Background: Mutations in SAMHD1 cause Aicardi-Goutières syndrome. SAMHD1 is the HIV-1 restriction factor preventing replication in myeloid cells.
Results: The SAMHD1 HD domain contains a dGTP-regulated deoxynucleotide triphosphohydrolase.
Conclusion: SAMHD1 catalytic activity reduces deoxynucleotide precursor levels, restricting HIV-1 replication.
Significance: The HIV-2 Vpx accessory protein targets SAMHD1 for degradation in a viral strategy to control cellular deoxynucleotide levels for efficient replication.
Keywords: Autoimmune Diseases, Enzyme Catalysis, HIV-1, Nucleoside Nucleotide Metabolism, Viral Replication
Abstract
The SAMHD1 protein is an HIV-1 restriction factor that is targeted by the HIV-2 accessory protein Vpx in myeloid lineage cells. Mutations in the SAMHD1 gene cause Aicardi-Goutières syndrome, a genetic disease that mimics congenital viral infection. To determine the physiological function of the SAMHD1 protein, the SAMHD1 gene was cloned, recombinant protein was produced, and the catalytic activity of the purified enzyme was identified. We show that SAMHD1 contains a dGTP-regulated deoxynucleotide triphosphohydrolase. We propose that Vpx targets SAMHD1 for degradation in a viral strategy to control cellular deoxynucleotide levels for efficient replication.
Introduction
Viruses target specific host cell molecules that would otherwise restrict viral propagation. The HIV-1 and other lentiviral genomes encode different sets of accessory proteins directed at eluding host adaptive and innate immune mechanisms to promote viral replication in specific cell environments (1, 2). The Vpx and Vpr accessory proteins have been linked mostly to infection of myeloid lineage cells by HIVs (3). The Vpx accessory protein is found exclusively in HIV-2 and some simian immunodeficiency viruses, and Vpr is found in HIV-1 and HIV-2. Vpx targets a restriction factor that prohibits viral DNA synthesis in monocytic cells (4–6). Recently, the protein encoded by the SAMHD1 gene was identified as the HIV-1 restriction factor in cells of the myeloid lineage (7, 8). SAMHD1 is thought to inhibit an early step in the viral life cycle by interfering with efficient synthesis of viral DNA, but the mechanism is not known.
SAMHD1 was identified as an interferon-induced protein in human macrophages and dendritic cells (9–12), suggesting a function in the innate immune response. Mutations in SAMHD1 cause the severe neurodegenerative disorder Aicardi-Goutières syndrome (13), a genetic encephalopathy that closely mimics congenital viral infection and is characterized by inappropriate immune activation and aberrant interferon-α secretion (14–16). Likewise, mutations in the TREX1 exonuclease gene and in the three genes encoding the RNase H2 endonuclease cause Aicardi-Goutières syndrome, linking nucleic acid metabolism with this autoimmune disorder (17–19). TREX1 is the major 3′-exonuclease that degrades cellular single- and double-stranded DNA (20–26). In HIV-1-infected cells, TREX1 degrades nonproductive transcripts to clear excess viral cDNA, preventing innate immune activation (27, 28). The RNase H2 recognizes and cleaves ribonucleotides present in RNA/DNA duplexes (29, 30), and silencing RNASEH2A impairs HIV replication (31). This suggests a role in HIV infection. Thus, SAMHD1, TREX1, and RNase H2 participate in a cellular nucleic acid metabolic pathway that overlaps the interferon-mediated innate antiviral and inflammatory responses.
The catalytic activity of SAMHD1 is identified in this study. The SAMHD1 contains an N-terminal putative protein-protein interaction sterile α motif (SAM)4 domain and a C-terminal predicted phosphohydrolase HD (His···His-Asp···Asp) domain. The SAMHD1 was cloned, and bacterially expressed recombinant enzyme was generated in sufficient quantities and purity to perform biochemical studies. The enzymatic properties indicate that the SAMHD1 HIV-1 restriction factor is a dGTP-regulated deoxynucleotide triphosphohydrolase.
EXPERIMENTAL PROCEDURES
Materials
The nucleotides were from Sigma, and calf intestinal phosphatase was from Promega. The human and mouse SAMHD1 cDNAs (Invitrogen) were recovered by PCR and cloned into the pCDFDuet-1 (Novagen) plasmid. The HD domain constructs were prepared by PCR, and final constructs were verified by DNA sequencing.
Enzyme Preparation
In expression studies, a His6 tag was cloned at the N or C terminus of the complete human and mouse SAMHD1 cDNAs, and maximal yields of soluble enzymes were recovered using the mouse N-terminal His6 tag SAMHD1 constructs (data not shown). Mouse SAMHD1 plasmid constructs were transformed into Escherichia coli BL21 (DE3) Rosetta 2 cells (Novagen), grown to an A600 = 0.5 at 37 °C, and quickly cooled on ice to 17 °C. After induction with 1 mm isopropyl-β-d-thiogalactopyranoside, the cells were allowed to grow for 18 h at 17 °C. The SAMHD1 enzymes in cell extracts were bound to a nickel-nitrilotriacetic acid resin (Qiagen), washed, eluted, and purified to homogeneity using Mono S chromatography (supplemental Fig. S1). TREX1 and RNase H2 were purified as described (22, 30). Protein concentrations were determined by A280 using the molar extinction coefficient for complete mouse SAMHD1 ϵ = 63,800 m−1 cm−1 and for SAMHD1 HD domain (amino acids 117–627) ϵ = 58,050 m−1 cm−1.
Phosphohydrolase Assays
Reactions (100 μl) were performed at 25 °C in 96-well microplates initiated by enzyme addition, and activity was monitored continuously at A410 using a Tecan Safire2TM. Phosphatase reactions contained 50 mm HEPES (pH 7.0), 4 mm p-nitrophenyl phosphate, 5 mm of the indicated divalent metal ion, and 500 nm SAMHD1. Phosphodiesterase reactions (100 μl) contained 50 mm Tricine (pH 8.5), 4 mm bis-p-nitrophenyl phosphate or p-nitrophenyl 5′-thymidine monophosphate, 5 mm of the indicated divalent metal ion, and 500 nm SAMHD1 (32).
Nucleotide Phosphohydrolase Assays
The nucleotide phosphohydrolase reactions contained 20 mm Tris-HCl (pH 7.5), 2 mm dithiothreitol, 1 μm EDTA, 5 mm MgCl2, the indicated nucleotides, and SAMHD1 enzyme. Reactions were incubated for the indicated times at 25 °C and stopped by the addition of EDTA to 5 mm final concentration. Protein was removed using 10,000 molecular weight cut-off concentrators (Millipore), and samples (10 μl) of the filtered solution were analyzed by ion pair reverse-phase chromatography (33, 34) using a CAPCELL PAK C18 column (Shiseido Fine Chemicals) on a Waters HPLC system. The column was equilibrated with buffer (20 mm sodium phosphate (pH 7.0), 5 mm tetra n-butylammonium phosphate, and 5% methanol) and eluted with a linear gradient of methanol from 5 to 60%. Products and reactants were quantified by measuring the A254. Quantification of the absorbance peaks was performed using the Empower software (Waters).
RESULTS
SAMHD1 Is a Phosphohydrolase
The primary amino acid sequence of SAMHD1 identifies this protein as a member of the diverse superfamily of enzymes known or predicted to possess metal-dependent phosphohydrolase activities (35). The phosphohydrolase activity contained in SAMHD1 was detected using the purified recombinant mouse enzyme with an artificial chromogenic substrate bis-p-nitrophenyl phosphate. A robust metal-dependent catalytic activity was detected using the bis-p-nitrophenyl phosphate substrate with Mg2+ and Mn2+; modest activity was detected with Co2+; and no activity was detected with Zn2+, Ni2+, or Ca2+ (supplemental Fig. S1). Similar levels of activity were detected using the purified SAMHD1 HD domain, therefore demonstrating that the HD domain is necessary and sufficient for full catalytic function in the absence of the SAM domain. The SAMHD1 activity is ∼5-fold greater than the phosphohydrolase activities exhibited by the TREX1 and RNase H2 nucleases using the bis-p-nitrophenyl phosphate substrate (supplemental Fig. S1). The SAMHD1 exhibits no detectable activity using p-nitrophenyl phosphate and p-nitrophenyl 5′-thymidine monophosphate substrates, in contrast to the robust TREX1 activity with p-nitrophenyl 5′-thymidine monophosphate (data not shown). These results indicate the SAMHD1 preference for phosphoester bond cleavage. Therefore, SAMHD1 was incubated with a variety of DNA and RNA polynucleotide substrates with no nuclease activity detected (data not shown). Thus, SAMHD1 is a metal-dependent phosphohydrolase with considerably different substrate specificity when compared with the TREX1 and RNase H2 nucleases.
SAMHD1 Is a Deoxynucleotide Triphosphohydrolase
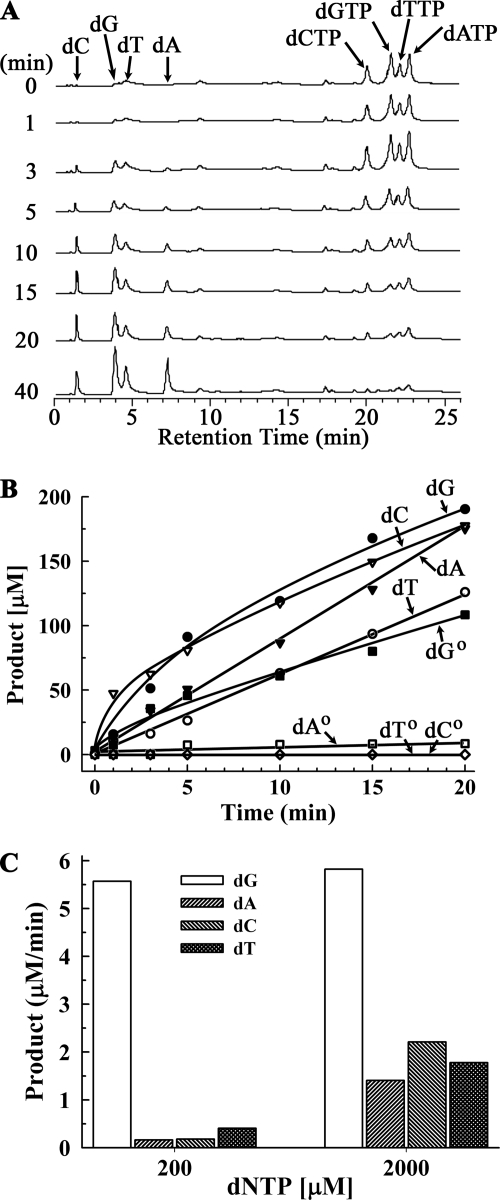
SAMHD1 cleaves deoxynucleoside triphosphates at the α-phosphate position generating the deoxynucleoside and triphosphate products. SAMHD1 was incubated in a time course reaction containing dGTP, dATP, dTTP, and dCTP with Mg2+, and the products were fractionated by HPLC chromatography (Fig. 1A). SAMHD1 triphosphohydrolase activity is apparent by the appearance of four product peaks eluting from the column at positions corresponding to the deoxynucleosides with the four products accumulating over time. Quantification of the deoxynucleoside products indicated that SAMHD1 hydrolyzed each of the four nucleotides at similar rates of ∼8 μm/min when all deoxynucleoside triphosphates were present in the reaction at 200 μm (Fig. 1B). In the presence of only dGTP, SAMHD1 activity was similar at a rate of ∼6 μm/min. However, the SAMHD1 activity was about 20-fold lower at ∼0.3 μm/min when only dATP, dTTP, or dCTP was present in reactions (Fig. 1B). SAMHD1 activity was also measured in reactions with the individual dNTP substrates at a 10-fold higher concentration of 2000 μm (Fig. 1C). The SAMHD1 triphosphohydrolase activity of ∼6 μm/min was similar for dGTP at both concentrations. The SAMHD1 activity in the presence of 2000 μm dNTPs resulted in measurable increases to ∼1.4 μm/min with dATP, ∼2.2 μm/min with dCTP, and ∼1.8 μm/min with dTTP. Together, these data indicate that SAMHD1 has a much higher affinity for dGTP relative to dATP, dTTP, and dCTP nucleotides and that efficient cleavage of deoxynucleotides is dependent upon the presence of dGTP. The dGTP dependence of SAMHD1 activity was demonstrated in reactions containing dATP, dTTP, or dCTP with dGTP (supplemental Fig. S2) and in reactions containing combinations of dATP, dTTP, and dCTP with dGTP (supplemental Fig. S3). Robust SAMHD1 hydrolysis was detected in all reactions containing 1–4 nucleotides only when dGTP was present in the reaction.
FIGURE 1.
SAMHD1 is a nucleotide triphosphohydrolase. A, time course reactions containing 400 nm SAMHD1, 200 μm dGTP, dATP, dTTP, and dCTP, and 5 mm MgCl2 were performed in triplicate for the indicated times, and products were fractioned by HPLC as described under “Experimental Procedures. ” A chromatogram at each time point from one of the reactions is shown. The indicated positions of elution of the substrates and products were determined in separate injections of each dNTP and nucleoside. B, the dG, dA, dC, and dT products generated at the indicated times from the three reactions in A and from triplicate time course reactions containing only dGTP (dG0), only dATP (dA0), only dTTP (dT0), and only dCTP (dC0) were quantified. C, the dG, dA, dT, and dC products from reactions containing 200 or 2000 μm of the indicated nucleotide were quantified, and rates were calculated.
The deoxynucleoside triphosphate specificity of SAMHD1 was tested. SAMHD1 exhibits a modest but detectable level of triphosphohydrolase activity using GTP when incubated in the presence of all four ribonucleoside triphosphates with Mg2+ as the activating metal. However, hydrolysis of ATP, UTP, and CTP is not detected (data not shown). There is no evidence for generation of the mono- or diphosphate nucleotides, indicating that SAMHD1 cleaves dNTPs to generate only the deoxynucleoside and triphosphate products. SAMHD1 exhibits no detectable activity using dGMP or dGDP. Also, SAMHD1 does not exhibit triphosphohydrolase activity of dATP using dGMP, dGDP, or dG as the activator molecule.
SAMHD1 Is a dGTP-regulated Deoxynucleotide Triphosphohydrolase
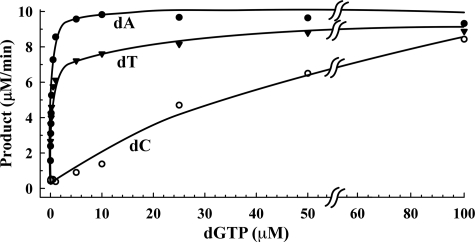
Binding of dGTP regulates SAMHD1 deoxynucleotide cleavage activity. The dGTP regulation of SAMHD1 was demonstrated by incubating with 200 μm dATP, dTTP, or dCTP in the presence of increased concentrations of dGTP and quantifying the dA, dT, and dC products (Fig. 2). The SAMHD1 exhibits dramatically increased rates of dATP, dTTP, and dCTP hydrolysis upon the addition of increased dGTP concentrations. The addition of only 150 nm dGTP resulted in half-maximal SAMHD1 activation to ∼4.5 μm/min for hydrolysis of dATP and dTTP. Complete activation of SAMHD1 for dATP and dTTP was detected at 5 and 25 μm dGTP, respectively. The activation of SAMHD1 for dCTP hydrolysis required higher levels of dGTP with the half-maximal activity of ∼4.7 μm/min detected at 25 μm dGTP and complete activation detected at 100 μm dGTP. These results demonstrate the differential activation of SAMHD1 by dGTP. The relatively low concentrations of dGTP required in these reactions to activate SAMHD1 triphosphohydrolase activity, particularly for dATP and dTTP, indicate that dGTP is an essential activator and suggest the presence of a dGTP-specific ligand-binding site. The SAMHD1 activity exhibits a hyperbolic response to increased concentrations of dGTP (Fig. 2). The dramatic increase in SAMHD1 deoxynucleotide triphosphohydrolase activity at relatively low dGTP concentrations points to a guanine-specific control mechanism.
FIGURE 2.
SAMHD1 is a dGTP-regulated triphosphohydrolase. Reactions containing 400 nm SAMHD1, 200 μm dATP, dTTP, or dCTP, 5 mm MgCl2, and the indicated concentrations of dGTP were performed for 20 min at 25 °C. The reactions were fractioned by HPLC, and the dA (●), dT (▾), and dC (○) products were quantified as described under “Experimental Procedures.” Curved symbols indicate a change in the x-axis scale.
DISCUSSION
SAMHD1 is an HIV-1 restriction factor that prohibits efficient viral DNA synthesis. Our investigations of SAMHD1 catalytic function have revealed its deoxynucleotide triphosphohydrolase activity, uncovering the mechanism of viral restriction. The NMR structure of the SAMHD1 SAM domain has been determined (Protein Data Bank (PDB) 2E8O), confirming the presence of this protein-protein interaction domain in the N-terminal region. Sequence alignments identified SAMHD1 genes in diverse eukaryotes, but the lack of biochemical studies limited insight into physiological function. To initiate SAMHD1 functional studies, a structural model was generated using ensemble fold recognition in the program Phyre (36, 37). Using the complete human and mouse SAMHD1 sequences, the apparent homologue Enterococcus faecalis EF1143 protein (PDB 3IRH) was identified (38). The Efa EF1143 protein is a deoxynucleotide triphosphohydrolase, and the SAMHD1 HD domain model suggests a high degree of sequence conservation at functional residues throughout the HD domain (supplemental Fig. S4). The likely SAMHD1 active site metal-binding residues His-167, His-206, Asp-207, and Asp-311 (His-66, His-110, Asp-111, and Asp-183 in Efa EF1143 protein) are identified. Also, the proposed nucleophile-generating residues His-210, Asp-218, and His-233 (His-114, Glu-122, and His-129 in Efa EF1143 protein) are conserved. The EF1143 x-ray structure is a tetramer with four dGTP-specific regulatory sites identified (38). Importantly, the SAMHD1 model indicates that residues Lys-116, Asp-137, Gln-142, Arg-145, Phe-165, Arg-451, Lys-455, and Lys-556 (Lys-14, Asn-36, Gln-41, Arg-44, Phe-64, Arg-326, Lys-330, and Lys-422 in Efa EF1143 protein) are in direct contact with the dGTP ligand. The model also predicts that residues Gln-149, His-162, Arg-164, Asp-319, His-370, and Tyr-374 (Gln-48, His-61, Arg-63, Asp-191, Tyr-239, and Tyr-243 in Efa EF1143 protein) are likely responsible for conferring deoxyribose substrate specificity. A high-resolution structure of SAMHD1 HD domain will be required to validate this model.
The HIV-2 Vpx targeting of SAMHD1 deoxynucleotide triphosphohydrolase to permit viral replication has a striking parallel in bacteriophage T7 restriction in bacteria. The E. coli mutant strain OptA1 is unable to support replication of bacteriophage T7 gene 1.2 mutants (39). The OptA1 strain is mutated in the upstream region of the dgt gene, causing overproduction of the encoded dGTPase and a reduced cellular level of dGTP (40, 41). Thus, the high triphosphohydrolase activity in the OptA1 strain resulted in cellular nucleotide precursor levels insufficient to support bacteriophage T7 replication. The gene 1.2 protein inhibits the E. coli dGTPase, which raises nucleotide levels and permits T7 bacteriophage replication (42, 43). Thus, targeting key nucleotide metabolism enzymes to control replication precursor molecule levels represents a mechanistic strategy in viral restriction that is perhaps more common than currently appreciated.
The finding of SAMHD1 deoxynucleotide triphosphohydrolase activity now links this Aicardi-Goutières syndrome enzyme with TREX1 and RNase H2 to various aspects of nucleic acid metabolism. Dysfunction of SAMHD1, TREX1, and RNase H2 enzymes in the same immune activation disease illustrates the critical nature of proper nucleotide metabolism, polynucleotide degradation, and RNA/DNA hybrid processing. Establishing SAMHD1 as an HIV-1 restriction factor and TREX1 in the degradation of HIV-1 transcripts to prevent innate immune activation places these enzymes at the interface of type I interferon-mediated autoimmunity and the antiviral response. Understanding SAMHD1, TREX1, and RNase H2 in cellular nucleic acid metabolism could provide new therapeutic strategies in autoimmune and viral disease.
Supplementary Material
This work was supported, in part, by National Institutes of Health Grant GM069962 (to F. W. P.). This work was also supported by Alliance for Lupus Research Grant 179222 (to F. W. P.) and American Heart Association Grant 10GRNT3650033 (to T. H.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- SAM
- sterile α motif
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Malim M. H., Emerman M. (2008) Cell Host Microbe 3, 388–398 [DOI] [PubMed] [Google Scholar]
- 2. Gramberg T., Sunseri N., Landau N. R. (2009) Curr. HIV/AIDS Rep. 6, 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ayinde D., Maudet C., Transy C., Margottin-Goguet F. (2010) Retrovirology 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wolfrum N., Mühlebach M. D., Schüle S., Kaiser J. K., Kloke B. P., Cichutek K., Schweizer M. (2007) Virology 364, 330–341 [DOI] [PubMed] [Google Scholar]
- 5. Goujon C., Arfi V., Pertel T., Luban J., Lienard J., Rigal D., Darlix J. L., Cimarelli A. (2008) J. Virol. 82, 12335–12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaushik R., Zhu X., Stranska R., Wu Y., Stevenson M. (2009) Cell Host Microbe 6, 68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li N., Zhang W., Cao X. (2000) Immunol. Lett. 74, 221–224 [DOI] [PubMed] [Google Scholar]
- 10. Crow M. K., Kirou K. A., Wohlgemuth J. (2003) Autoimmunity 36, 481–490 [DOI] [PubMed] [Google Scholar]
- 11. Liao W., Bao Z., Cheng C., Mok Y. K., Wong W. S. (2008) Proteomics 8, 2640–2650 [DOI] [PubMed] [Google Scholar]
- 12. Zhao D., Peng D., Li L., Zhang Q., Zhang C. (2008) Virol. J. 5, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., Bertini E., Bodemer C., Brockmann K., Brueton L. A., Corry P. C., Desguerre I., Fazzi E., Cazorla A. G., Gener B., Hamel B. C., Heiberg A., Hunter M., van der Knaap M. S., Kumar R., Lagae L., Landrieu P. G., Lourenco C. M., Marom D., McDermott M. F., van der Merwe W., Orcesi S., Prendiville J. S., Rasmussen M., Shalev S. A., Soler D. M., Shinawi M., Spiegel R., Tan T. Y., Vanderver A., Wakeling E. L., Wassmer E., Whittaker E., Lebon P., Stetson D. B., Bonthron D. T., Crow Y. J. (2009) Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aicardi J., Goutières F. (1984) Ann. Neurol. 15, 49–54 [DOI] [PubMed] [Google Scholar]
- 15. Goutières F. (2005) Brain Dev. 27, 201–206 [DOI] [PubMed] [Google Scholar]
- 16. Goutières F., Aicardi J., Barth P. G., Lebon P. (1998) Ann. Neurol. 44, 900–907 [DOI] [PubMed] [Google Scholar]
- 17. Crow Y. J., Hayward B. E., Parmar R., Robins P., Leitch A., Ali M., Black D. N., van Bokhoven H., Brunner H. G., Hamel B. C., Corry P. C., Cowan F. M., Frints S. G., Klepper J., Livingston J. H., Lynch S. A., Massey R. F., Meritet J. F., Michaud J. L., Ponsot G., Voit T., Lebon P., Bonthron D. T., Jackson A. P., Barnes D. E., Lindahl T. (2006) Nat. Genet. 38, 917–920 [DOI] [PubMed] [Google Scholar]
- 18. Crow Y. J., Leitch A., Hayward B. E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., Baumann C., Baxter P., Bertini E., Chandler K. E., Chitayat D., Cau D., Déry C., Fazzi E., Goizet C., King M. D., Klepper J., Lacombe D., Lanzi G., Lyall H., Martínez-Frías M. L., Mathieu M., McKeown C., Monier A., Oade Y., Quarrell O. W., Rittey C. D., Rogers R. C., Sanchis A., Stephenson J. B., Tacke U., Till M., Tolmie J. L., Tomlin P., Voit T., Weschke B., Woods C. G., Lebon P., Bonthron D. T., Ponting C. P., Jackson A. P. (2006) Nat. Genet. 38, 910–916 [DOI] [PubMed] [Google Scholar]
- 19. Crow Y. J., Rehwinkel J. (2009) Hum. Mol. Genet. 18, R130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fye J. M., Orebaugh C. D., Coffin S. R., Hollis T., Perrino F. W. (2011) J. Biol. Chem. 286, 32373–32382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Höss M., Robins P., Naven T. J., Pappin D. J., Sgouros J., Lindahl T. (1999) EMBO J. 18, 3868–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lehtinen D. A., Harvey S., Mulcahy M. J., Hollis T., Perrino F. W. (2008) J. Biol. Chem. 283, 31649–31656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mazur D. J., Perrino F. W. (1999) J. Biol. Chem. 274, 19655–19660 [DOI] [PubMed] [Google Scholar]
- 24. Orebaugh C. D., Fye J. M., Harvey S., Hollis T., Perrino F. W. (2011) J. Biol. Chem. 286, 40246–40254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stetson D. B., Ko J. S., Heidmann T., Medzhitov R. (2008) Cell 134, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang Y. G., Lindahl T., Barnes D. E. (2007) Cell 131, 873–886 [DOI] [PubMed] [Google Scholar]
- 27. Yan N., Lieberman J. (2011) Curr. Opin. Immunol. 23, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan N., Regalado-Magdos A. D., Stiggelbout B., Lee-Kirsch M. A., Lieberman J. (2010) Nat. Immunol. 11, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coffin S. R., Hollis T., Perrino F. W. (2011) J. Biol. Chem. 286, 16984–16991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perrino F. W., Harvey S., Shaban N. M., Hollis T. (2009) J. Mol. Med. 87, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Genovesio A., Kwon Y. J., Windisch M. P., Kim N. Y., Choi S. Y., Kim H. C., Jung S., Mammano F., Perrin V., Boese A. S., Casartelli N., Schwartz O., Nehrbass U., Emans N. (2011) J. Biomol. Screen. 16, 945–958 [DOI] [PubMed] [Google Scholar]
- 32. Kuznetsova E., Proudfoot M., Sanders S. A., Reinking J., Savchenko A., Arrowsmith C. H., Edwards A. M., Yakunin A. F. (2005) FEMS Microbiol. Rev. 29, 263–279 [DOI] [PubMed] [Google Scholar]
- 33. Kondo N., Kuramitsu S., Masui R. (2004) J. Biochem. 136, 221–231 [DOI] [PubMed] [Google Scholar]
- 34. Stocchi V., Cucchiarini L., Canestrari F., Piacentini M. P., Fornaini G. (1987) Anal. Biochem. 167, 181–190 [DOI] [PubMed] [Google Scholar]
- 35. Aravind L., Koonin E. V. (1998) Trends Biochem. Sci. 23, 469–472 [DOI] [PubMed] [Google Scholar]
- 36. Bennett-Lovsey R. M., Herbert A. D., Sternberg M. J., Kelley L. A. (2008) Proteins 70, 611–625 [DOI] [PubMed] [Google Scholar]
- 37. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 38. Vorontsov II, Minasov G., Kiryukhina O., Brunzelle J. S., Shuvalova L., Anderson W. F. (2011) J. Biol. Chem. 286, 33158–33166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saito H., Richardson C. C. (1981) J. Virol. 37, 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beauchamp B. B., Richardson C. C. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seto D., Bhatnagar S. K., Bessman M. J. (1988) J. Biol. Chem. 263, 1494–1499 [PubMed] [Google Scholar]
- 42. Kornberg S. R., Lehman I. R., Bessman M. J., Simms E. S., Kornberg A. (1958) J. Biol. Chem. 233, 159–162 [PubMed] [Google Scholar]
- 43. Wurgler S. M., Richardson C. C. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2740–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.