Abstract
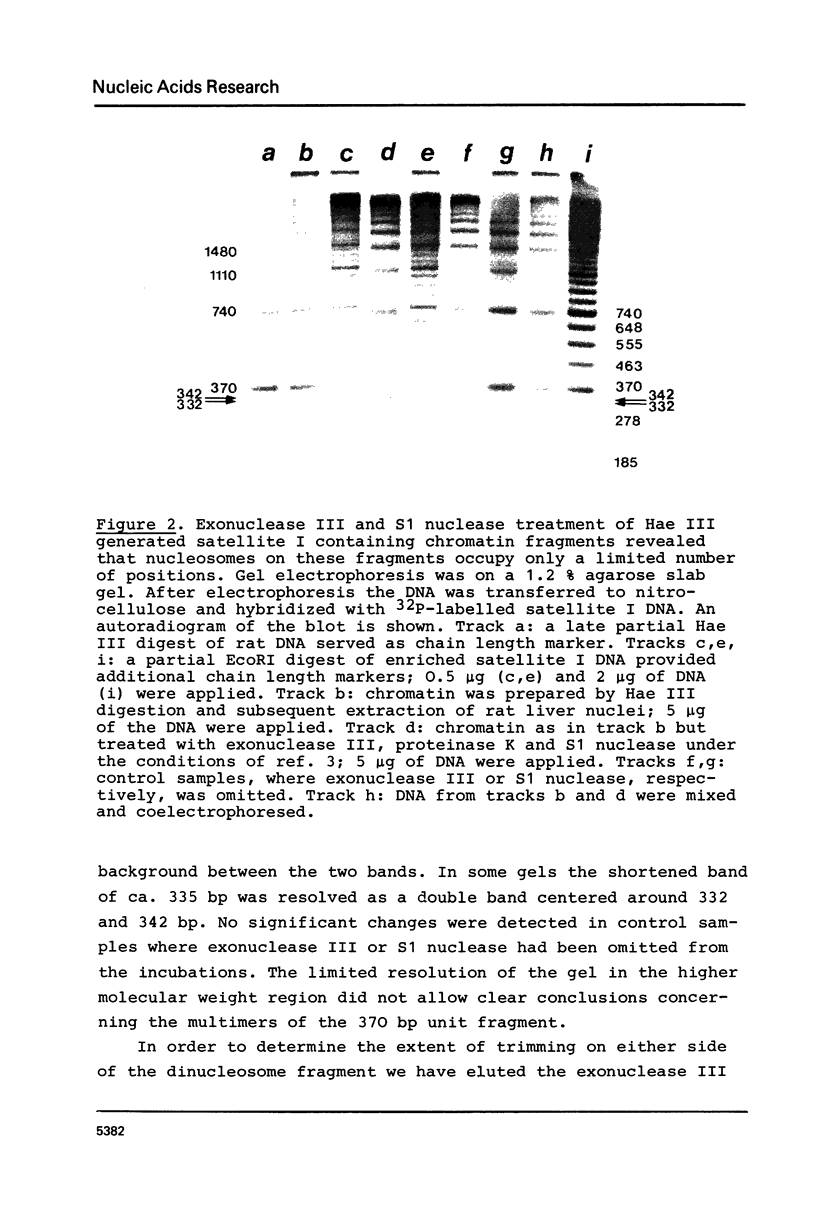
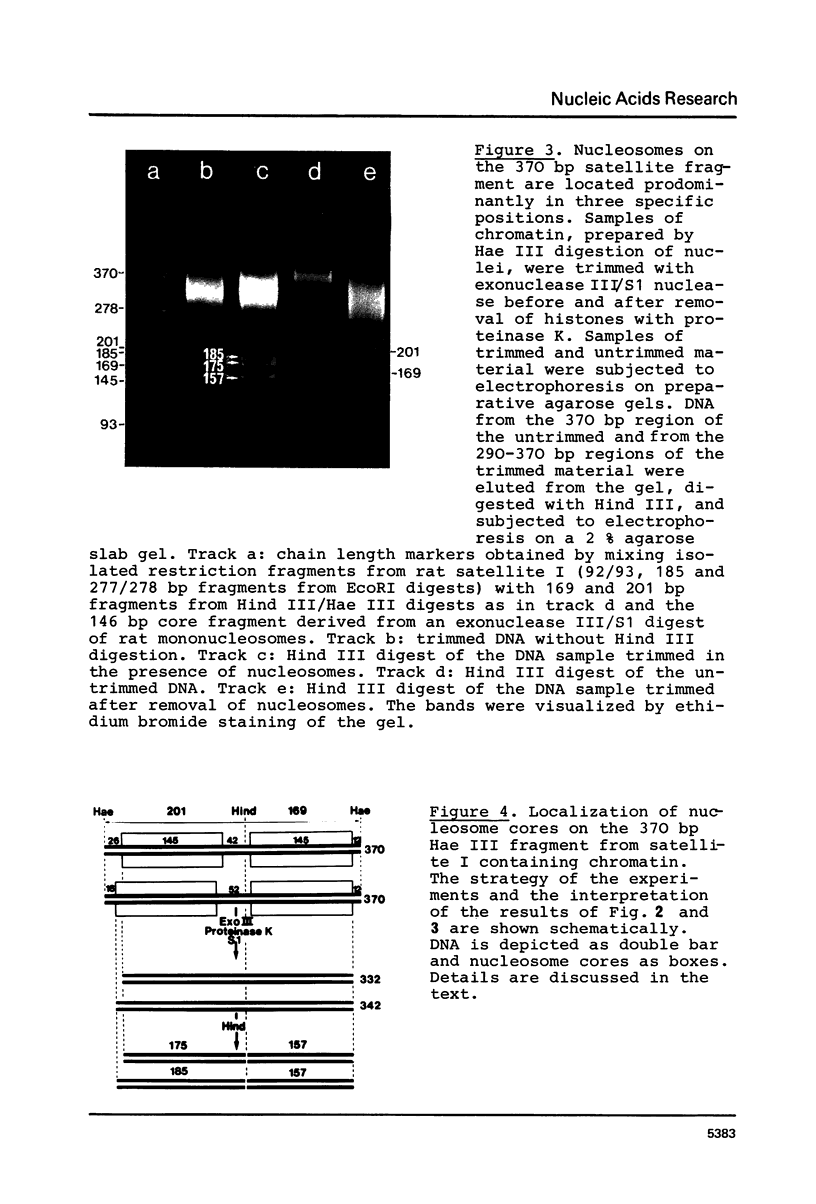
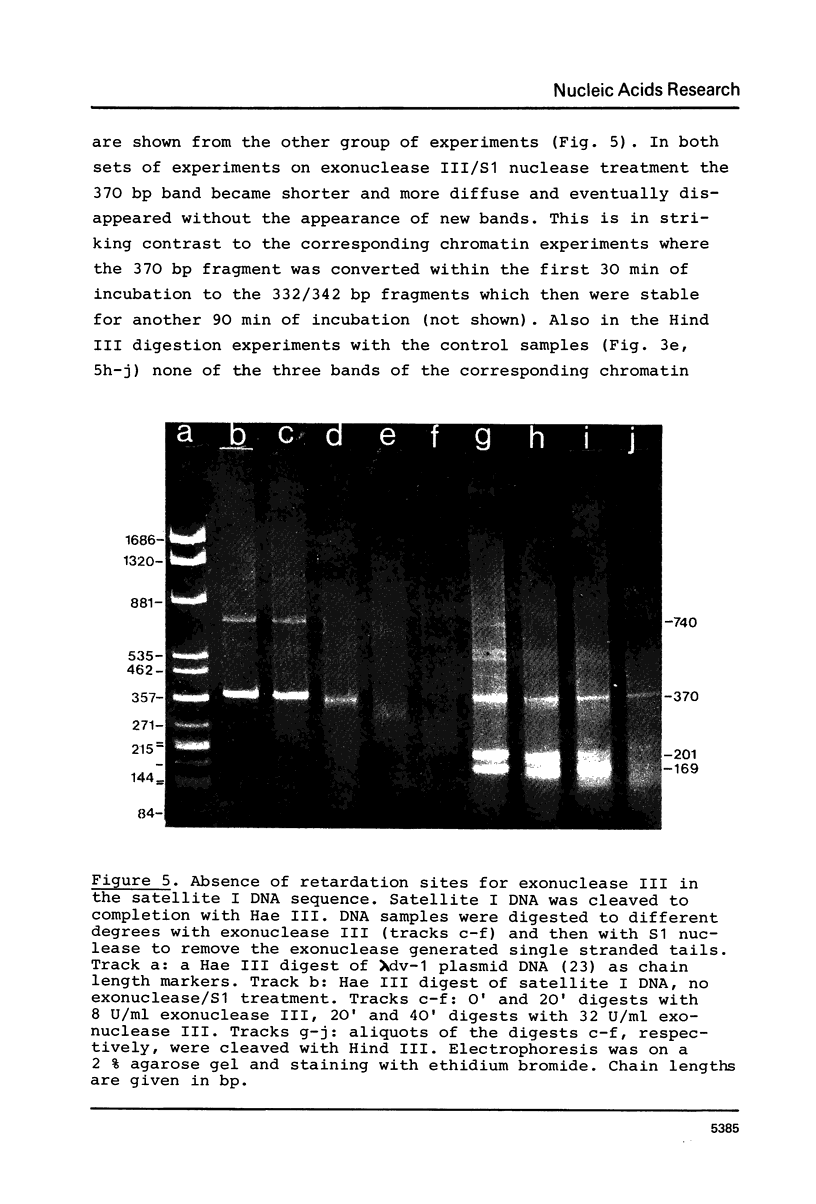
The location of nucleosomes on the nucleotide sequence of rat satellite I DNA was investigated using micrococcal nuclease, exonuclease III, and restriction nucleases as tools. Hae III cleaved the satellite DNA containing chromatin very preferentially in the linker region. Nucleosomes were found predominantly in three defined positions on the 370 bp satellite I monomer unit. This type of arrangement occurs on not more than half of the satellite DNA containing chromatin while the rest of this chromatin is arranged differently. The arrangement of nucleosomes with high probability in preferred frames and with low probability in less preferred frames may be a general phenomenon which can be discussed as a possible mechanism to modulate sequence recognition.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenburger W., Hörz W., Zachau H. G. Comparative analysis of three guinea pig satellite DNA's by restriction nucleases. Eur J Biochem. 1977 Mar 1;73(2):393–400. doi: 10.1111/j.1432-1033.1977.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Baer B. W., Kornberg R. D. Random location of nucleosomes on genes for 5 S rRNA. J Biol Chem. 1979 Oct 10;254(19):9678–9681. [PubMed] [Google Scholar]
- Crémisi C., Pignatti P. F., Yaniv M. Random location and absence of movement of the nucleosomes on SV 40 nucleoprotein complex isolated from infected cells. Biochem Biophys Res Commun. 1976 Dec 6;73(3):548–554. doi: 10.1016/0006-291x(76)90845-7. [DOI] [PubMed] [Google Scholar]
- Fittler F., Zachau H. G. Subunit structure of alpha-satellite DNA containing chromatin from African green monkey cells. Nucleic Acids Res. 1979 Sep 11;7(1):1–13. doi: 10.1093/nar/7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Melton D. A. The length of nucleosome-associated DNA is the same in both transcribed and nontranscribed regions of chromatin. Nature. 1978 May 25;273(5660):317–319. doi: 10.1038/273317a0. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Greil W., Zachau H. G. Prepartation of soluble chromatin and specific chromatin fractions with restriction nucleases. Nucleic Acids Res. 1977 Oct;4(10):3387–3400. doi: 10.1093/nar/4.10.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musich P. R., Maio J. J., Brown F. L. Subunit structure of chromatin and the organization of eukaryotic highly repetitive DNA: indications of a phase relation between restriction sites and chromatin subunits in African green monkey and calf nuclei. J Mol Biol. 1977 Dec 15;117(3):657–677. doi: 10.1016/0022-2836(77)90063-8. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Omori A., Igo-Kemenes T., Zachau H. G. Different repeat lengths in rat satellite I DNA containing chromatin and bulk chromatin. Nucleic Acids Res. 1980 Nov 25;8(22):5363–5375. doi: 10.1093/nar/8.22.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Igo-Kemenes T., Zachau H. G. Nucleotide sequence of a highly repetitive component of rat DNA. Nucleic Acids Res. 1979 Sep 25;7(2):417–432. doi: 10.1093/nar/7.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B., McCarthy B. Location of histones on simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2895–2899. doi: 10.1073/pnas.72.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder B. A., Crawford L. V. The arrangement of nucleosomes in nucleoprotein complexes from polyoma virus and SV40. Cell. 1977 May;11(1):35–49. doi: 10.1016/0092-8674(77)90315-4. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to DNA sequences. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):103–108. doi: 10.1101/sqb.1978.042.01.011. [DOI] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Nucleosomal DNA is digested to repeats of 10 bases by exonuclease III. Cell. 1978 Feb;13(2):281–293. doi: 10.1016/0092-8674(78)90197-6. [DOI] [PubMed] [Google Scholar]
- Schachat F. H., Hogness D. S. Repetitive sequences in isolated Thomas circles from Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1974;38:371–381. doi: 10.1101/sqb.1974.038.01.040. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Singer D. S. Arrangement of a highly repeated DNA sequence in the genome and chromatin of the African green monkey. J Biol Chem. 1979 Jun 25;254(12):5506–5514. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Streeck R. E., Zachau H. G. A long-range and two short-range periodicities are superimposed in the 1.706-g/cm3 satellite DNA from calf thymus. Eur J Biochem. 1978 Aug 15;89(1):267–279. doi: 10.1111/j.1432-1033.1978.tb20923.x. [DOI] [PubMed] [Google Scholar]
- Thomas C. A., Jr, Pyeritz R. E., Wilson D. A., Dancis B. M., Lee C. S., Bick M. D., Huang H. L., Zimm B. H. Cyclodromes and palindromes in chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:353–370. doi: 10.1101/sqb.1974.038.01.039. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Sequence-dependent deformational anisotropy of chromatin DNA. Nucleic Acids Res. 1980 Sep 11;8(17):4041–4053. doi: 10.1093/nar/8.17.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Waldeck W., Föhring B., Chowdhury K., Gruss P., Sauer G. Origin of DNA replication in papovavirus chromatin is recognized by endogenous endonuclease. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5964–5968. doi: 10.1073/pnas.75.12.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Oudet P., Chambon P. Preferential in vitro assembly of nucleosome cores on some AT-rich regions of SV40 DNA. Nucleic Acids Res. 1979 Oct 10;7(3):705–713. doi: 10.1093/nar/7.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig B., Wittig S. A phase relationship associates tRNA structural gene sequences with nucleosome cores. Cell. 1979 Dec;18(4):1173–1183. doi: 10.1016/0092-8674(79)90230-7. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]