Abstract
Background
The model plant Arabidopsis thaliana (Arabidopsis) shows a wide range of genetic and trait variation among wild accessions. Because of its unparalleled biological and genomic resources, the potential of Arabidopsis for molecular genetic analysis of this natural variation has increased dramatically in recent years.
Scope
Advanced genomics has accelerated molecular phylogenetic analysis and gene identification by quantitative trait loci (QTL) mapping and/or association mapping in Arabidopsis. In particular, QTL mapping utilizing natural accessions is now becoming a major strategy of gene isolation, offering an alternative to artificial mutant lines. Furthermore, the genomic information is used by researchers to uncover the signature of natural selection acting on the genes that contribute to phenotypic variation. The evolutionary significance of such genes has been evaluated in traits such as disease resistance and flowering time. However, although molecular hallmarks of selection have been found for the genes in question, a corresponding ecological scenario of adaptive evolution has been difficult to prove. Ecological strategies, including reciprocal transplant experiments and competition experiments, and utilizing near-isogenic lines of alleles of interest will be a powerful tool to measure the relative fitness of phenotypic and/or allelic variants.
Conclusions
As the plant model organism, Arabidopsis provides a wealth of molecular background information for evolutionary genetics. Because genetic diversity between and within Arabidopsis populations is much higher than anticipated, combining this background information with ecological approaches might well establish Arabidopsis as a model organism for plant evolutionary ecology.
Key words: Arabidopsis, natural genetic variation, natural trait variation, QTL mapping, LD mapping, plant development, plant evolution, molecular ecology
INTRODUCTION
Natural variation within species has been the main subject of evolutionary genetics, because it is considered to be the main resource for evolutionary change and for the adaptive potential of a species to environments that vary in space and time. The model plant Arabidopsis thaliana shows a wide range of genetic and trait variation among wild-type lines collected in the field. These lines are often called ecotypes, but are now commonly referred to by the more neutral term accessions, which does not inherently imply but also not exclude local adaptation. In addition, because of the unparalleled availability of genomic resources, the potential of A. thaliana for studies of natural genetic variation is increasingly recognized. Accordingly, research articles that aim to comprehend the molecular mechanisms underlying phenotypic variation between accessions are now published almost every month. Importantly, the molecular analysis of natural genetic variation has not only led to the correlation of allelic variation of known genes with phenotypic variation, but also to the discovery of novel genes. This identification of genes that account for natural phenotypic variation is and will remain one of the principal goals in this field. However, beyond this goal, the analysis of natural genetic variation also offers an excellent opportunity to overcome the often perceived dichotomy between molecular and organismal biology. This is because molecular genetic analyses provide an excellent basis for an ecological assessment of accessions and for the experimental investigation of the functional significance of trait variation. The feasibility of this approach is currently increasing, based on large sets of accessions collected worldwide and on fine-scale genotyping of their genome sequence. This merging of molecular genetics and ecology will enable researchers to address fundamental questions of evolution and ecology in a more targeted manner. For instance, although the molecular genetic analyses will help to determine the molecular mechanisms that maintain phenotypic variation in the wild, the complementing ecological analyses can tell whether this phenotypic variation is critical for adaptation to different natural environments.
Here we review the latest research and methodology in analysing natural variation in the model plant A. thaliana. We begin by pointing out the general aspects of the genetics and biology of this plant, followed by a discussion of useful methods for the analysis of natural variation. We will then exemplify the potential of analysing natural variation for dissecting the variation in two different traits, pathogen defence and the control of flowering time. Finally, the potential of A. thaliana to become a model organism for evolutionary ecology is discussed.
GENETIC RESOURCES OF A. THALIANA
Arabidopsis thaliana (L.) Heyhn (2n = 10), commonly known as mouse ear cress or wild thale, belongs to the mustard family (Brassicaceae, formerly Cruciferae). The genus Arabidopsis comprises nine species and eight subspecies (Al-Shehbaz and O'Kane, 2002). Among them, A. thaliana can be distinguished by morphological characteristics such as fruit and seed shape. The nine species of the genus Arabidopsis are mainly found in Europe. Two species are also found in Asia and North America, but only A. thaliana has a worldwide distribution. In fact, A. thaliana can be found in diverse habitats, for instance in open or disturbed habitat, on sandy soils or on river banks, at sea level or at high altitude, up to 4000 m a.s.l. (Al-Shehbaz and O'Kane, 2002). The rapid expansion of habitat colonization by A. thaliana implies that this species has a huge capacity to adapt to a wide range of ecological niches. This is why subspecies or local populations of A. thaliana have been conventionally referred to as ecotypes. However, the rapid post-glacial spread of A. thaliana as compared with the other species of the genus could be a result of its self-compatibility and very high selfing rate, rather than the result of a particular capacity to adapt to ecological niches. Thus, the more neutral term accessions, which does not necessarily imply local adaptation (Mitchell-Olds, 2001), is now preferred for germplasm collections.
The current boom of natural variation studies of A. thaliana (hereafter Arabidopsis) is in large part driven by the availability of natural populations. Hundreds of accessions or samples from natural populations collected from diverse worldwide locations are currently available through public sources (Scholl et al., 2000; Koornneef et al., 2004). Their number is still increasing as a result of the deposit of new accessions by many research groups. By March 2006, a total of 1494 accessions comprising original lines and their bulked or single-seed descendants were available at the Arabidopsis Biological Resource Center (ABRC) at Ohio State University, USA (http://www.arabidopsis.org/abrc/catalog/natural_accession_1). These lines are also distributed by the Nottingham Arabidopsis Stock Centre (NASC) at Nottingham University, UK (http://arabidopsis.info/). Beyond geographical information about the collection locations, for many lines morphological characteristics such as leaf shape or plant height are described in this catalogue. From these descriptions alone it is already evident that Arabidopsis accessions show an extraordinarily wide phenotypic variation. Thus far, significant natural variation has been reported for every phenotypic trait investigated (Koonneef et al., 2004). Some developmental traits, such as flowering time or seed dormancy, have drawn particular attention, partly because they are of applied interest to crop breeding, and partly because they are easy to investigate. However, in addition to visually obvious phenotypes, natural variation has also been observed in genetic mechanisms such as cytosine methylation (Riddle and Richards, 2002). Moreover, assays of metabolite profiles by large-scale unbiased metabolomics methods have uncovered natural variation at the level of small molecules, suggesting that they reflect physiological phenotypes that could be under selection in nature (Keurentjes et al., 2006).
Finally, the natural variation resources of Arabidopsis are complemented by the annotated genome sequence, which for instance enables high-density genotyping, and by collections of knockout mutants, which provide a powerful tool to verify the prospective roles of genes involved in natural trait variation by independent means.
GENETIC STRUCTURE AND BIOGEOGRAPHY OF ARABIDOPSIS POPULATIONS
Although Arabidopsis has been found in the southern hemisphere including Africa, South America and Australia, all literature discussing its biogeography consistently concludes that the native range of Arabidopsis is western Eurasia. Because Arabidopsis crossed the oceans, it is assumed that human mobility and disturbance significantly contributed to its recent expansion.
Several molecular phylogenetic studies have been performed to determine the relationship between genetic diversity and biogeography among Arabidopsis accessions. Genotyping of molecular markers in 142 accessions suggested that Arabidopsis colonization of Europe might have started from populations in Mediterranean Pleistocene refugia (mainly the Iberian peninsula) and Central Asia (Sharbel et al., 2000). Subsequently, admixture of these populations occurred in Central and Eastern Europe. In agreement with this, a positive correlation between genetic variation and geographical origin of accessions, i.e. isolation by distance, has also been shown by the latest molecular phylogenetic analyses of 96 Arabidopsis accessions based on genome-wide polymorphism genotyping, suggesting the existence of population structure at a global geographical scale (Nordborg et al., 2005). Further support has been obtained from the genotyping of 351 accessions, which also suggests that Central and Eastern European accessions represent admixed populations in which genomes were reshuffled by recombination events (Schmid et al., 2006). Finally, several additional molecular phylogeny studies refute the hypothesis that Arabidopsis is native to North America and East Asia, supporting the idea that it has spread there in the wake of human activity (Bergelson et al., 1998; Breyne et al., 1999; Miyashita et al., 1999; Vander Zwan et al., 2000).
In addition to these phylogeographical approaches, Arabidopsis distribution has also been analysed with respect to the climatic range model, which considers climate as the main factor limiting a species distribution range (Hoffmann, 2002, 2005). By comparing the distribution frequency of Arabidopsis with the climatic parameters of the collection sites, a significant correlation between geographical distribution and the gradients of temperature and precipitation was demonstrated. In brief, low temperature in spring and autumn may limit the distribution range in Northern Europe, whereas high temperature and low precipitation throughout the year or the summer may determine the range boundaries in North Africa and South-West Asia, and in Middle Asia, respectively.
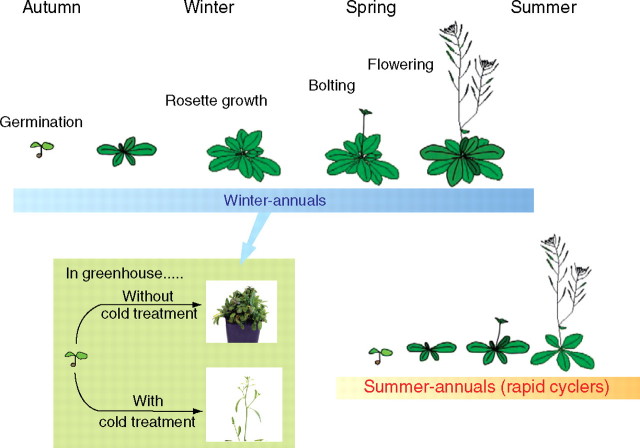
The biogeographical analyses demonstrate that Arabidopsis occupies a larger distribution range than its close relatives (Hoffmann, 2005). The high capacity of Arabidopsis to colonize a broad geographical spectrum is probably connected to its life cycle strategy, especially the timing of seed germination and flowering initiation. In Europe, Arabidopsis accessions generally flower in spring and early summer, and the mature seeds are available from May to July, occasionally also in late summer up to early autumn (Lawrence, 1976; Koornneef et al., 2004). As an annual weed, Arabidopsis accessions are principally classified into summer-annual and winter-annual types with regard to flowering (Napp-Zinn, 1985; Fig. 1). The genetic mechanisms controlling seed dormancy and vernalization requirement (the degree of dependence on low temperatures to initiate flowering) account for this differentiation (Donohue, 2002). Accessions behaving as winter-annual types germinate during late summer or autumn, over-winter as rosettes and grow to initiate the reproductive phase in spring. In greenhouse conditions, the flowering of winter-annual types is delayed, but can be accelerated by extended low temperature that is equivalent to over-wintering in nature. The summer-annual types are able to complete all steps of the life cycle during the same summer season, as long as the climatic conditions are appropriate for seed production. Accessions of this type, also referred to as rapid cyclers, over-winter as seeds and therefore display increased seed dormancy. However, it should be noted that most summer-annual types can also follow the winter-annual strategy of over-wintering as rosettes if conditions for flowering and seed set are adverse during winter (Pigliucci, 2003; Koornneef et al., 2004). In general, accessions from Southern Europe are either winter- or summer-annual types, whereas the majority of Northern European accessions are typically winter-annual. The strong winter-annuals, whose flowering initiation exclusively depends on vernalization, are observed mainly in Northern Europe above 45°N (Shindo et al., 2005). However, a clear geographical pattern of flowering time has not been observed (Nordborg and Bergelson, 1999; Shindo et al., 2005), although a latitudinal cline has been predicted (Stinchcombe et al., 2004). This may suggest that natural variation of the life cycle is continuous and potentially subject to complex selection and constraints, such as trade-offs.
Fig. 1.
Arabidopsis accessions are classified into summer-annual and winter-annual types. Accessions behaving as winter-annual types germinate during late summer or autumn, over-winter as rosettes and grow to initiate the reproductive phase in spring. Under greenhouse conditions, the flowering of winter-annual types is delayed, but can be accelerated by extended low temperature that is equivalent to over-wintering in nature. The summer-annual types are able to complete all steps of the life cycle during the same summer season, as long as the climatic conditions are appropriate for seed production. Accessions of this type, also referred to as rapid cyclers, over-winter as seeds and therefore display increased seed dormancy.
It is commonly assumed that Arabidopsis is a completely, or nearly completely, self-fertilizing species, owing to its characteristic flowering morphology, which is typical for inbreeding plants: the flowers are small, lack strong scent and the anthers are positioned close to the stigmata (Charlesworth and Vekemans, 2005). Indeed, the selfing rate in natural environments has been estimated in some studies to be greater than 95 % (Abbott and Gomes, 1989; Charlesworth and Vekemans, 2005; Stenøien et al., 2005). Thus, local Arabidopsis populations are generally regarded to consist of a single inbred sibship. However, despite inbreeding, an unexpected amount of genetic variation has been found within local populations (Nordborg et al., 2005; Bakker et al., 2006b), suggesting gene flow between populations, which might be mediated to a substantial extent through exchange of pollen rather than by seed dispersal (Bakker et al., 2006b). These molecular population analyses provide an answer to a classical question with regard to the eco-physiology of Arabidopsis: is Arabidopsis absolutely self-fertilizing in the field? Based on the latter studies, the answer is clearly no. On the other hand, Arabidopsis out-crossing is rarely observed under laboratory conditions. Thus, it appears likely that at the protogynous stage (when anthers are immature and the stigma protrudes from the flower), stigmata could be cross-pollinated by a vector. In fact, insects, such as solitary bees, diptera and thrips, have been suggested as candidate transporters of pollen between Arabidopsis plants and populations (Hoffmann et al., 2003). It would be interesting to monitor the extent of cross-pollination under field conditions, for instance by genotyping maternal plants and the seeds within naturally pollinated fruits at co-dominant markers to infer the number and genetic identity of pollen donors. In addition, fluorescent staining of pollen could be employed to estimate the probability of outcross pollen receipt on focus plants, and to examine the consequences of preventing pollinator access (through bagging of inflorescences) for offspring number and vigour.
ANALYSIS OF NATURAL GENETIC VARIATION IN ARABIDOPSIS
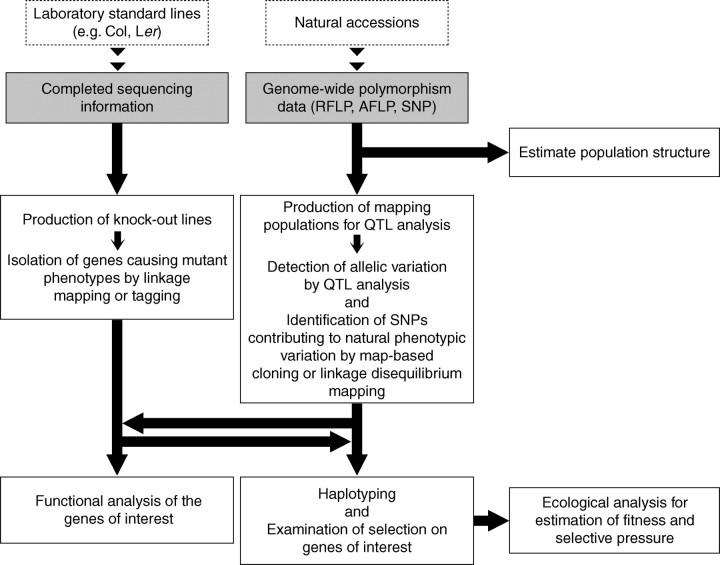
Thus far, studies of natural variation in Arabidopsis have pursued either of two major goals: (1) the identification of allelic variation and/or the isolation of novel genes that contribute to natural phenotypic variation, or (2) the search for footprints of selection by genome-wide genotyping of populations (Fig. 2). Both aspects fully exploit the huge and highly advanced genomic information for Arabidopsis. In particular, this information has rendered high-resolution quantitative trait loci (QTL) mapping possible.
Fig. 2.
Schematic methodologies for analysing natural variation in Arabidopsis accessions. In addition to the conventional genetic analysis with laboratory standard lines, a series of genetic analyses utilizing natural accessions are expected to allow the evaluation of the evolutionary significance of a given gene. AFLP, amplified fragment length polymorphism; QTL, quantitative trait loci; RFLP, restriction fragment length polymorphism; SNP, single nucleotide polymorphism.
QTL mapping
QTL mapping is an important tool for studies of natural variation, because most physiological or morphological traits exhibit a continuous phenotypic distribution within or among populations and are thus quantitative. These traits are frequently controlled by multiple loci, the QTL, which contribute to the variation in the trait to varying degrees. Therefore, even if two parental lines with a strong quantitative difference in the trait of interest are crossed, the F2 progeny will often display a continuous distribution of the trait, because the multiple parental loci are largely randomly mixed by recombination, giving rise to novel allele combinations with quantitatively different effects. QTL analysis traces the location and the quantitative impact of the parental alleles based on phenotypic and genotypic data. In principle, this can already be done at the F2 generation, although, to reduce genetic complexity and increase the reliability of trait measurements, commonly recombinant inbred lines (RILs) derived from the F2 generation by repeated selfing are used. These lines represent individual homozygous mosaics of the original parental genomes. Routine protocols for QTL mapping have been established during the past 20 years for many organisms in both the plant and the animal kingdoms, mainly for breeding purposes. However, although QTL analysis has been traditionally employed to follow loci of interest in breeding programmes, its feasibility for isolating these loci at the molecular level has been limited by biological and technical obstacles. On the biological side, the contribution of many QTL to the overall quantitative trait variation is often too small to allow their molecular identification. On the technical side, QTL analysis has been hampered by a lack of molecular markers and missing information on the genome structure and sequence. In Arabidopsis, these technical obstacles have been overcome by the readily available genomic resources. This now even enables the molecular identification of relatively minor QTL that contribute as little as 12 % to the observed phenotypic variance (Bentsink et al., 2006; Sergeeva et al., 2006). Moreover, there is increasing evidence that so-called major QTL are more common than expected. Major QTL have a large effect on the trait variation that can be easily detected in isolation, i.e. when the respective allele is introgressed into a different genetic background, or in a respective mutant. Lines in which a QTL allele of one parent is introgressed into the other parental background by repeated back-crossing are referred to as near isogenic lines (NILs). The phenotype of such NILs can then be compared with mutants in candidate genes within the region the QTL has been mapped to, aiding in the final identification. Importantly, extensive mutant collections covering most Arabidopsis genes are publicly available, strengthening the status of Arabidopsis as an ideal model organism for the isolation of genes and alleles with a role in natural variation (Table 1) (Alonso-Blanco and Koornneef, 2000; Borevitz and Nordborg, 2003; Maloof, 2003; Borevitz and Ecker, 2004; Alonso-Blanco et al., 2005; Weigel and Nordborg, 2005).
Table 1.
A list of examples of genes that contribute to natural phenotypic variation in Arabidopsis
| Gene | Function/phenotype | Methods by which the gene was originally identified | Reference* |
|---|---|---|---|
| PHYA | Photoreceptor/hypocotyl length | Genomic library and mutant screening | [I] Sharrock and Quail (1989) |
| [N] Maloof et al. (2001) | |||
| PHYC | Photoreceptor/hypocotyl length | Genomic library screening | [I] Sharrock and Quail (1989) |
| [N] Balasubramanian et al. (2006) | |||
| PHYD | Photoreceptor/hypocotyl length, flowering time | Genomic library screening | [I] Clack et al. (1994) |
| [N] Aukerman et al. (1997) | |||
| CRY2 | Photoreceptor/Flowering time | cDNA library screening | [I] Lin et al. (1996) |
| [N] El-Assal et al. (2001) | |||
| FLC | MADS TF/flowering time | Map-based cloning | [I] Michaels and Amasino (1999) |
| [N] Gazzani et al. (2003) | |||
| FLM | MADS TF/flowering time | Phylogenetic analysis and cDNA library screening | [I] Scortecci et al. (2001) |
| [N] Werner et al. (2005) | |||
| FRI | Unknown/flowering time | QTL analysis followed by map-based cloning | [I] [N] Johanson et al. (2000) |
| BRX | Unknown/primary root length | QTL analysis followed by map-based cloning | [I] [N] Mouchel et al. (2004) |
| CAL | MADS TF/inflorescence morphology | Genomic library screening | [I] [N] Kempin et al. (1994) |
| AOP2 & 3 | Glucosinolates biosynthesis/secondary metabolism | QTL analysis followed by map-based cloning | [I] [N] Kliebenstein et al. (2001) |
| ESM1 | Myrosinase-associated protein/glucosinolate hydrolysis and insect resistance | QTL analysis followed by map-based cloning | [I] [N] Zhang et al. (2006) |
| ESP | Epithiospecifier protein/glucosinolate hydrolysis | QTL analysis followed by map-based cloning | [I] [N] Lambrix et al. (2001) |
| MAM1 & 2 | Methylthioalkylmalate synthase/glucosinolate biosynthesis | QTL analysis followed by map-based cloning | [I] Kroymann et al. (2001) |
| [N] Kroymann et al. (2003) | |||
| RTM1 | Jacalin-like protein/virus resistance | Map-based cloning | [I] [N] Chisholm et al. (2000) |
| RPM1 | LRR protein/resistance to pseudomonads | Map-based cloning using natural accessions | [I] Grant et al. (1995) |
| [N] Stahl et al. (1999) | |||
| RPS2 | LRR protein/resistance to pseudomonads | Map-based cloning using natural accessions | [I] [N] Mindrinos et al. (1994) |
* [I] Reference reporting the gene isolation and [N] the identification of its natural allelic variation.
Recombinant inbred line populations
Arabidopsis is perfectly suited to establish RIL mapping populations for fine-scale QTL analysis, owing to its small size, short generation time, self-fertilization, high fecundity and relatively high recombination rate. An important aspect of RIL populations is that the lines are practically homozygous for every locus and therefore represent a homogeneous genetic resource that can be maintained indefinitely. Thus, the initial genotype map of an RIL population is valid indefinitely and can be used for the QTL mapping of different phenotypes, which can be assayed in multiple replicates. This benefit of RILs increases the accuracy of QTL mapping and is particularly useful for minimizing the environmental component in trait measurements, especially if the trait of interest is highly plastic. Repeated measurement of the same phenotype in different environments also enables the identification of QTL with pleiotropic effects (Alonso-Blanco and Koornneef, 2000). Currently more than 60 RIL populations have been produced in different laboratories and some of them are publicly available (see http://www.inra.fr/internet/Produits/vast/).
Published studies have thus far focused on a few, well-characterized RIL populations, notably one derived from the Columbia (Col-0) and Landsberg erecta (Ler) accessions, and one derived from the Cape Verde Islands (Cvi) and Ler accessions. In particular, exploitation of the Cvi × Ler population has proven to be a treasure for the identification of novel allelic variation. A well-known example is the isolation of the EDI allele of the CRY2 photoreceptor (El-Assal et al., 2001). In total, the Cvi × Ler RILs have been analysed for over 40 traits, revealing numerous highly significant QTL (Koornneef et al., 2004). Although it should be noted that Cvi-0 appears to represent a unique genetic linage as compared with other accessions, as judged from genome-wide polymorphism genotyping (Nordborg et al., 2005), the multiple analyses of the Cvi × Ler cross already offer a glimpse of the potential of natural genetic variation for the isolation of differentially active alleles. The presence of QTL for many different traits appears to be the norm rather than the exception, as evident from the many loci mapped in other RIL populations, notably those derived from the Col × Ler and Bayreuth-0 × Shahdara crosses (Loudet et al., 2002; Koornneef et al., 2004).
The use of novel RIL populations will surely increase as molecular marker information becomes more abundant and genotyping techniques become more affordable. Several techniques have been employed to construct high-density polymorphism maps for multiple accessions, creating a wealth of marker data (Jander et al., 2002; Borevitz et al., 2003; Schmid et al., 2003; Borevitz and Ecker, 2004; Comai et al., 2004; Nordborg et al., 2005). Notably, the polymorphism information identified by these projects is freely available on the Web (e.g. http://walnut.usc.eduor http://msqt.weigelworld.org), enabling researchers to choose polymorphic markers for their cross of interest in silico. Moreover, the actual genotyping work can be outsourced to ever cheaper genotyping service companies, rendering a high-density genotype map of a standard RIL population affordable for most laboratories. Beyond RIL characterization, the huge amount of genome-wide genotyping information increases the feasibility of an alternative approach for QTL mapping in Arabidopsis: association mapping.
Association mapping
Association mapping, also known as linkage disequilibrium (LD) mapping, is a population-based survey with the aim to identify trait–marker relationships based on LD, i.e. based on the non-random association of alleles and phenotypes (Flint-Garcia et al., 2003). Unlike linkage analysis, which analyses mapping populations derived from crosses of parental lines, LD mapping identifies candidate genes by analysis of populations of unrelated (i.e. without recent common ancestry) individuals. Given abundant sequence information, this method has the potential to identify single polymorphisms underlying phenotypic variation.
Arabidopsis is very well suited for LD mapping for two main reasons. First, the high level of inbreeding of accessions decreases heterozygosity, thus facilitating haplotype analysis by directly genotyping accessions. Second, LD in Arabidopsis has been estimated to decay within 50–250 kb, depending on the locus and population (Hagenblad and Nordborg, 2002; Nordborg et al., 2002, 2005). Although novel data suggest that LD might decay more rapidly than this, approaching 5–10 kb (M. Nordborg, pers. comm.), this value is still higher than in other model organisms, such as Drosophila. Therefore, considering the small genome size, comparably few markers are required for whole-genome LD mapping in Arabidopsis. This offers the prospect of whole-genome association studies in natural population samples with relatively little effort.
Despite these benefits, LD mapping has so far not been a major strategy for the investigation of the genetics underlying natural variation in Arabidopsis. Although several pilot analyses have successfully demonstrated the enormous potential of LD mapping in Arabidopsis (Nordborg et al., 2002; Hagenblad et al., 2004; Olsen et al., 2004; Aranzana et al., 2005; Kim et al., 2006), the same studies point out that some issues need to be resolved before launching association mapping as a standard approach. The most critical concern is the elimination of spurious (false positive) association between markers and phenotypes, which is caused by population structure, and thus unequal allele distribution. This problem could probably be overcome however by inference of genetic population structure in Arabidopsis accessions with large sets of genetic markers (Nordborg et al., 2005; Schmid et al., 2006), and by comparing the results with those obtained from standard linkage mapping (Borevitz and Nordborg, 2003). Given the increasing number of established mapping populations, the latter option is becoming increasingly feasible.
APPROACHES TO ASSESS THE EVOLUTIONARY SIGNIFICANCE OF NATURAL VARIATION
The identification of alleles that are responsible for natural phenotypic variation immediately evokes the question of whether this allelic variation has been maintained by selection pressure. Genome-wide single nucleotide polymorphism (SNP) data are useful in detecting the signature of natural selection by comparing the actual extent of nucleotide diversity in a gene to the extent expected under the standard neutral model. A popular statistical indicator of such comparison is the mean value of Tajima's D (Tajima, 1989). A value around zero is expected under simple neutral models, whereas clearly positive and negative values are attributed to the long-term maintenance of high levels of variability (balancing selection) or fixation of a favourable mutation (positive selection). Published molecular population analyses have reported that at least 18 genes and an estimated 15–30 % of Arabidopsis genes have been subject to natural selection (Nordborg et al., 2005; Wright and Gaut, 2005; Schmid et al., 2006). In most cases, however, the loci identified by this strategy have not been analysed in fitness assays and thus it remains an open question whether the selective forces acting on those genes contribute to adaptive phenotypes in natural populations (Kawecki and Ebert, 2004). Because this topic lies at the crossroads of genetics and ecology, it has recently increasingly attracted the attention of evolutionary ecologists who wish to identify the selection mechanisms underlying ecologically important phenotypes (Tonsor et al., 2005; Mitchell-Olds and Schmitt, 2006). Published studies in this area have concentrated on traits of potentially high adaptive value, such as disease resistance or flowering time.
Selection in pathogen defence
Intensive molecular biology and genetic studies on resistance to herbivores and pathogens provide an excellent basis to understand selection mechanisms in adaptive evolution. Mechanisms of plant defence against insects and pathogens are generally divided into three conceptually distinct phases: attack recognition, signal transduction and defence development. Plant resistance genes (R-genes) are important in the response to many different plant pathogens. The R-proteins monitor the plant cell for the presence of pathogen-secreted proteins. Upon their detection, they then rapidly activate robust plant defence pathways, including local cell death, cell-wall reinforcement, production of secondary compounds and systemic acquired resistance.
A prototypical R-gene, RPM1 of Arabidopsis, encodes a peripheral plasma membrane protein that recognizes an attack by Pseudomonas syringae pathogens (Grant et al., 1995). Both resistance and susceptibility alleles, which are defined by the absence and presence of respective polymorphisms, frequently occur together within natural populations and are common across the Arabidopsis range (Stahl et al., 1999). Consistent with this observation, analysis of RPM1 nucleotide diversity suggests that evolutionarily stable polymorphisms are probably maintained by balancing selection. Other R-genes (RPS2 and RPS5) have also been shown to maintain high levels of nucleotide polymorphism due to balancing selection (Caicedo et al., 1999; Tian et al., 2002). The latest study (Bakker et al., 2006a) estimated that RPP13 has been targeted by a long-lived balancing selection, although several other loci could be identified as candidates for recent selective sweeps. From an ecological–evolutionary point of view, these examples provoke the question of what mechanisms prevent the resistance alleles from being fixed by natural selection despite their obvious benefits? In fact, two ecological approaches using isogenic lines for different RPM1 and RPS2 alleles propose an at least partial explanation of this phenomenon: the resistant plants produce fewer progeny than the susceptible plants, revealing that a trade-off between benefit and cost of resistance contributes to the maintenance of polymorphisms in R-genes (Tian et al., 2003; Korves and Bergelson, 2004). Alternatively, frequency-dependent selection may maintain variability: rare pathogen variants may increase in frequency, therefore imposing selection on hosts for specific resistance, which results in an increase in frequency of resistant hosts and thus again selection favouring novel and yet rare pathogen variants.
Adaptive variation in flowering time
Flowering time is a critical life-history trait that significantly contributes to plant fitness (Stearns, 1992), as it is essential for plants to complete flower development, pollination and seed production in favourable conditions. Hence, plants have developed sophisticated mechanisms to integrate information from environmental cues such as light, temperature, water availability and biotic factors, resulting in presumable optimization of timing of flowering. Most importantly, information regarding seasonal changes is inferred from daylength (photoperiod) and ambient temperature. In particular for winter-annual accessions (Fig. 1), flowering time strongly depends on vernalization, i.e. the acceleration of flowering by exposure to prolonged cold. Both photoperiod and average temperature not only vary according to season but also according to geographical location. Consistent with this, flowering time varies widely among natural Arabidopsis accessions, reflecting their wide range of habitat from the equator to the arctic circle. This variation mainly reflects genetic variation in both photoperiod and vernalization pathways (Henderson et al., 2003; Balasubramanian et al., 2006).
A large effort focused on mutant analysis has identified more than 80 genes that regulate flowering in response to different environmental and endogenous cues (Simpson and Dean, 2002). Notably, natural allelic variation has only been identified in few of these genes thus far. Among them, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) are pivotal regulators of the vernalization pathway. Molecular analysis of FRI and FLC by haplotyping and phenotyping of a large set of natural accessions revealed that polymorphisms in these genes are the key factors in the extensive natural variation of flowering time. The data also suggest that early-flowering types have evolved from late-flowering ancestral types through independent loss-of-function mutations in FRI and/or FLC (Gazzani et al., 2003; Michaels et al., 2003; Shindo et al., 2005). Thus, FRI and FLC appear to be major targets for natural selection. In particular, non-functional FRI alleles probably play a critical role for adaptive divergence of flowering time, as FRI activity determines the extent of vernalization requirement by activating FLC expression. Several molecular analyses support this hypothesis and have succeeded in detecting the trace of selection acting on FRI, for instance by comparing statistical values of population differentiation at phenotypic traits (QST) and at neutral molecular markers (FST) within and among populations (Le Corre, 2005; Evanno et al., 2006). QST for flowering time without vernalization was significantly higher than FST for neutral markers, and FST for functional and non-functional haplotypes was significantly higher than that for markers, suggesting adaptive divergence of flowering time through selection for loss of FRI function. Furthermore, selective sweep signature and haplotype sharing around FRI suggests recent selection for early flowering (Hagenblad et al., 2004; Aranzana et al., 2005; Toomajian et al., 2006).
Although allelic variation of FRI is apparently the major player for adaptive variation of flowering time in Arabidopsis, natural allelic variation has also been reported in other flowering-time genes. Notably, in most cases, naturally occurring mutations result in an early-flowering life cycle (Roux et al., 2006). Under which conditions would early flowering be adaptive as compared with late flowering? If plants are growing under benign conditions, late flowering leads to a greater accumulation of resources due to longer vegetative growth, which should result in increased numbers of offspring with optimal seed provisioning. Early flowering implies a shorter vegetative growth phase and thus lower biomass accumulation. However, in specific habitats or environments, early flowering is more advantageous than late flowering because it enables the plant to avoid stressful conditions that may damage seed production, such as drought or elevated temperature later in summer. In fact, directional selection favouring early flowering has been reported under conditions making it necessary to avoid herbivores, shade or drought (Callahan and Pigliucci, 2002; McKay et al., 2003; Pigliucci, 2003).
Although molecular data clearly show that genes responsible for flowering-time variation in nature have been under selection, proof of this from an ecological point of view has not been straightforward, especially in the case of natural variation in vernalization. For instance, one might expect a latitudinal cline in this trait, i.e. late-flowering types or winter-annuals should inhabit predominantly northern areas, where both winter temperature and period are optimal to accelerate their flowering. Indeed, a latitudinal cline depending on functional FRI alleles of differential activity has been reported (Stinchcombe et al., 2004). However, no such cline has been detected for accessions that contain loss-of-function FRI alleles. In another analysis of a set of accessions including several accessions from northern and southern Sweden, no significant geographical gradient was detected, although very late-flowering accessions were non-randomly distributed in specific areas of the Scandinavian Peninsula (Shindo et al., 2005). The lack of a strong latitudinal cline in flowering time is somewhat surprising and might indicate that variation in flowering time correlates with specific environmental conditions in local habitats rather than geographical position. A reciprocal transplant field experiment might be able to confirm this notion (Kawecki and Ebert, 2004; Mitchell-Olds and Schmitt, 2006).
ASSESSING ADAPTIVE POTENTIAL OF NATURAL VARIATION IN MORPHOLOGICAL TRAITS
The above examples demonstrate the general difficulty of linking molecular analyses with field observations or experiments, even if the phenotypes investigated are primarily thought to be under the influence of macro-environmental factors. Therefore, one could expect that adaptation to micro-environmental factors will turn out to be even more difficult to prove. This would, for instance, apply to the majority of morphological traits, which at best might represent local adaptation.
Generally speaking, natural variation in morphological traits such as leaf or flower shape and root architecture is considered to correlate tightly with speciation (Shepard and Purugganan, 2002). Indeed, molecular population genetics has detected the signature of selection in Arabidopsis MADS-box genes involved in flower development (Purugganan and Suddith, 1998; Moore et al., 2005). However, to determine whether natural variation in morphological phenotypes resulted from local adaptation among Arabidopsis accessions would require a substantial effort in ecological analysis. For instance, the micro-environmental factors that are responsible for selective pressure in the original habitat are usually unknown. Even if known, the complex genetic make up observed to underlie many trait differences between accessions makes it difficult to assess the adaptive contribution of individual loci. This challenge somewhat resembles the problem of deciphering the mechanisms of plant development through analysis of wild-type plants, without making use of the mutagenesis approach. Thus, in analogy, a reasonable starting point for evolutionary–ecological analyses would be to assess the adaptive potential of individual alleles in isolation, in a common genetic background, as for instance represented by NILs for different alleles of a single gene. Careful meta-analysis that combines complementing evidence from different experiments (Arnqvist and Wooster, 1995) might prove to be a powerful approach to assess the adaptive potential of allelic variants.
For such analyses, several independent NILs for the alleles in question are necessary to correct for the effect of any unknown linked modifiers carried over from the different genetic backgrounds in which the original alleles were detected. As an indicator of fitness, the ultimate parameters of reproductive success should be assayed: the number of progeny per individual of a certain genotype, and the survival and recruitment rate of this next generation into the reproductive stage. To measure how allelic variation influences relative fitness, we propose competitive experimental plots as one of the most promising strategies for ecological analysis. This approach measures the relative fitness of alternative phenotypes and/or genotypes in a range of environments, and has for instance been successfully employed to demonstrate adaptive plasticity in shade avoidance (Schmitt et al., 1995). The experimental plots can be in the field or in the greenhouse. Importantly, designs for greenhouse experiments should take into account that common laboratory conditions for growing Arabidopsis (i.e. temperature above approx. 20 °C and sufficient moisture) are highly artificial for most accessions (Hoffmann, 2002). Ideally, the experimental conditions should encompass a more realistic range of variation as encountered in natural environments, e.g. for temperature, moisture and/or light regimes, with the factors chosen to vary depending on the presumed adaptive value of the trait of interest. Finally, the approach should be rounded up by field data from the original collection site. This would be important for several reasons. First, accessions deposited in the stock centre represent only a small, isogenized subset of the Arabidopsis collected in a certain location. Therefore, sampling and genotyping of the extant population at the original collection site can give important insight into the genetic variation within this population and hint towards possible founder effects or genetic sweeps. Second, accurate field data regarding the collection sites of individual genotypes, such as soil composition and characteristics, human disturbance, presence/absence of natural enemies, density or competition levels, might aid in identifying adaptive advantages of genotypes that are not evident from greenhouse or standard field plot experiments. They might also allow us to relate genotype frequencies to environmental gradients at a fine scale. In our laboratories, we have initiated a series of competition analyses to estimate the relative fitness of a naturally occurring null mutant in the BREVIS RADIX (BRX) gene (Mouchel et al., 2004; Briggs et al., 2006), which strongly influences root system architecture. This study consists of the elements indicated above and might become a test case for the feasibility of this combined approach.
CONCLUSIONS
The analysis of natural genetic variation in Arabidopsis is in full bloom and on its way to become a standard approach for the discovery of novel genes and alleles, and in addressing questions in evolutionary developmental biology. Without the substantial contributions from genomics and mutant analysis, this would not have been possible. Those resources put Arabidopsis in a unique position with regard to evolutionary ecology. First, the detailed analyses of developmental processes provide a wealth of background information that aids in understanding the relative importance of genes and their synergy in shaping given traits, with important implications for genetic mechanisms of ecological and evolutionary relevance, such as epistatic relationships. Second, biological resources of Arabidopsis are plentiful, easy to maintain in the laboratory, and easy to analyse for phenotypes and genotypes. Third, Arabidopsis is found in many different habitats across a wide geographical space and therefore must presumably possess either a strong capacity for rapid adaptation or a very high phenotypic and physiological plasticity. Finally, recent analyses clearly demonstrate that Arabidopsis populations are by far not as genetically uniform as previously assumed, which was one of the main arguments for the unsuitability of Arabidopsis for population genetics and evolutionary ecology. Thus, in summary Arabidopsis might very well become the model organism for plant evolutionary ecology, thereby bridging the dichotomy between the molecular and organismal disciplines of biology.
ACKNOWLEDGEMENTS
We would like to thank Professor M. Nordborg for helpful comments on the manuscript, and the University of Lausanne for financial support through an Interdisciplinary Grant of the Faculty of Biology and Medicine (to C.S.H. and G.B.). G.B. also acknowledges support from the Swiss NSF (PPOOA-102944/1).
LITERATURE CITED
- Abbott RJ, Gomes MF. Population genetic structure and outcrossing rate of Arabidopsis thaliana (L.) Heynh. Heredity. 1989;62:411–418. [Google Scholar]
- Alonso-Blanco C, Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends in Plant Science. 2000;5:22–29. doi: 10.1016/s1360-1385(99)01510-1. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M. From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. The International Journal of Developmental Biology. 2005;49:717–732. doi: 10.1387/ijdb.051994ca. [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz IA, O'Kane SL. The Arabidopsis Book. 2002. Taxonomy and phylogeny of Arabidopsis (Brassicaceae) doi: 10.1199/tab.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, et al. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genetics. 2005;1:e60. doi: 10.1371/journal.pgen.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G, Wooster D. Meta-analysis: synthetizing research findings in ecology and evolution. Trends in Ecology and Evolution. 1995;10:236–240. doi: 10.1016/S0169-5347(00)89073-4. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for Phytochrome D in red/far-red light sensing. The Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EG, Toomajian C, Kreitman M, Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. The Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EG, Stahl EA, Toomajian C, Nordborg M, Kreitman M, Bergelson J. Distribution of genetic variation within among local populations of Arabidopsis thaliana over its species range. Molecular Ecology. 2006;15:1405–1418. doi: 10.1111/j.1365-294X.2006.02884.x. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, et al. The PHOTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nature Genetics. 2006;38:711–715. doi: 10.1038/ng1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proceedings of the National Academy of Sciences of the USA; 2006. pp. 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Stahl E, Dudek S, Kreitman M. Genetic variation within and among populations of Arabidopsis thaliana. Genetics. 1998;148:1311–1323. doi: 10.1093/genetics/148.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Nordborg M. The impact of genomics on the study of natural variation in Arabidopsis. Plant Physiology. 2003;132:718–725. doi: 10.1104/pp.103.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Ecker JR. Plant genomics: The third wave. Annual Review of Genomics and Human Genetics. 2004;5:443–477. doi: 10.1146/annurev.genom.5.061903.180017. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, et al. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Research. 2003;13:513–523. doi: 10.1101/gr.541303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P, Rombaut D, Van Gysel A, Van Montagu M, Gerats T. AFLP analysis of genetic diversity within and between Arabidopsis thaliana ecotypes. Molecular and General Genetics. 1999;261:627–634. doi: 10.1007/s004380050005. [DOI] [PubMed] [Google Scholar]
- Briggs GC, Mouchel CF, Hardtke CS. Characterization of the plant-specific BREVIS RADIX gene family reveals limited genetic redundancy despite high sequence conservation. Plant Physiology. 2006;140:1306–1316. doi: 10.1104/pp.105.075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Schaal BA, Kunkel BN. Diversity and molecular evolution of the Rps2 resistance gene in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA; 1999. pp. 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan HS, Pigliucci M. Shade-induced plasticity and its ecological significance in wild populations of Arabidopsis thaliana. Ecology. 2002;83:1965–1980. [Google Scholar]
- Charlesworth D, Vekemans X. How and when did Arabidopsis thaliana become highly self-fertilising. BioEssays. 2005;27:472–476. doi: 10.1002/bies.20231. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Mahajan SK, Whitham SA, Yamamoto ML, Carrington JC. Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proceedings of the National Academy of Sciences of the USA; 2000. pp. 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded be five genes: the sequences and expression of PHYD and PHYE. Plant Molecular Biology. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Comai L, Young K, Till BJ, Reynolds SH, Greene EA, Codomo CA, et al. Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. The Plant Journal. 2004;37:778–786. doi: 10.1111/j.0960-7412.2003.01999.x. [DOI] [PubMed] [Google Scholar]
- Donohue K. Germination timing influences natural selection on life-history characters in Arabidopsis thaliana. Ecology. 2002;83:1006–1016. [Google Scholar]
- El-Assal SE, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nature Genetics. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- Evanno G, Castella E, Goudet J. Evolutionary aspects of population structure for molecular and quantitative traits in the freshwater snail Radix balthica. Journal of Evolutionary Biology. 2006;19:1071–1082. doi: 10.1111/j.1420-9101.2006.01098.x. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Annual Review of Plant Biology. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiology. 2003;132:1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Hagenblad J, Nordborg M. Sequence variation and haplotype structure surrounding the flowering time locus FRI in Arabidopsis thaliana. Genetics. 2002;161:289–298. doi: 10.1093/genetics/161.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad J, Tang C, Molitor J, Werner J, Zhao K, Zheng H, et al. Haplotype structure and phenotypic associations in the chromosomal regions surrounding two Arabidopsis thaliana flowering time loci. Genetics. 2004;168:1627–1638. doi: 10.1534/genetics.104.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Shindo C, Dean C. The need for winter in the switch to flowering. Annual Review of Genetics. 2003;37:371–392. doi: 10.1146/annurev.genet.37.110801.142640. [DOI] [PubMed] [Google Scholar]
- Hoffmann MH. Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae) Journal of Biogeography. 2002;29:125–134. [Google Scholar]
- Hoffmann MH. Evolution of the realized climatic niche in the genus Arabidopsis (Brassicaceae) Evolution. 2005;59:1425–1436. [PubMed] [Google Scholar]
- Hoffmann MH, Bremer M, Schneider K, Burger F, Stolle E, Moritz G. Flower visitors in a natural population of Arabidopsis thaliana. Plant Biology. 2003;5:491–494. [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. Arabidopsis map-based cloning in the post-genome era. Plant Physiology. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1994;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- Keurentjes JB, Fu J, Ric de Vos CH, Lommen A, Hall RD, Bino RJ, et al. The genetics of plant metabolism. Nature Genetics. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Kim S, Zhao K, Jiang R, Molitor J, Borevitz JO, Nordborg M, Marjoram P. Association mapping with single-feature polymorphisms. Genetics. 2006;173:1125–1133. doi: 10.1534/genetics.105.052720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. The Plant Cell. 2001;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annual Review of Plant Biology. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Korves T, Bergelson J. A novel cost of R gene resistance in the presence of disease. The American Naturalist. 2004;163:489–504. doi: 10.1086/382552. [DOI] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiology. 2001;127:1077–1088. [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proceedings of the National Academy of Sciences of the USA; 2003. pp. 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MJ. Variations in natural populations of Arabidopsis thaliana (L.) Heynh. In: Vaughan JG, Macleod AJ, editors. The biology and chemistry of the Cruciferae. London/New York/San Francisco: Academic Press; 1976. pp. 167–190. [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J. The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell. 2001;13:2793–2807. doi: 10.1105/tpc.010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre V. Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Molecular Ecology. 2005;14:4181–4192. doi: 10.1111/j.1365-294X.2005.02722.x. [DOI] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Chan J, Cashmore AR. CRY2: a second member of the Arabidopsis cryptochrome gene family (accession No. U43397) (PGR 96-001) Plant Physiology. 1996;110:1047. [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theoretical and Applied Genetics. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- Maloof JN. Genomic approaches to analyzing natural variation in Arabidopsis thaliana. Current Opinion in Genetics & Development. 2003;13:576–582. doi: 10.1016/j.gde.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevits JO, Dabi T, Lutes J, Nehring RB, Redfern JL, et al. Natural variation in light sensitivity of Arabidopsis. Nature Genetics. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Mitchell-Olds T. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Molecular Ecology. 2003;12:1137–1151. doi: 10.1046/j.1365-294x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as repressor of flowering. The Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2003;100:10102–10107. doi: 10.1073/pnas.1531467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends in Ecology & Evolution. 2001;16:693–700. [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Miyashita NT, Kawabe A, Innan H. DNA variation in the wild plant Arabidopsis thaliana revealed by amplified fragment length polymorphism analysis. Genetics. 1999;152:1723–1731. doi: 10.1093/genetics/152.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Grant SR, Purugganan MD. Molecular population genetics of redundant floral-regulatory genes in Arabidopsis thaliana. Molecular Biology and Evolution. 2005;22:91–103. doi: 10.1093/molbev/msh261. [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes and Development. 2004;18:700–714. doi: 10.1101/gad.1187704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napp-Zinn K. In: Arabidopsis thaliana. Halevy AH, editor. Boca Raton, FL: CRC Press; 1985. pp. 492–503. CRC handbook of flowering. [Google Scholar]
- Nordborg M, Bergelson J. The effect of seed and rosette cold treatment on germination and flowering time in some Arabidopsis thaliana (Brassicaceae) ecotypes. American Journal of Botany. 1999;86:470–475. [PubMed] [Google Scholar]
- Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, et al. The extent of linkage disequilibrium in Arabidopsis thaliana. Nature Genetics. 2002;30:190–193. doi: 10.1038/ng813. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biology. 2005;3 doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Halldorsdottir SS, Stinchcombe JR, Weinig C, Schmitt J, Purugganan MD. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics. 2004;167:1361–1369. doi: 10.1534/genetics.103.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. Selection in a model system: ecological genetics of flowering time in Arabidopsis thaliana. Ecology. 2003;84:1700–1712. [Google Scholar]
- Purugganan MD, Suddith JI. Molecular population genetics of the Arabidopsis CAULIFLOWER regulatory gene: nonneutral evolution and naturally occurring variation in floral homeotic function. Proceedings of the National Academy of Sciences of the USA; 1998. pp. 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Richards EJ. The control of natural variation in cytosine methylation in Arabidopsis. Genetics. 2002;162:355–363. doi: 10.1093/genetics/162.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F, Touzet P, Cuguen J, Le Corre V. How to be early flowering: an evolutionary perspective. Trends in Plant Science. 2006;11:375–381. doi: 10.1016/j.tplants.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Research. 2003;13:1250–1257. doi: 10.1101/gr.728603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Torjek O, Meyer R, Schmuths H, Hoffmann MH, Altmann T. Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theoretical and Applied Genetics. 2006;112:1104–1114. doi: 10.1007/s00122-006-0212-7. [DOI] [PubMed] [Google Scholar]
- Schmitt KJ, McCormac AC, Smith H. A test of the adaptive plasticity hypothesis using transgenic and mutant plants disabled in Phytochrome-mediated elongation response to neighbors. The American Naturalist. 1995;146:937–953. [Google Scholar]
- Scholl RL, May ST, Ware DH. Seed and molecular resources for Arabidopsis. Plant Physiology. 2000;124:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant Journal. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJ, Bentsink L, Vonk J, van der Plas LH, Koornneef M, Vreugdenhil D. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proceedings of the National Academy of Sciences of the USA; 2006. pp. 2994–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel TS, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Molecular Ecology. 2000;9:2109–2118. doi: 10.1046/j.1365-294x.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes and Development. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shepard KA, Purugganan MD. The genetics of plant morphological evolution. Current Opinion in Plant Biology. 2002;5:49–55. doi: 10.1016/s1369-5266(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C. Role of FRIGIDA and FLOWEING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiology. 2005;138:1163–1173. doi: 10.1104/pp.105.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C. Arabidopsis, the rosetta stone of flowering time? Science. 2002;296:285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Stearns SC. The evolution of life histories. Oxford: Oxford University Press; 1992. [Google Scholar]
- Stenøien HK, Fenster CB, Tonteri A, Savolainen O. Genetic variability in natural populations of Arabidopsis thaliana in northern Europe. Molecular Ecology. 2005;14:137–148. doi: 10.1111/j.1365-294X.2004.02359.x. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, et al. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proceedings of the National Academy of Sciences of the USA; 2004. pp. 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Araki H, Stahl E, Bergelson J, Kreitman M. Signature of balancing selection in Arabidopsis. Proceedings of the National Academy of Sciences of the USA; 2002. pp. 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Tonsor SJ, Alonso-Blanco C, Koornneef M. Gene function beyond the single trait: natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana. Plant, Cell and Environment. 2005;28:2–20. [Google Scholar]
- Toomajian C, Hu TT, Aranzana MJ, Lister C, Tang C, Zheng H, et al. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biology. 2006;4:e137. doi: 10.1371/journal.pbio.0040137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Zwan C, Brodie S, Campanella J. The intraspecific phylogenetics of Arabidopsis thaliana in worldwide populations. Systematic Botany. 2000;25:47–59. [Google Scholar]
- Weigel D, Nordborg M. Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiology. 2005;138:567–568. doi: 10.1104/pp.104.900157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proceedings of the National Academy of Sciences of the USA; 2005. pp. 2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Gaut BS. Molecular population genetics and the search for adaptive evolution in plants. Molecular Biology and Evolution. 2005;22:506–519. doi: 10.1093/molbev/msi035. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ober JA, Kliebenstein The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistant in Arabidopsis. The Plant Cell. 2006;18:1524–1536. doi: 10.1105/tpc.105.039602. [DOI] [PMC free article] [PubMed] [Google Scholar]