Abstract
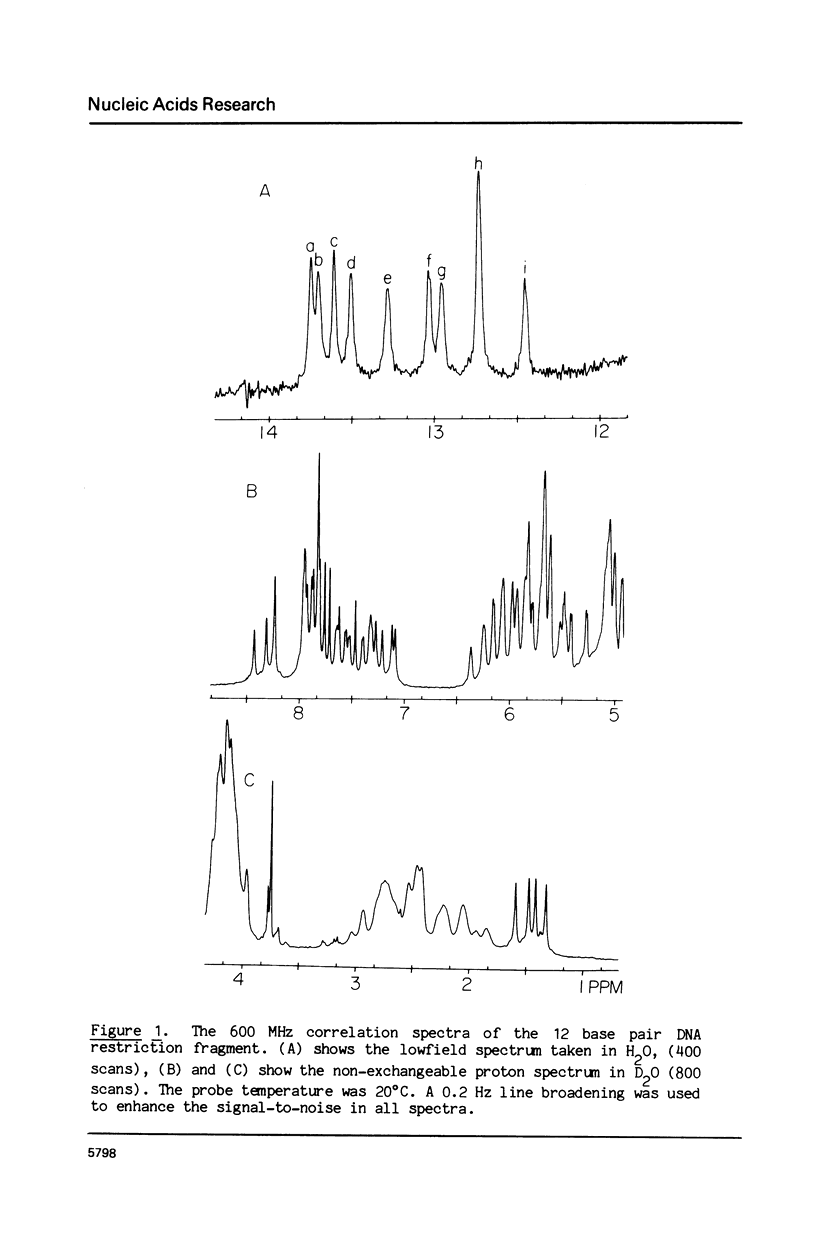
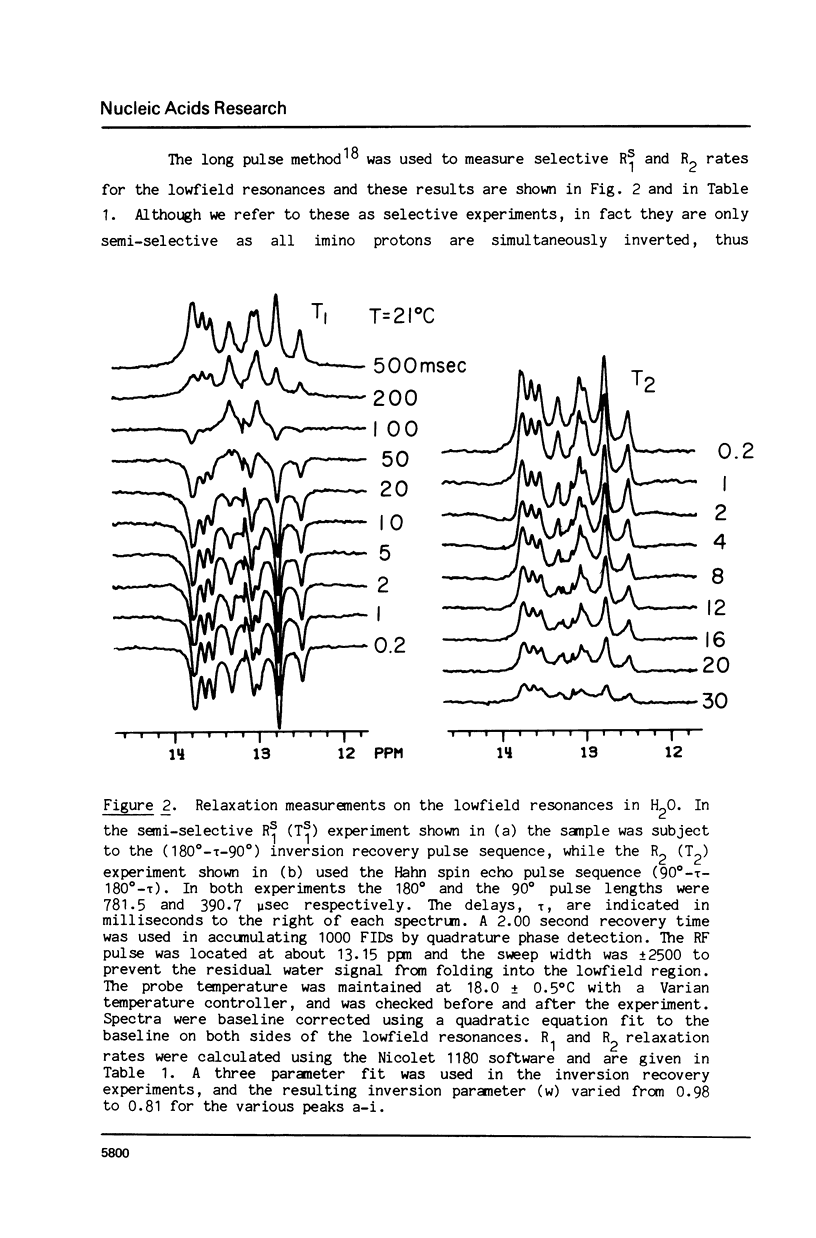
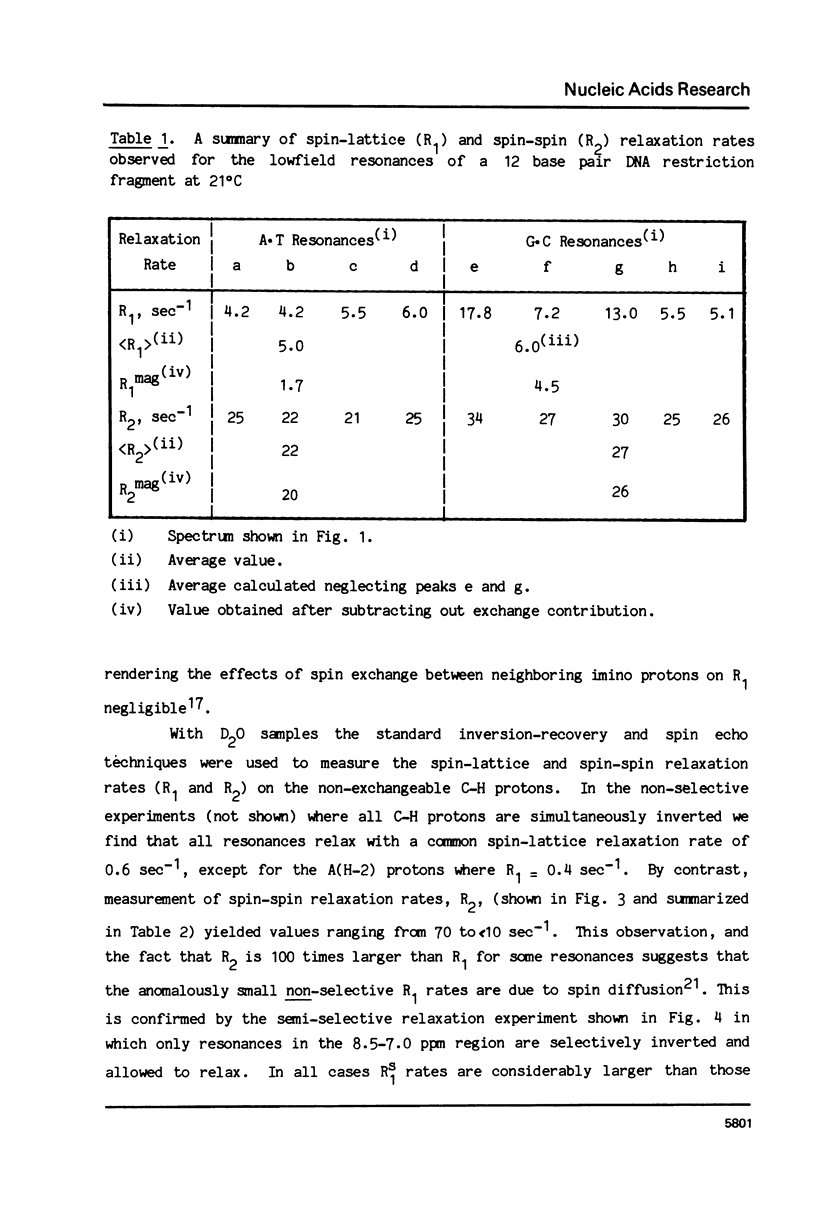
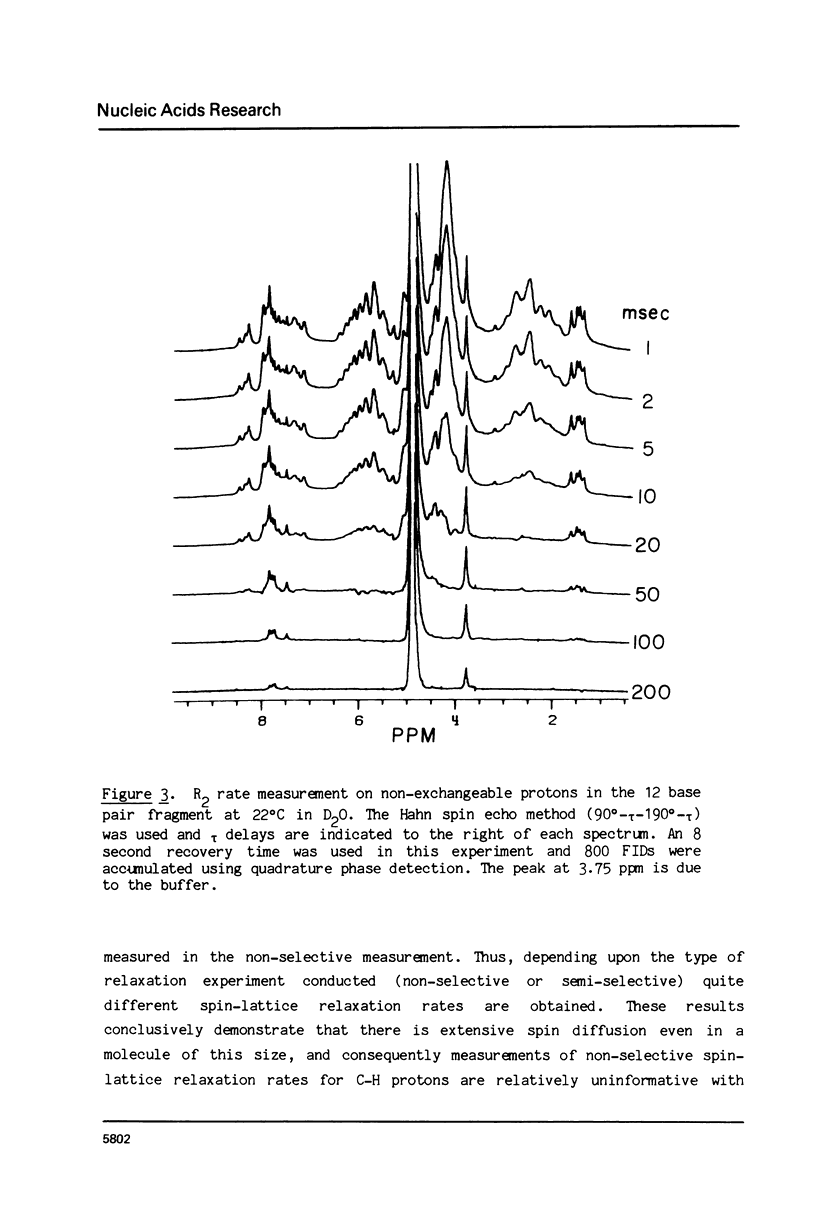
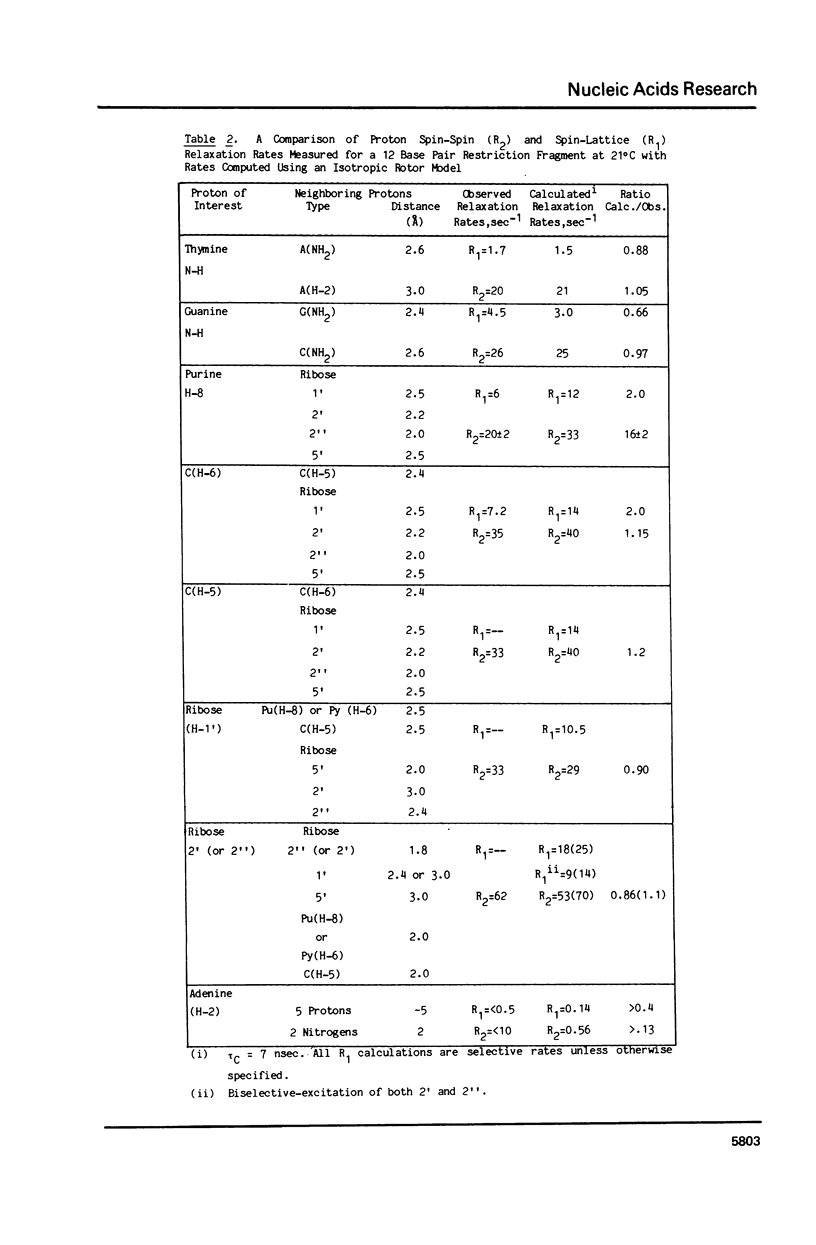
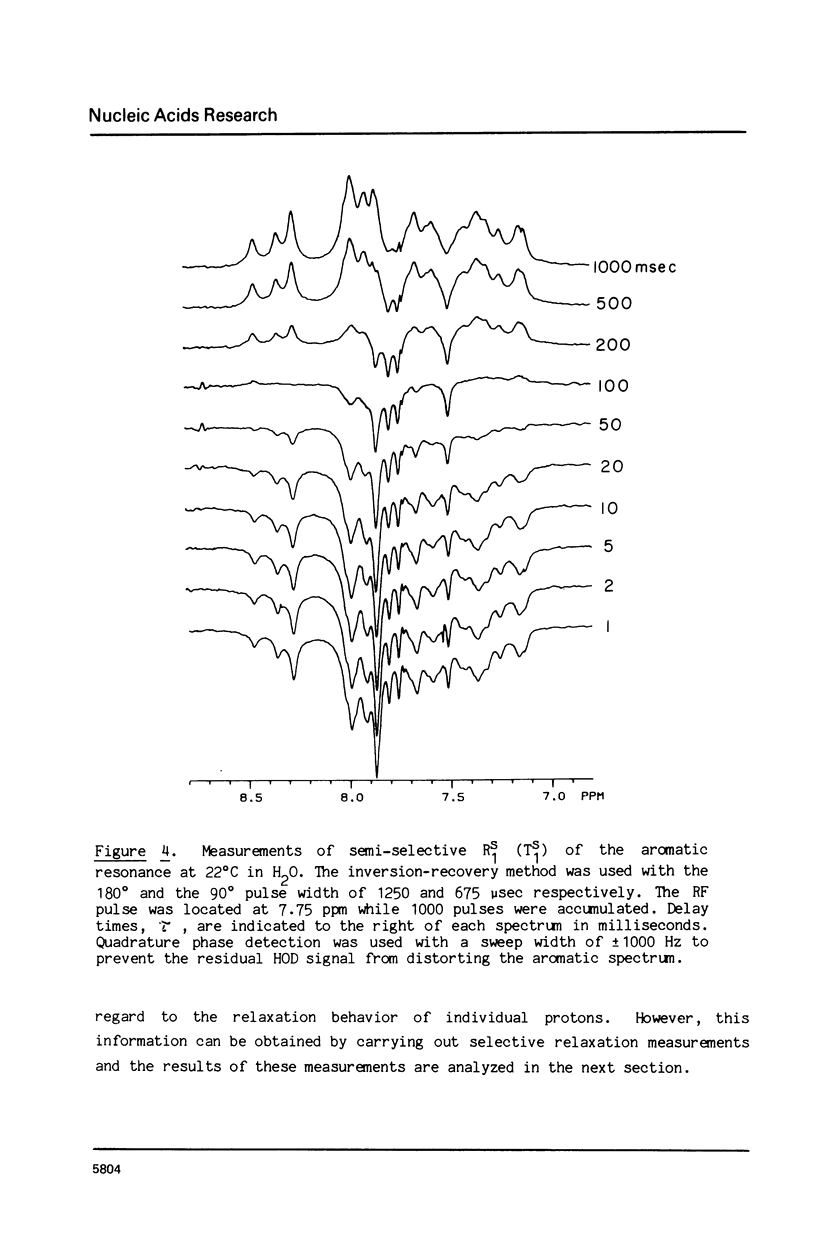
The 1H NMR spectrum of a 12 base pair DNA restriction fragment has been measured at 300 and 600 MHz and resonances from over 70 protons are individually resolved. Relaxation rate measurements have been carried out at 300 MHz and compared with the theoretical predictions obtained using an isotropic rigid rotor model with coordinates derived from a Dreiding model of DNA. The model gives results that are in excellent agreement with experiment for most protons when a 7 nsec rotational correlation time is used, although agreement is improved for certain base protons by using a shorter correlation time for the sugar group, or by increasing the sugar-base interproton distances. A comparison of non-selective and selective spin-lattice relaxation rates for carbon bound protons indicates that there is extensive spin diffusion even in this short DNA fragment. Examination of the spin-spin relaxation rates for the same type of proton on different base pairs reveals little sequence effect on conformation.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beetz C. P., Jr, Asearelli G. The low-frequency vibrational modes of an RNA: poly(I)-poly(C). Biopolymers. 1976 Nov;15(11):2299–2301. doi: 10.1002/bip.1976.360151118. [DOI] [PubMed] [Google Scholar]
- Bolton P. H., James T. L. Conformational mobility of deoxyribonucleic acid, transfer ribonucleic acid, and poly(adenylic acid) as monitored by carbon-13 nuclear magnetic resonance relaxation. Biochemistry. 1980 Apr 1;19(7):1388–1392. doi: 10.1021/bi00548a019. [DOI] [PubMed] [Google Scholar]
- Brunner W. C., Maestre M. F. Circular dichroism of films of polynucleotides. Biopolymers. 1974;13(2):345–357. doi: 10.1002/bip.1974.360130210. [DOI] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R. 1H nuclear magnetic resonance investigation of flexibility in DNA. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4165–4169. doi: 10.1073/pnas.76.9.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Burd J. F., Larson J. E., Wells R. D. High resolution proton nuclear magnetic resonance investigation of the structural and dynamic properties of d(C15A15)-d(T15G15). Biochemistry. 1977 Feb 8;16(3):541–551. doi: 10.1021/bi00622a031. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Harrison S. C. DNA arrangement in isometric phage heads. Nature. 1977 Aug 18;268(5621):598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- Erfurth S. C., Bond P. J., Peticolas W. L. Characterization of the A in equilibrium B transition of DNA in fibers and gels by laser Raman spectroscopy. Biopolymers. 1975 Jun;14(6):1245–1257. doi: 10.1002/bip.1975.360140613. [DOI] [PubMed] [Google Scholar]
- Godfrey J. E., Eisenberg H. The flexibility of low molecular weight double-stranded DNA as a function of length. I. Light scattering measurements and the estimation of persistence lengths from light scattering, sedimentation and viscosity. Biophys Chem. 1976 Sep;5(3):301–318. doi: 10.1016/0301-4622(76)80042-7. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Morgan A. R., Ratliff R. L. A comparison of the circular dichroism spectra of synthetic DNA sequences of the homopurine . homopyrimidine and mixed purine- pyrimidine types. Nucleic Acids Res. 1978 Oct;5(10):3679–3695. doi: 10.1093/nar/5.10.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M. E., Jardetzky O. Internal motions in deoxyribonucleic acid II. Biochemistry. 1980 Jul 22;19(15):3460–3468. doi: 10.1021/bi00556a009. [DOI] [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transient electric dichroism of rod-like DNA molecules. Proc Natl Acad Sci U S A. 1978 Jan;75(1):195–199. doi: 10.1073/pnas.75.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inners L. D., Felsenfeld G. Conformation of polyribouridylic acid in solution. J Mol Biol. 1970 Jun 14;50(2):373–389. doi: 10.1016/0022-2836(70)90199-3. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Borer P. N., Ts'o P. O. Conformation and interaction of short nucleic acid double-stranded helices. II. Proton magnetic resonance studies on the hydrogen-bonded NH-N protons of ribosyl ApApGpCpUpU helix. Biochemistry. 1975 Nov 4;14(22):4864–4869. doi: 10.1021/bi00693a013. [DOI] [PubMed] [Google Scholar]
- Kearns D. R. High-resolution nuclear magnetic resonance studies of double helical polynucleotides. Annu Rev Biophys Bioeng. 1977;6:477–523. doi: 10.1146/annurev.bb.06.060177.002401. [DOI] [PubMed] [Google Scholar]
- LUZZATI V., MATHIS A., MASSON F., WITZ J. SUTURE TRANSITIONS OBSERVED IN DNA AND POLY A IN SOLUTION AS A FUNCTION OF TEMPERATURE AND PH. J Mol Biol. 1964 Oct;10:28–41. doi: 10.1016/s0022-2836(64)80025-5. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. S., Wagner T. E. Origins of the differences between the circular dichroism of DNA and RNA: theoretical calculations. Biopolymers. 1973;12(1):201–221. doi: 10.1002/bip.1973.360120119. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L., Pohl F. M. "Alternating B-DNA" conformation for the oligo(dG-dC) duplex in high-salt solution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2508–2511. doi: 10.1073/pnas.76.6.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. Nuclear magnetic resonance studies of the helix-coil transition of poly (dA-dT) in aqueous solution. Proc Natl Acad Sci U S A. 1976 Mar;73(3):674–678. doi: 10.1073/pnas.73.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Hilbers C. W. Proton nuclear magnetic resonance investigations of fraying in double-stranded d-ApTpGpCpApT in H2O solution. Biochemistry. 1975 Jun 17;14(12):2651–2656. doi: 10.1021/bi00683a014. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Tonelli A. E. Assignment of the proton Nmr chemical shifts of the T-N3H and G-N1H proton resonances in isolated AT and GC Watson-Crick base pairs in double-stranded deoxy oligonucleotides in aqueous solution. Biopolymers. 1974;13(10):1943–1964. doi: 10.1002/bip.1974.360131003. [DOI] [PubMed] [Google Scholar]
- Pilet J., Blicharski J., Brahms J. Conformations and structural transitions in polydeoxynucleotides. Biochemistry. 1975 May 6;14(9):1869–1876. doi: 10.1021/bi00680a011. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Woodbury C. P., Inman R. B. Characterization of rodlike RNA fragments. Biopolymers. 1975 Feb;14(2):393–408. doi: 10.1002/bip.1975.360140212. [DOI] [PubMed] [Google Scholar]
- Selsing E., Wells R. D., Early T. A., Kearns D. R. Two contiguous conformations in a nucleic acid duplex. Nature. 1978 Sep 21;275(5677):249–250. doi: 10.1038/275249a0. [DOI] [PubMed] [Google Scholar]
- Small E. W., Peticolas W. L. Conformational dependence of the Raman scattering intensities from polynucleotides. Biopolymers. 1971;10(1):69–88. doi: 10.1002/bip.360100107. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Trifonov E. N. Possibility of nonkinked packing of DNA in chromatin. Proc Natl Acad Sci U S A. 1978 Jan;75(1):103–107. doi: 10.1073/pnas.75.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]


