Abstract
Sequence homology indicates the existence of three human cytosolic acyl protein thioesterases, including APT1 that is known to depalmitoylate H- and N-Ras. One of them is the lysophospholipase-like 1 (LYPLAL1) protein that on the one hand is predicted to be closely related to APT1 but on the other hand might also function as a potential triacylglycerol lipase involved in obesity. However, its role remained unclear. The 1.7 Å crystal structure of LYPLAL1 reveals a fold very similar to APT1, as expected, but features a shape of the active site that precludes binding of long-chain substrates. Biochemical data demonstrate that LYPLAL1 exhibits neither phospholipase nor triacylglycerol lipase activity, but rather accepts short-chain substrates. Furthermore, extensive screening efforts using chemical array technique revealed a first small molecule inhibitor of LYPLAL1.
Keywords: lysophospholipase, α/β hydrolase, chemical array screening, inhibitor
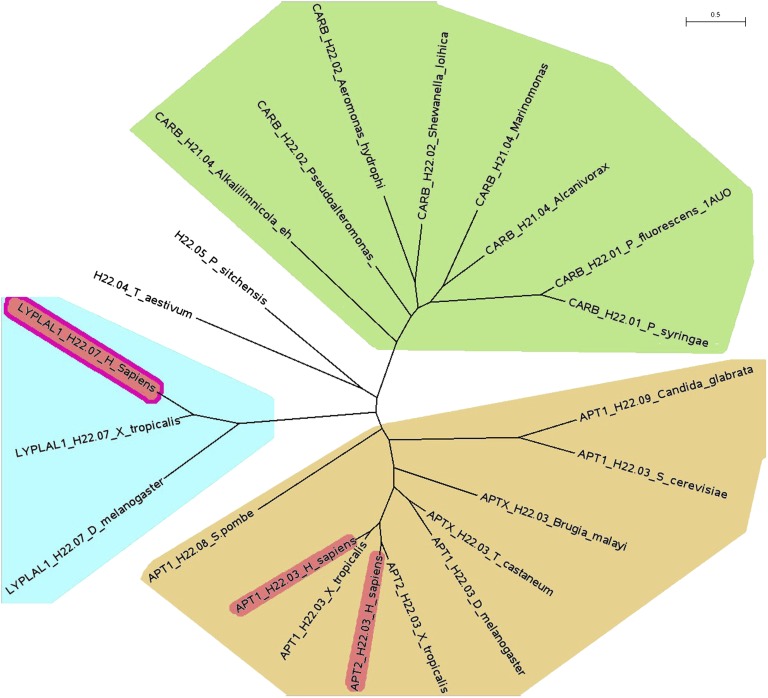
The human acyl protein thioesterases APT1, APT2, and the protein lysophospholipase-like 1 (LYPLAL1) belong, according to their amino acid sequence identities, to two different subclasses of the lysophospholipase family (Fig. 1), which is in turn a subclass of the α/β hydrolases superfamily (1). A BLAST search with LYPLAL1 finds acyl protein thioesterases as the closest relatives. All three proteins possess a catalytic triad consisting of a serine, an aspartate, and a histidine. The published crystal structure of APT1 (PDB ID 1fj2) showed that the protein has indeed a canonical α/β hydrolase fold and presented the protein in its dimeric form (2). The only other crystal structures known among the lysophospholipase family are carboxylesterase from two different bacteria (e.g., PDB ID 1auo, Fig. 1 includes the corresponding sequence labeled “CARB_H21.01_P_fluorescens_1AUO”) and two uncharacterized proteins (PDB ID 3b5e and 2r8b, SCOP database, Carboxylesterase/thioesterase 1 family) (3). APT1 is the best-characterized protein of the family and is known to depalmitoylate Gα-proteins as well as Ras proteins, and thus is involved in the modulation of the Ras signaling pathway that plays an important role in various types of cancer. The cellular localization of H-Ras and N-Ras is maintained by a palmitoylation/depalmitoylation cycle (4) that can be disturbed by APT1 inhibitors in vivo (5), leading to a reversal of the transforming effects induced by active Ras. The close relationship between the LYPLAL1 and APT1 families and the presence of the catalytic triad suggested a cellular function comparable to APT1. However, LYPLAL1 was also proposed to act as a triglyceride lipase in adipose tissue (6), and several studies indicated a significant relationship between the lyplal1 gene locus and the waist-hip ratio of study participants (7, 8) as well as fat distribution (9) and nonalcoholic fatty liver disease (10). Yet, the biological function and significance of LYPLAL1 remained barely investigated, because no structural or biochemical information was available, so we set out to solve the crystal structure of human LYPLAL1 and characterize its enzymatic properties.
Fig. 1.
Phylogenetic tree (23) of LYPLAL1, the closely related acyl protein thioesterases (APT) and representatives of the most similar carboxylesterases. Proteins are labeled with the group identifier, the family name used in the “lipase engineering database” (1) and the species. The LYPLAL1 group (family H22.07) is highlighted in light blue, the APTs (family H22.03) in ochre and the carboxylesterases (several different families) in green. The proteins highlighted in red are the only human representatives in the superfamily H22, the superfamily H21 does not comprise any human proteins. The only available structures in the PDB database are human APT1 (highlighted in red, PDB ID 1fj2), carboxylesterase from Pseudomonas fluorescens (PDB ID 1auo, 2nd from right in the green area) and human LYPLAL1 (highlighted red, from this work).
Materials and Methods
Molecular cloning
Human lyplal1 cDNA (imaGenes clone IRAUp969E0370D) was used as a template for PCR amplification. The lyplal1 full-length gene was gateway cloned into a pGEX 4T1 expression vector (GE Healthcare), primer modified to encode a PreScission protease cleavage site immediately upstream to the lyplal1 start codon, leaving the two amino acids GP preceding the N-terminal start methionine as cloning artifact.
Protein expression and purification
Escherichia coli BL21 Codon +RIL cells were transformed using the cloned construct. Cells were grown at 25°C and protein expression was induced overnight at 20°C using 0.1 mM IPTG. Cells were harvested, lysed using a high pressure cell disruptor, cell debris was removed by centrifugation at 100,000 g, and the supernatant was loaded on a GSH affinity column. The column was washed with 20 column volumes of buffer. PreScission protease (GE Healthcare) was then loaded on the column and slowly circulated over the column overnight. Afterwards, all cleaved protein was washed from the column. The entire GSH affinity chromatography was carried out under the following buffer conditions: 20 mM Tris-HCl pH 8.5, 150 mM NaCl, 5% Glycerol, 5 mM DTE. The protein was subjected to subsequent gel filtration, using the following buffer: 20 mM Tris-HCl pH 8.0, 30 mM NaCl, 1 mM TCEP-HCl. Finally, the protein was concentrated using 10,000 kDa MWCO spin filters to a concentration of 17 mg ml−1, and flash-frozen. GST-LYPLAL1 fusion protein was purified the same way but eluted from the GSH column using 20 mM glutathione.
Protein crystallization and structure solution
Protein crystallization was carried out in 200 nl sitting drops, using a 1:1 ratio of crystallization buffer (0.1 M Bis-Tris pH 5.5, 25% (w/v) PEG 3350) and protein solution. The crystal was a thin needle and grew within 5 days. Data was collected at the SLS PXII beamline in Villigen, Switzerland and processed with XDS (11). The structure was solved using molecular replacement with PHASER [CCP4 suite (12)] and APT1 (PDB code 1fj2) as template. The structure was refined using REFMAC (CCP4 suite) and COOT (13) to a final R factor of 18.20 and an Rfree of 21.99 with excellent geometries (Ramachandran plot values 97.3%, 2.7%, and 0.0%) and visualized with PyMOL (14). The coordinates have been deposited in the Protein Data Bank under the accession code 3u0v.
Microarray screening
The slides of Natural Products Depository (NPDepo) (15) were prepared and analyzed as reported before (16–18). The microarray screening was performed using the following buffer: 20 mM Hepes, 150 mM NaCl, pH 7.53-7.55. For protein and antibody incubation, the array slide was covered by a glass slide from Matsunami Glass Ind., Ltd (microscope cover glass, 24 × 60 mm) and incubated using 50 μl of GST-protein solution (1 μM) in buffer containing 1% skim milk at 30°C for 1 h. After washing, array slides were incubated with anti-GST antibody (rabbit IgG fraction, Invitrogen, 30 μg ml−1) in buffer containing 1% skim milk at 30°C for 1 h. This incubation was followed by another wash step and incubation with a second antibody (Millipore, goat anti-rabbit IgG, Cy5 conjugate, 50 μg ml−1) at 30°C for 1 h. After the final wash step, slides were scanned at 635 nm on a GenePix 4200AL microarray scanner (Molecular Devices). The fluorescence signals were quantified using GenePix 6.1 software with local background subtraction and data analyzed.
Substrate screening
Parameters of steady state kinetics were measured at a final concentration of 300 nM LYPLAL1 using either a colorimetric (para-nitrophenyl acetate, PNPA) or fluorimetric detection (6,8-difluoro-4-methylumbelliferyl acetate, DiFMUA). In the colorimetric assay, the release of yellow para-nitrophenol (PNP) from PNPA was monitored by recording the increase of absorbance at 405 nm at 37°C over 40 min at 30 s intervals using a Tecan Infinite M200 microplate reader. The reaction was measured at 300 nM LYPLAL1 and 500 μM of the PNP-ester in assay buffer [20 mM Hepes, 150 mM NaCl, pH 7.53-7.55, 0.01% (v/v) Triton-X100] with final PNPA concentrations ranging from 2400 μM to 37.5 μM. The resulting absorbance was referenced to a linear PNP absorbance-concentration relationship. For the comparison of the other PNP-esters to PNP-acetate (Fig. 3), 300 nM LYPLAL1 in the above assay buffer and 500 μM of each compound was used to measure the initial velocities (Table 1). In the fluorimetric assay, the increase of fluorescence intensity at 358/455 nm (excitation/emission) with final DiFMUA concentrations ranging from 40 μM to 1.25 μM in assay buffer were recorded at 37°C over 40 min at 30 s intervals. The resulting fluorescence was referenced to a linear 6,8-difluoro-4-methylumbelliferol (DiFMU) fluorescence-concentration relationship. The specific activity of the LYPLAL1 protein preparation under the described assay conditions was 1.3 U/mg for PNPA and 0.021 U/mg for DiFMUA. Km and Vmax values were obtained by nonlinear least square fitting of Michaelis-Menten equation parameters to the data using XL Fit. The determined constants are: PNPA-assay (Km = 782 ± 130 μM; vmax = 168 nmol s−1; Kcat = 0.56 s−1; Kcat/Km = 0.7 s−1mM−1) and DiFMUA-assay (Km = 12.6 ± 5.6 μM; vmax = 2.72 nmol s−1; Kcat = 0.009 s−1; Kcat/Km = 0.7 s−1mM−1).
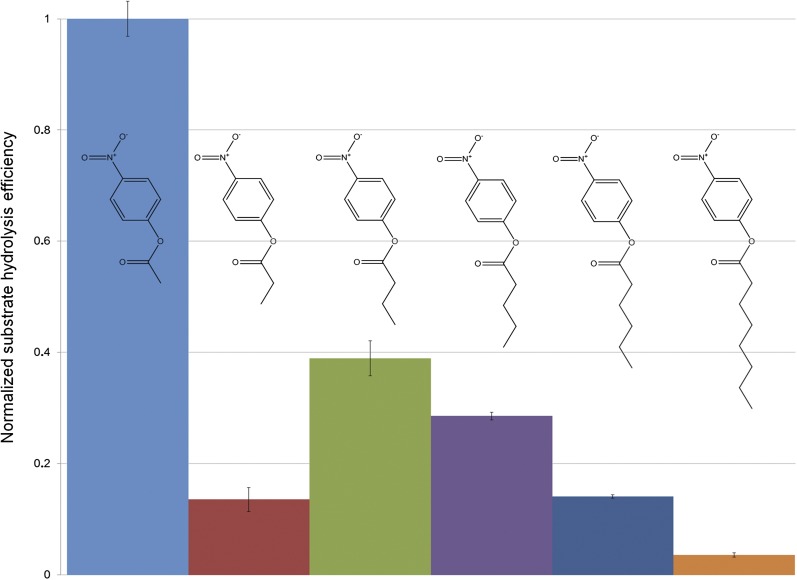
Fig. 3.
Normalized hydrolysis of different substrates by LYPLAL1. Release of 4-nitrophenol upon hydrolysis of different PNP esters (each at a concentration of 500 μM) by 150 nM LYPLAL1 was measured in triplicates, the slopes of the regression lines were averaged and then normalized against the slope of the most efficiently hydrolyzed compound (PNPA). The error bars represent the standard deviation of the results. The following compounds were used as drawn above the bars (from left to right): PNP acetate, PNP propionate, PNP butyrate, PNP valerate, PNP hexanoate, and PNP octanoate. The specific activity of the LYPLAL1 preparation was 1.3 U/mg for PNP acetate.
TABLE 1.
List of tested substrates/inhibitors
| 1 | 4-nitrophenyl acetatev @ 500 μM = 65.5 2.1 nmol/s, Km = 782 μM, kcat 0.56 s , Vmax = 168 nmol/s |
| 2 | 4-nitrophenyl propionatev @ 500 μM = 8.9 1.4 nmol/s |
| 3 | 4-nitrophenyl butyratev @ 500 μM = 25.5 0.7 nmol/s |
| 4 | 4-nitrophenyl valeratev @ 500 μM = 18.7 0.5 nmol/s |
| 5 | 4-nitrophenyl hexanoatev @ 500 μM = 9.2 0.2 nmol/s |
| 6 | 4-nitrophenyl octanoatev @ 500 μM = 2.4 0.2 nmol/s |
| 7 | 4-nitro-phenoxyacetic acid |
| 8 | Resorufin oleate |
| 9 | 4-(Trifluoromethy)umbelliferyl oleate |
| 10 | 8-Octanoyloxypyrene-1,3,6-trisulfonic acid |
| 11 | 1,2-Dioleoyl-3-(pyren-1-yl)decanoyl-rac-glycerol |
| 12 | Dansyl-11-aminoundecanoic acid |
| 13 | D,L-2-amino-5-phosphonovaleric acid |
| 14 | Propylphosphoric acid |
| 15 | Phosphonoacetic acid |
| 16 | 3-Aminopropylphosphonic acid |
| Substrates from Invitrogen phospholipase kits: | |
| 17 | A1: Glycero-phosphoethanolamine (dye-labeled at the acyl chain sn-1 position, quencher at the glycerophosphoethanolamine head group, noncleavable ether linkage in the sn-2 position), full name: N-((6-(2,4-DNP)amino)hexanoyl)-1-(BODIPY® FL C5)-2-hexyl-sn-glycero-3-phosphoethanolamine |
| 18 | A2: Glycero-phosphocholine (red/green dye-labeled at the acyl chain sn-1 and sn-2 positions, noncleavable ether linkage in the sn-1 position) full name: 1-O-(6-BODIPY® 558⁄568-aminohexyl)-2-BODIPY® FL C5-sn-glycero-3-phosphocholine |
| 19 | C: Glycero-phosphoethanolamine (dye-labeled at the acyl chain sn-2 position, proprietary substrate) |
| 20 | D: Phosphatidylcholine |
v @ 500 μM, initial velocities at 500 μM substrate concentration; protein concentration was 300 nM.
Synthesis of fluorescent substrate DiFMUA
To a solution of DiFMU (19) (100 mg, 0.47 mmol, 1 eq.) in dichloromethane (5 ml), was added triethylamine (99 μl, 0.71 mmol, 1.5 eq.) followed by acetyl chloride (47 μl, 0.47 mmol, 1.1 eq.). The reaction mixture was stirred at room temperature overnight. Solvents were evaporated and the product purified via flash chromatography to give a white solid (78 mg, 65%). m.p. 129-131°C; Rf = 0.39 (petroleum ether/ethyl acetate, 3:1); 1H-NMR (400 MHz, DMSO): δ = 2.41 (s, 3H, COCH3), 2.46 (s, 3H, CH3), 6.55 (s, 3H, CH), 7.75 (dd, 1H, J1 = 10.5, J2 = 2.3, CHar); 13C-NMR (101 MHz, DMSO): δ = 18.3 (CH3), 19.8 (COCH3), 107.2 (d, J = 4, CHar), 115.7 (CH), 118.5-118.6 (m, Cq), 128.7 (dd, J1 = 18, J2 = 13, Cq), 138.7 (dd, J1 = 10, J2 = 3, Cq), 141.8 (dd, J1 = 253, J2 = 5, Cq), 150.0 (dd, J1 = 242, J2 = 4, Cq), 152.3-152.4 (m, Cq), 157.9 (CO), 167.2 (CO); HRMS (ESI): calcd for [M-H]+ C12H9O4F2 255.04634, found 255.04658.
Enzymatic assay
The enzyme activity was determined by measuring the release of fluorescent DiFMU by LYPLAL1 hydrolysis of DiFMUA in a Tecan Infinite M200 microplate reader in a 96-well format. The final enzyme concentration was 300 nM and the final substrate concentration was 40 μM. In the assay, 30 μl inhibitor solutions (at varying concentrations) in assay buffer were mixed with 20 μl LYPLAL1 solution (1.5 μM) in assay buffer. The mixture was incubated for 20 min. at 37°C. Subsequently, 50 μl of a solution of DiFMUA (80 μM) in assay buffer was added to the reaction mixture and after 2 min lag time the formation of fluorescent DiFMU was recorded (λex = 358 nm, λem = 455 nm) over 40 min at 60 s intervals at 37°C. During the incubation and in between measurements, the reaction mixture was shaken. The reaction rate was determined by linear regression analyses (r2 ≥ 0.98) of the fluorescence emission increase over time. The background reaction rate with no enzyme present was subtracted and the reaction rates were normalized to the reaction rate with no inhibitor present (100%).
Results
LYPLAL1 features an active site shape different from APT1
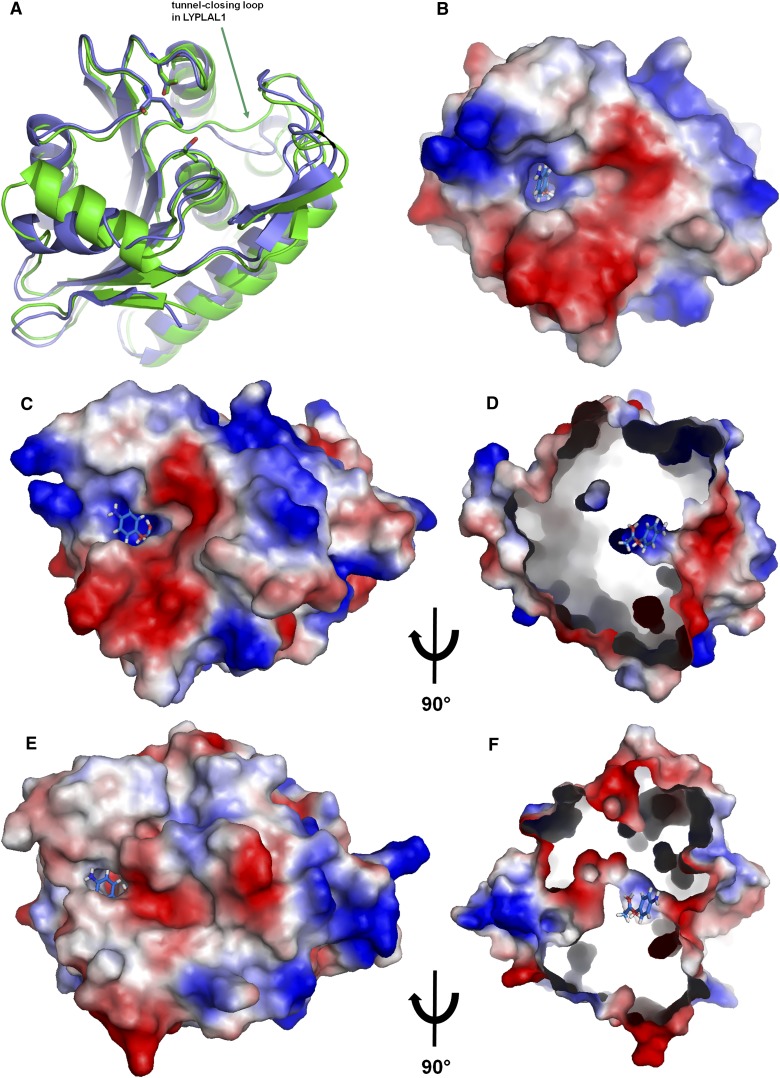
The crystal structure of human LYPLAL1 revealed a typical α/β hydrolase fold and features the classical catalytic triad of serine, aspartate, and histidine. Despite a relatively low sequence identity of 33% to APT1, the latter is picked up as the most similar fold in the protein database by a DALI search (20). The electron density with a resolution of 1.72 Å (Table 2) is very well defined throughout starting with the residue Leu9; the preceding residues of the N-terminus are disordered. Remarkable differences to APT1 are apparent in the electrostatic surface potential surrounding the active site and in the shape of the active site itself: In LYPLAL1 the typical hydrophobic tunnel of the acyl protein thioesterases that accommodates the lipid moiety of the substrate is closed due to a movement of the loop and adjacent α-helix around Asn152 (Fig. 2). This region is strongly conserved in the lysophospholipase-like subfamily (abH22.07 as defined in the Lipase Engineering Database (1), but less conserved in the lysophospholipase-subfamily (including the APTs, abH22.03 in the database). In addition, the side chain of Arg80 that sits in place of Ile70 in APT1 would block the center of a potential tunnel and is also totally conserved in the LYPLAL-subfamily. These conformational differences between APT1 and LYPLAL1 lead to the closing of the rear-end of the APT1 tunnel (Fig. 2 C–F) by a tightly packed network of side and main chains with relatively low temperature factors, making a rearrangement leading to a tunnel opening highly unlikely. LYPLAL1 therefore misses the structural basis for delipidation of large substrates like palmitoylated proteins as well as long chain lipid degradation in general. Indeed, the follow-up experiments confirmed that LYPLAL1 is neither able to hydrolyze long-chain esters (see below) nor to depalmitoylate N-Ras protein in vitro (unpublished observations) so that a potential induced-fit mechanism is very unlikely.
TABLE 2.
X-ray statistics
| Data collection | |
| Space group | P212121 |
| a,b,c (Å) | 47.78 60.78 76.62 |
| α, β, γ (°) | 90.0 90.0 90.0 |
| Resolution (Å) | 1.72 (1.76-1.72)* |
| Rsym | 5.7 (34.6) |
| I / σI | 17.54 (3.10) |
| Completeness (%) | 98.4 (94.3) |
| Redundancy | 4.77 (2.95) |
| Refinement | |
| Resolution (Å) | 47.62-1.72 |
| No. reflections | 25449 |
| Rwork / Rfree | 17.3 / 21.1 |
| No. atoms | 1942 |
| Protein | 1797 |
| Water | 145 |
| B-factors average | 19.7 |
| R.m.s. deviations: | |
| Bond lengths (Å) | 0.032 |
| Bond angles (°) | 2.65 |
Values in parentheses are for highest resolution shell.
Fig. 2.
Structural representations of LYPLAL1. A: Crystal structure of LYPLAL1 compared with APT1 (PDB ID 1fj2). The catalytic triad is shown as sticks. B: Active site of LYPLAL1 with a docked 4-nitrophenyl acetate molecule in the same orientation as A. C–F: Comparison of the active site shape between LYPLAL1 [top (C and D)] and APT1 in the same orientation [bottom (E and F), PDB ID 1fj2]. D and F are shown in a side view and the surfaces are cut open to show the active site tunnel of APT1 extending through the protein, in contrast to the shallower active site of LYPLAL1 that does not form a tunnel. The 4-nitrophenyl acetate molecule docked into LYPLAL1 is shown in all four views for comparison. The surfaces are colored according to the electrostatic surface properties. Red represents negative, blue positive potential.
The electrostatic surface potential around the active site shows distinct positive charge in the immediate vicinity of the catalytic serine. (Fig. 2–D). The active site is completely solvent accessible, and judging from the temperature factors, there is no indication of a potential lid-like movement of the adjacent loops. The crystal packing suggests that LYPLAL1 is a monomer in solution, in contrast to APT1 where a dimer is found in the asymmetric unit. Thus, despite having the most similar fold to APT1 among all known PDB structures, LYPLAL1 has an open, solvent-accessible active site reminiscent of α/β hydrolases that hydrolyze smaller substrates (like e.g., the closely related carboxylesterases, Fig. 1). The activity assays described in the following confirmed this hypothesis.
LYPLAL1 prefers short-chain substrates
Because it was originally proposed that LYPLAL1 is a phospholipase, the protein was subjected to activity tests for the four different types of phospholipases A1, A2, C and D (Invitrogen EnzChek phospholipase assays, compounds 17–20 in Table 1). As already suggested by the crystal structure, LYPLAL1 did not display any measurable phospholipase activity (data not shown). Furthermore, substrates detecting triacylglycerol lipase and lipase activity in general (compounds 8–12 in Table 1) did not result in any observable enzymatic activity. Subsequently, guided by the crystal structure, other substrates were tested as potential substrates of LYPLAL1. 4-nitrophenyl esters differing in length were used to check for the presence of free 4-nitrophenol upon hydrolysis of the compound's ester bond. Among all tested 4-nitrophenyl esters, 4-nitrophenyl acetate (PNPA) was the one being most efficiently hydrolyzed by LYPLAL1 (Fig. 3), corroborating the already suspected preference for small substrates. Also, the published APT1 inhibitor Palmostatin B (5) did not show any detectable inhibitory effect on LYPLAL1 at 50 μM inhibitor concentration. For substrates with increasing chain lengths, LYPLAL1 showed reduced activity, consistent with the shallow shape of the active site (Fig. 2B–D). Interestingly, 4-nitrophenyl propionate is an exception to that rule, and showed (reproducibly) less hydrolysis than 4-nitrophenyl butyrate by the enzyme (Fig. 3).
In order to develop a more sensitive assay that allows for lower substrate concentrations, PNPA was substituted by DiFMUA in the activity assay. Michaelis-Menten kinetics showed that LYPLAL1 hydrolyzed DiFMUA with the same catalytic efficiency (kcat/Km) as PNPA (see Materials and Methods section). Virtual screening using the software GOLD (21) with a subset of the ZINC library (22) identified compounds with phosphate groups (related to compounds 13-16 in Table 1) or carboxyl groups (like compound 1, see below) buried in the positively charged part of the active site. However, the compounds containing a phosphate group (13-16 in Table 1) did not show inhibitory activity when tested in the assay with DiFMUA as a substrate.
In order to get an idea about potential physiological substrates, it would be helpful to know the detailed binding modes of at least some compounds. Therefore, cocrystallization attempts with a nonhydrolysable PNPA analog (compound 7 in Table 1) were made. As no electron density could be found in the active site, probably due to insufficient affinity, a high-throughput screening was performed as described below.
Identification of a small-molecule inhibitor
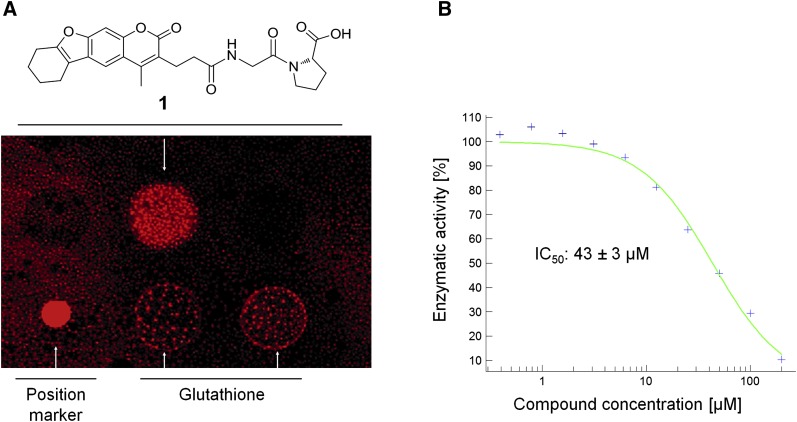
To obtain lead structures for potential LYPLAL1 binders, we screened small molecules from the RIKEN NPDepo library (15) by chemical array. In total, 12,356 compounds from NPDepo were immobilized on glass microarrays as bait for a GST-LYPLAL1-fusion protein and a GST-specific antibody was used for detection of bound protein. From the screen, small molecule 1 could be identified showing a positive binding signal on the array glass slide (Fig. 4A). In a follow-up, it was shown that compound 1 not only binds to LYPLAL1 in the microarray screening, but that it also inhibits its enzymatic activity (Fig. 4B). Compound 1 constitutes a novel scaffold that could serve as starting point for the development of more potent inhibitors. Attempts to cocrystallize LYPLAL1 with compound 1 failed, probably due to a combination of insufficient binding affinity and solubility problems of the compound.
Fig. 4.
Detection and evaluation of the LYPLAL1 inhibitor. A: Chemical array showing that compound 1 binds to GST-LYPLAL1. B: IC50 value determination of LYPLAL1 inhibition by compound 1.
Discussion
The crystal structure of LYPLAL1 in combination with the biochemical assays has revealed an unexpected substrate preference compared with the closely related enzyme APT1: LYPLAL1 has a shallow active site whereas APT1 features a large hydrophobic tunnel that most likely accommodates the large lipid substituents of its natural substrates, namely prenylated proteins. Despite its close relationship to APT1, LYPLAL1 does not cleave thioester bonds of prenylated N-Ras nor does it have any phospholipase activity as initially assumed (6). Corroborating this finding, the known APT1 inhibitor Palmostatin B, also containing a large fatty acid chain, does not exhibit any inhibition of LYPLAL1. The sequence around the crucial residue Asn152 that is mainly responsible for the different shapes of the active sites of the two enzymes is highly conserved in evolution among the lysophospholipase-like proteins, thus suggesting that the lack of a large substrate binding site is a feature of all members of the lysophospholipase-like subfamily, which in turn, interestingly, only seems to be present in metazoans.
The assay developed in order to further investigate the substrate specificity of LYPLAL1 showed that small nitrophenyl esters are readily hydrolyzed. The positively charged patch close to the active site of the LYPLAL1 structure indicates that a substrate with an at least partial negative charge is most likely to bind to the enzyme. However, compounds containing a phosphate group did not inhibit the protein activity, most likely because this group is sterically not preferred to bind in the active site of the enzyme. On the other hand, compounds with carboxyl groups showed (inhibitory) activity, suggesting a selectivity for smaller functional groups. 4-nitrophenyl acetate turned out to be the best substrate for LYPLAL1, confirming the preference for small substrates. Compared with acetate and butyrate, the relatively low efficiency in hydrolysis of 4-nitrophenyl propionate might suggest an alternative binding mode in the active site for this molecule, e.g., by the propionate binding deeper into the pocket than the butyrate, thus positioning the ester bond that is to be cleaved in a nonoptimal position relative to the catalytic serine. Substitution of PNPA by DiFMUA enhanced the sensitivity of the assay and did not affect the hydrolysis efficiency significantly, indicating that the fluorophore does not sterically hinder the hydrolysis of the ester bond, or at least not more than the PNP does. These data also give a hint about how the substrate is most likely oriented in the active site, because the fluorophore would not fit into the deepest part of the active site close to the catalytic serine (Fig. 1A,B).
A further search for the natural substrate of LYPLAL1 guided by the X-ray structure and virtual screening trials was not successful, but a small molecule binder with an IC50 value of 43 μM was identified with the help of a microarray screen of the RIKEN NP Depo library. This compound has a carboxyl group that could potentially interact with the positive patch in the active site, but no defined electron density was visible in the crystals grown in the presence of this inhibitor, either because of a nondefined binding mode of the inhibitor or due to solubility problems of the small molecule.
Because the physiological role of LYPLAL1 is still unknown, a high-affinity inhibitor that can be developed with the aid of our lead compound and the X-ray structure could be used to probe the cellular function of LYPLAL1 by. e.g., applying it to (adipocyte) cell cultures or whole organisms. Existing covalent inhibitors for serine hydrolases could then be modified into covalently attached inhibitors that are specific for LYPLAL1 and would allow for e.g., activity-based protein profiling to test for up- or down-regulation of the protein or help to elucidate the natural substrate(s) of LYPLAL1 via metabolite profiling.
In summary, we have determined the first crystal structure of human LYPLAL1 and revealed some of its enzymatic properties. These results have shown that this protein, despite its close relationship to acyl protein thioesterases, neither cleaves thioesterase bonds of prenylated proteins, in agreement with the differently shaped active site revealed by the X-ray structure, nor is it a triglyceride lipase as initially assumed (6), and thus apparently has a metabolic role that is different than previously proposed. Our data not only provide the structural basis for further studies to clarify LYPLAL1’s detailed physiological function; we also developed an assay and presented the first small molecule inhibiting the enzyme's activity, a compound that will facilitate further research of this protein. As this molecule allows for potent inhibition of LYPLAL1, it will be easier to probe the enzyme's exact function in future investigations by using this compound (or optimized molecules derived from the lead compound) in biochemical and cell-based assays.
Acknowledgments
X-ray data collection was performed at the Swiss Light Source, beamline X10SA, Paul Scherrer Institute, Villigen, Switzerland. The authors thank the beamline staff and A. Meinhart, D. Frey and I. Schlichting for help with the data collection. We thank the RIKEN-Max Planck Joint Research Center for support, T. Saito for supplying compounds from the NPDepo, and K. Honda and M. Uchida for technical support.
Footnotes
Abbreviations:
- APT
- acyl protein thioesterase
- DiFMU
- 6,8-difluoro-4-methylumbelliferol
- DiFMUA
- DiFMU acetate
- LYPLAL
- lysophospholipase-like
- DALI
- Distance mAtrix aLIgnment
- NPDepo
- Natural Products Depository
- PNP
- para-nitrophenol
- PNPA
- para-nitrophenyl acetate
This study was supported by SFB 642 from the Deutsche Forschungsgemeinschaft to M.B. and the International Max Planck Research School (IMPRS) as well as Kekulé fellowship to T.Z.
REFERENCES
- 1.Fischer M., Pleiss J. 2003. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acids Res. 31: 319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devedjiev Y., Dauter Z., Kuznetsov S. R., Jones T. L., Derewenda Z. S. 2000. Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A. Structure. 8: 1137–1146. [DOI] [PubMed] [Google Scholar]
- 3.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247: 536–540. [DOI] [PubMed] [Google Scholar]
- 4.Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. H. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 307: 1746–1752. [DOI] [PubMed] [Google Scholar]
- 5.Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Schölermann B., et al. 2010. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6: 449–456. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg G. R., Kemp B. E., Watt M. J. 2007. Adipocyte triglyceride lipase expression in human obesity. Am. J. Physiol. Endocrinol. Metab. 293: E958–E964. [DOI] [PubMed] [Google Scholar]
- 7.Heid I. M., Jackson A. U., Randall J. C., Winkler T. W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M. C., Speliotes E. K., Mägi R., et al. 2010. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42: 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamin A. M., Suchindran S., Pearce K., Rowell J., Lien L. F., Guyton J. R., McCarthy J. J. 2011. Gene by sex interaction for measures of obesity in the Framingham Heart Study. J. Obes. 2011: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindgren C. M., Heid I. M., Randall J. C., Lamina C., Steinthorsdottir V., Qi L., Speliotes E. K., Thorleifsson G., Willer C. J., Herrera B. M., et al. 2009. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 5: e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speliotes E. K., Yerges-Armstrong L. M., Wu J., Hernaez R., Kim L. J., Palmer C. D., Gudnason V., Eiriksdottir G., Garcia M. E., Launer L. J., et al. 2011. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 7: e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- 13.Emsley P., Lohkamp B., Scott W. G., Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC [Google Scholar]
- 15.Osada H. 2010. Introduction of new tools for chemical biology research on microbial metabolites. Biosci. Biotechnol. Biochem. 74: 1135–1140. [DOI] [PubMed] [Google Scholar]
- 16.Hagiwara K., Kondoh Y., Ueda A., Yamada K., Goto H., Watanabe T., Nakata T., Osada H., Aida Y. 2010. Discovery of novel antiviral agents directed against the influenza A virus nucleoprotein using photo-cross-linked chemical arrays. Biochem. Biophys. Res. Commun. 394: 721–727. [DOI] [PubMed] [Google Scholar]
- 17.Kanoh N., Asami A., Kawatani M., Honda K., Kumashiro S., Takayama H., Simizu S., Amemiya T., Kondoh Y., Hatakeyama S., et al. 2006. Photo-cross-linked small-molecule microarrays as chemical genomic tools for dissecting protein-ligand interactions. Chem. Asian J. 1: 789–797. [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki I., Simizu S., Ichimiya H., Kawatani M., Osada H. 2008. Robust and systematic drug screening method using chemical arrays and the protein library: identification of novel inhibitors of carbonic anhydrase II. Biosci. Biotechnol. Biochem. 72: 2739–2749. [DOI] [PubMed] [Google Scholar]
- 19.Sun W-C., Gee K. R., Haugland R. P. 1998. Synthesis of novel fluorinated coumarins: Excellent UV-light excitable fluorescent dyes. Bioorg. Med. Chem. Lett. 8: 3107–3110. [DOI] [PubMed] [Google Scholar]
- 20.Dietmann S., Park J., Notredame C., Heger A., Lappe M., Holm L. 2001. A fully automatic evolutionary classification of protein folds: Dali Domain Dictionary version 3. Nucleic Acids Res. 29: 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones G., Willett P., Glen R. C., Leach A. R., Taylor R. 1997. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267: 727–748. [DOI] [PubMed] [Google Scholar]
- 22.Irwin J. J., Shoichet B. K. 2005. ZINC - a free database of commercially available compounds for Virtual Screening. J. Chem. Inf. Model. 45: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouy M., Guindon S., Gascuel O. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224. [DOI] [PubMed] [Google Scholar]