC3 in those locations were counterbalanced by opposing factors or that HDAC3 was not functional at these regions and confirming the specific role of HDAC3 in gene repression. These data demonstrate that HDAC3 functions at enhancers near genes characteristic of IL-4-induced alternative activation to limit histone acetylation and thereby contribute to repression of gene expression.
Figure 4.
HDAC3 regulates alternatively activated macrophage signature genes by deacetylation of nearby enhancer regions. (A) Average signal profiling of H3K9Ac around regions of HDAC3 enrichment near IL-4-induced genes that are up-regulated by deletion of HDAC3. (B) Genome browser image demonstrating H3K9Ac at a select region of chromosome 6 in the presence and absence of HDAC3 and/or IL-4; regions of HDAC3 enrichment are represented by blue bars. (C) ChIP-qPCR interrogating select HDAC3 regions used in A for changes in H3K9Ac; regions at the Alb and Arbp transcriptional start sites served as controls. Statistically significant differences between either untreated controls and MacHD3 knockout or IL-4-treated controls and treated MacHD3 knockout are reported (n = 4). (D) Average signal profiling of H3K9Ac around regions of HDAC3 enrichment near IL-4 down-regulated genes that are also decreased by deletion of HDAC3. Error bars, SEM; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Th2 cytokine-driven pulmonary inflammation is limited by deletion of macrophage HDAC3
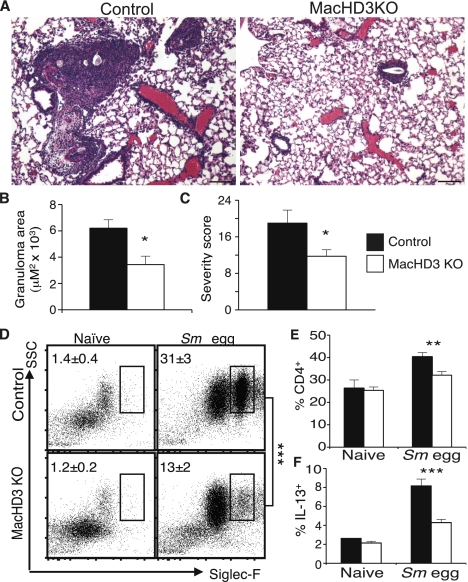
The results thus far strongly suggested that HDAC3 functions to suppress macrophage alternative activation. We observed evidence of increased alternatively activated macrophages in vivo (Supplemental Fig. 5A,B) and therefore tested the physiological relevance of this altered macrophage polarization using a pulmonary challenge with ova from the helminth S. mansoni, a model in which alternatively activated macrophages limit excessive pulmonary inflammation (Herbert et al. 2004, 2010; Nair et al. 2009; Pesce et al. 2009). Eight days after intravenous challenge with the ova, large inflammatory granulomas formed around the eggs in the lungs of control mice (Fig. 5A). In comparison, the granulomas observed in the MacHD3 knockout lungs were much smaller in size, and the overall lung inflammation was markedly less (Fig. 5B,C). In concordance with this, eosinophilia was markedly reduced in the ova-challenged MacHD3 knockout lungs (Fig. 5D), and there were fewer CD4+ T cells (Fig. 5E). Furthermore, when the draining parathymic lymph nodes from challenged mice were removed and restimulated, the frequency of IL-13-producing CD4+ T cells was greatly reduced in the MacHD3 knockout mice, consistent with the suppressed T-cell response and decreased disease severity (Fig. 5F). Thus, deletion of HDAC3 in macrophages has a protective, anti-inflammatory effect on S. mansoni ova challenge, reflecting the enhanced alternative activation of HDAC3 knockout macrophages in vivo.
Figure 5.
Reduced Th2 cytokine-driven pulmonary inflammation by deletion of macrophage HDAC3. (A) Hematoxylin and eosin-stained sections of lungs from S. mansoni egg-challenged mice. Representative sections from control mice (left) and MacHD3 knockout (right) are shown. (B) Quantitation of the size of granulomas observed histologically. (C) Histopathological score of lung. (D) Representative dot plots demonstrating frequency of Siglec-F+ eosinophils among CD45+ hematopoietic cells within the lungs; averages are reported in the top left of the plots. (E) Frequency of CD4+ cells within the CD45+ hematopoietic cells of the lungs. (F) IL-13-producing CD4+ T cells in the draining parathymic lymph node poststimulation with anti-CD3 and anti-CD28 antibodies. n = 3–6; error bars, SEM; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Discussion
We showed here that HDAC3 limits basal and cytokine-stimulated alternative activation of macrophages in vitro and in vivo and has a marked influence on a living organism's ability to cope with inflammatory stimuli. HDAC3 binds to the macrophage genome and deacetylates histone tails at a subset of sites bound by lineage-specific transcription factor Pu.1, which is the major determinant of the macrophage epigenome (Ghisletti et al. 2010; Heinz et al. 2010; Natoli 2010; Natoli et al. 2011). The macrophage HDAC3 cistrome is remarkably different from that in liver, where similar unbiased motif analysis of genomic sites of HDAC3 binding revealed a preference for the nuclear receptor half-site motif, with the heme receptor Rev-erbα playing a major role in delivering HDAC3 to the genome (Feng et al. 2011). Moreover, whereas dysregulation of lipid metabolism is the major consequence of HDAC3 deletion in liver (Knutson et al. 2008; Feng et al. 2011) and in heart (Montgomery et al. 2008; Sun et al. 2011), we report here that the main function of HDAC3 in macrophages is to regulate inflammatory gene expression and function.
Several transcription factors cooperate with Pu.1 to activate macrophage gene expression in a loci- and signal-specific manner (Barish et al. 2010; Heinz et al. 2010; Lefterova et al. 2010; Natoli et al. 2011; Sullivan et al. 2011). This has been most extensively studied in the context of classical activation or proinflammatory response to LPS. For example, LPS stimulates transcription factor NF-κB to bind to the genome in cooperation with cofactor p300 while simultaneously evicting transcriptional repressor Bcl-6, thereby promoting expression of a subset of the inflammatory gene program (Barish et al. 2010; Ghisletti et al. 2010). Much less is understood about the transcriptional networks that dictate alternative activation via modification of Pu.1-driven enhancers. Recently, the histone demethylase Jmjd3 was shown to remove a repressive chromatin mark at a single site at the Irf4 gene during alternative activation induced by chitin, but not by the classical alternative activation inducer IL-4 (Satoh et al. 2010). In contrast, our findings reveal a key genome-wide role for HDAC3, acting at a subset of Pu.1-guided enhancers, in restricting activating histone marks at many IL-4 target genes. Loss of HDAC3 removes this brake and thereby promotes the IL4-induced alternatively activated phenotype. To our knowledge, this is the first report of a chromatin remodeling enzyme restricting alternative activation in a macrophage-autonomous manner. It remains possible, however, that the function of HDAC3 in alternative activation also involves deacetylation of nonhistone proteins.
The epigenomic landscape of the macrophage is a promising immunomodulatory target (Ishii et al. 2009; Satoh et al. 2010), and shifting the balance of polarization in favor of alternative activation is predicted to have beneficial effects in multiple inflammatory conditions, including helminth infection, obesity-related insulin resistance, and atherosclerosis (Vats et al. 2006; Kang et al. 2008; Odegaard et al. 2008; Martinez et al. 2009; Gordon and Martinez 2010; Tabas 2010; Odegaard and Chawla 2011). HDACs are attractive targets due to the availability and existing clinical use of HDAC inhibitors (Huang 2006; Haberland et al. 2009; Shakespear et al. 2011). However, existing pan-HDAC inhibitors did not phenocopy the effects of genetic loss of HDAC3 on macrophage polarization and had idiosyncratic effects, which could be due to their variable potency in inhibiting multiple HDACs as well as off-target effects (Shakespear et al. 2011). Increased acetylation at HDAC3-marked enhancers upon the deletion of HDAC3 strongly supports the notion that HDAC3 is functioning enzymatically to regulate alternative activation. However, our results do not completely eliminate the possibility that the function of HDAC3 in ameliorating alternative activation is independent of HDAC activity, perhaps related to the nuclear receptor corepressors, transducin β-like proteins, and other factors that are normally present in complex with HDAC3 (Li et al. 2000; Guenther et al. 2001; Zhang et al. 2002; Privalsky 2004). Targeting HDAC3 to release the brake on alternative activation and alter the progression of inflammatory disorders affected by shifts in macrophage polarization will therefore require innovative and specific approaches that mimic depletion of HDAC3.
Materials and methods
Generation of MacHD3 knockout mice
A recombineering strategy was used to generate a targeting vector with loxP sites flanking exons 4–7 of Hdac3. The targeting vector was linearized and electroporated into C57Bl/6 embryonic stem (ES) cells. ES cell clones that had undergone homologous recombination were injected into C57Bl/6 blastocysts and implanted in C57Bl/6 pseudopregnant female mice. Chimeric mice were bred with C57Bl/6 mice, and offspring with confirmed germline transmission of the floxed Hdac3 allele were bred with C57Bl/6 ACT-FLPe mice to excise the FRT-flanked NeoR cassette, generating floxed HDAC3 mice (HDAC3f/f). C57Bl/6 mice expressing Cre-recombinase expressed under the control of the Lysozyme M promoter (Jackson Laboratories) (Clausen et al. 1999) were bred to HDAC3f/f to generate MacHD3 knockout mice.
BMDMs
Bone marrow cells were cultured in 30% L929 conditioned medium or 20 ng/mL purified M-CSF (Peprotech) for 6 d, changing the medium on the third day of culture. On day 6, macrophages were recovered from the Petri dishes, counted, plated, and rested overnight. Cells were then treated with vehicle (bovine serum albumin [BSA]), 10 ng/mL IL-4, or 40 ng/mL IL-13 for 24 h. Urea production representing arginase activity was measured as described previously (Classen et al. 2009). For HDAC inhibitor studies, a 4-h pretreatment of either 30 nM TSA or 1 μM MS-275 (Sigma) prior to the addition of IL-4 for an additional 24 h was performed.
Antibodies, ChIP, Western blotting, and flow cytometry
The following antibodies were used: HDAC3 (Abcam, ab7030), Ran (BD Transduction Laboratories), Pu.1 (Santa Cruz Biotechnologies, sc-352X), H3K9Ac (Upstate Biotechnology, 06-942), and H3K27Ac (Abcam, ab4729). Antibodies for flow cytometry were from BD Biosciences (CD11b, CD45, Siglec-F, IL-13), AbDSerotec (Mgl), Ebiosciences (CD4, F480), and Chi3l3 (R&D Systems). ChIP was performed as described previously (Lefterova et al. 2010; Feng et al. 2011). Briefly, Cells were fixed in 1% PFA for 15 min and quenched with glycine. Nuclei were harvested and lysed. Extracts were sonicated using a Bioruptor (Diagenode). Immunoprecipitated DNA was analyzed by qPCR using Power SYBR Green PCR Master mix on the 7900HT Fast Real-Time PCR system (Applied Biosystems). All primer sequences used can be found in Supplemental Table 3. Analyses were performed using a standard curve, and enrichments as percent of total input were determined. Surface and intracellular cell staining for flow cytometry was performed as described previously (Nair et al. 2009).
ChIP-seq
ChIP DNA was prepared from BMDMs for sequencing according to the amplification protocol from Illumina. DNA for the HDAC3 ChIP-seq was obtained by pooling DNA from two independent ChIPs, each using a pool of four mice for each condition, while the DNA for the H3K9Ac ChIP-seq was obtained from a pool of four mice for each condition. The H3K9Ac ChIP-seq was repeated on a biologically independent group of mice, and similar results were obtained. Sequencing was performed by the Functional Genomics Core at the University of Pennsylvania. Sequence reads of 36 base pairs (bp) were obtained using the Solexa Analysis Pipeline and were mapped to the mouse genome (mm8). All but one of the identically aligning reads were removed to minimize PCR bias. Regions that overlapped with peaks in the input data were discarded. GLITR (Tuteja et al. 2009) was used to identify regions of HDAC3 enrichment at a false discover rate (FDR) of 0.5%. Only regions with a peak height of 15 or greater were considered unless otherwise noted. Nearest gene assignments, genomic distribution, and average profiles were determined using CEAS (Shin et al. 2009). GO analysis was performed using DAVID (Dennis et al. 2003). Sequence motifs were discovered de novo using MDscan (Liu et al. 2002). Comparisons with Pu.1-binding sites were completed after converting the HDAC3 regions to mm9. ChIP-seq data have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE33609.
Gene expression
RNA was isolated from cells or tissue using the RNeasy kit (Qiagen). cDNA was generated using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). qPCR was performed using Power SYBR Green PCR Master mix on the 7900HT Fast Real-Time PCR system (Applied Biosystems). All data were analyzed using a standard curve and normalized to Arbp expression. For specific primer sequences, see Supplemental Table 3.
Microarray analysis
Total RNA from four biological replicates for each condition (control, HDAC3 knockout, control treated with IL-4, and HDAC3 knockout treated with IL4) were prepared and submitted to the Functional Genomics Core at the University of Pennsylvania for array hybridization, scanning, and statistical analysis. Briefly, 50 ng of RNA was amplified and labeled using the Ovation V2 Amplification kit (NuGEN) and hybridized to the Whole-Mouse Genome Oligo Microarray (Agilent). Bad hybridizations and samples were discarded, leaving at least three replicates for all conditions. Differentially expressed probes were identified with SAM (Tusher et al. 2001) using quantile-normalized data. Detailed microarray data have been deposited in GEO and are accessible through GEO series accession number GSE33609. GO analysis was performed using DAVID (Dennis et al. 2003). Agilent Microarray data for LPS treatment (Ghigo et al. 2010) were downloaded through GEO (GSE20207). The log2 fold change upon LPS and IL-4 treatment for each common probe was plotted. All probes that had processed signal values <100 in both the treatment and control conditions were automatically assumed to have no fold change in that experiment. If the probe significantly increased in the HDAC3 knockout macrophages, it was colored red, and the probe was colored green if it significantly decreased. Probes for which there was no fold change due to insufficient signal in either treatment were excluded from the quadrant counts.
Schistosoma ova challenge
The Schistosoma ova challenge was performed in two independent experiments as previously described (Nair et al. 2009). Mice were immunized intraperitoneally (i.p.) with 5000 Sm eggs, followed by intravenous (i.v.) challenge with 5000 eggs 14 d later. Mice were euthanized 8 d later. Lung tissue was recovered for gene expression, flow cytometry, or histology. Draining mediastinal lymph node cells were cultured with 1 μg/mL each α-CD3/α-CD28 (eBioscience) for 24 h.
Histology
Lungs were fixed in 4% paraformaldehyde, paraffin-embedded, sectioned, and stained in hematoxylin and eosin (H&E). Images of all granulomas present in one section from each mouse were captured, and the size of each granuloma was calculated using IVision. Average granuloma size per mouse was determined, and then the average per genotype was determined. Blind scoring of H&E-stained lung sections reflects severity of consolidation (1–5), bronchial/bronchiolar epithelial hyperplasia (1–5), arteritis (1–5), and recruitment of macrophages/multinucleated giant cells (1–5), granulocytes (1–5), and lymphocytes (1–5), for an overall maximum score of 30.
Statistics
For all comparisons of two groups, a Student's unpaired t-test was used to compute P-values. P-values <0.05 were considered statistically significant.
Acknowledgments
We thank the Functional Genomics Core of the Penn Diabetes and Endocrinology Research Center (DK19525) for ChIP-seq and microarray data generation and analyses, John Le Lay for guidance in generating the targeting vector, and the Morphology Core (DK49210) for histological assistance. This work was supported by DK43806 and DK43806S1 (to M.A.L.); AI61570 (to D.A.); the Penn Diabetes, Endocrinology, and Metabolism Training Grant (DK07314); the Picower Foundation; and the Cox Institute for Medical Research.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.175950.111.
References
- Anthony RM, Rutitzky LI, Urban JF Jr, Stadecker MJ, Gause WC 2007. Protective immune mechanisms in helminth infection. Nat Rev Immunol 7: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM 2010. Bcl-6 and NF-κB cistromes mediate opposing regulation of the innate immune response. Genes Dev 24: 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen A, Lloberas J, Celada A 2009. Macrophage activation: Classical versus alternative. Methods Mol Biol 531: 29–43 [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8: 265–277 [DOI] [PubMed] [Google Scholar]
- Clayton AL, Hazzalin CA, Mahadevan LC 2006. Enhanced histone acetylation and transcription: A dynamic perspective. Mol Cell 23: 289–296 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3 doi: 10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, et al. 2009. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J 28: 3341–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham PJ 2009. Insights from genomic profiling of transcription factors. Nat Rev Genet 10: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K 2010. Development of monocytes, macrophages, and dendritic cells. Science 327: 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo E, Barry AO, Pretat L, Al Moussawi K, Desnues B, Capo C, Kornfeld H, Mege JL 2010. IL-16 promotes T. whipplei replication by inhibiting phagosome conversion and modulating macrophage activation. PLoS ONE 5: e13561 doi: 10.1371/journal.pone.0013561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Gordon S, Martinez FO 2010. Alternative activation of macrophages: Mechanism and functions. Immunity 32: 593–604 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 21: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN 2009. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet 10: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halili MA, Andrews MR, Sweet MJ, Fairlie DP 2009. Histone deacetylase inhibitors in inflammatory disease. Curr Top Med Chem 9: 309–319 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert DR, Holscher C, Mohrs M, Arendse B, Schwegmann A, Radwanska M, Leeto M, Kirsch R, Hall P, Mossmann H, et al. 2004. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 20: 623–635 [DOI] [PubMed] [Google Scholar]
- Herbert DR, Orekov T, Roloson A, Ilies M, Perkins C, O'Brien W, Cederbaum S, Christianson DW, Zimmermann N, Rothenberg ME, et al. 2010. Arginase I suppresses IL-12/IL-23p40-driven intestinal inflammation during acute schistosomiasis. J Immunol 184: 6438–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L 2006. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases. J Cell Physiol 209: 611–616 [DOI] [PubMed] [Google Scholar]
- Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WFt, Cavassani KA, Li X, Lukacs NW, et al. 2009. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114: 3244–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Reilly SM, Karabacak V, Gangl MR, Fitzgerald K, Hatano B, Lee CH 2008. Adipocyte-derived Th2 cytokines and myeloid PPARδ regulate macrophage polarization and insulin sensitivity. Cell Metab 7: 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW 2008. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J 27: 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA 2010. Cell-specific determinants of peroxisome proliferator-activated receptor γ function in adipocytes and macrophages. Mol Cell Biol 30: 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J 19: 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS 2002. An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20: 835–839 [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Trievel RC 2009. Histone modifying enzymes: Structures, mechanisms, and specificities. Biochim Biophys Acta 1789: 58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Helming L, Gordon S 2009. Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol 27: 451–483 [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN 2008. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest 118: 3588–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP 2008. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8: 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair MG, Du Y, Perrigoue JG, Zaph C, Taylor JJ, Goldschmidt M, Swain GP, Yancopoulos GD, Valenzuela DM, Murphy A, et al. 2009. Alternatively activated macrophage-derived RELM-α is a negative regulator of type 2 inflammation in the lung. J Exp Med 206: 937–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G 2010. Maintaining cell identity through global control of genomic organization. Immunity 33: 12–24 [DOI] [PubMed] [Google Scholar]
- Natoli G, Ghisletti S, Barozzi I 2011. The genomic landscapes of inflammation. Genes Dev 25: 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Chawla A 2011. Alternative macrophage activation and metabolism. Annu Rev Pathol 6: 275–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, Vats D, Morel CR, Goforth MH, Subramanian V, Mukundan L, Ferrante AW, Chawla A 2008. Alternative M2 activation of Kupffer cells by PPARδ ameliorates obesity-induced insulin resistance. Cell Metab 7: 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA 2009. Arginase-1–expressing macrophages suppress Th2 cytokine–driven inflammation and fibrosis. PLoS Pathog 5: e1000371 doi: 10.1371/journal.ppat.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalsky ML 2004. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol 66: 315–360 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, Ding XC, Chanson AL, Reymond MK, Miconnet I, et al. 2011. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 117: 1205–1217 [DOI] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K 2007. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res 17: 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, et al. 2010. The Jmjd3–Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 11: 936–944 [DOI] [PubMed] [Google Scholar]
- Shakespear MR, Halili MA, Irvine KM, Fairlie DP, Sweet MJ 2011. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol 32: 335–343 [DOI] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS 2009. CEAS: Cis-regulatory element annotation system. Bioinformatics 25: 2605–2606 [DOI] [PubMed] [Google Scholar]
- Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM, Glass CK 2011. Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol Cell Biol 31: 861–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Singh N, Mullican SE, Everett LJ, Li L, Yuan L, Liu X, Epstein JA, Lazar MA 2011. Diet-induced lethality due to loss of HDAC3 in heart and skeletal muscle. J Biol Chem 286: 33301–33309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I 2010. Macrophage death and defective inflammation resolution in atherosclerosis. Nat Rev Immunol 10: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, White P, Schug J, Kaestner KH 2009. Extracting transcription factor targets from ChIP-seq data. Nucleic Acids Res 37: e113 doi: 10.1093/nar/gkp536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A 2006. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab 4: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG 2002. The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell 9: 611–623 [DOI] [PubMed] [Google Scholar]