Abstract
Background and Aims
Knowledge of host factors affecting plant–nematode interactions is scarce. Here, relevant interaction phenotypes between a nodulating model host, Lotus japonicus, and the endoparasitic root-knot nematode Meloidogyne incognita are assessed via a genetic screen.
Methods
Within an alpha experimental design, 4-week-old replicate plants from 60 L. japonicus ecotypes were inoculated with 1000 nematodes from a single egg mass population, and evaluated for galling and nematode egg masses 6 weeks after inoculation.
Key Results
Statistical analysis of data for 57 ecotypes showed that ecotype susceptibilities ranged from 3·5 to 406 galls per root, and correlated strongly (r = 0·8, P < 0·001, log scale) with nematode reproduction (ranging from 0·6 to 34·5 egg masses per root). Some ecotypes, however, showed a significant discrepancy between disease severity and nematode reproduction. Necrosis and developmental malformations were observed in other infected ecotypes.
Conclusions
The first evidence is provided of significant variability in the interactions between L. japonicus and root-knot nematodes that may have further implications for the genetic dissection and characterization of host pathways involved in nematode parasitism and, possibly, in microbial symbiosis.
Key words: Genetics, Lotus japonicus, nematode parasitism, plant–nematode interactions, susceptibility factors
INTRODUCTION
The parasitic relationships between plant nematodes and their hosts result in significant yield losses in a wide variety of crops (Sasser and Freckman, 1987). Most plant nematodes infect specific tissues, and known nematode resistance genes are typically strong, cell autonomous and durable when elicited (Williamson and Gleason, 2003; Cabrera Poch et al., 2006). Thus, root-knot nematode (RKN, Meloidogyne spp.) parasitism is an example of an extreme interaction that has developed during coevolution with their host plants (Davis et al., 2000).
The cellular adaptations necessary for compatibility between plant hosts and their endoparasitic nematodes must depend on finely tuned recognition processes and on molecular coordination between plant and parasite (Bird, 2004). Preparasitic, infective second-stage juveniles (J2) of RKNs cause nematode-induced root hair waviness and branching (Weerasinghe et al., 2005). The J2 then penetrate the root, migrate intercellularly and trigger the formation of a complex structure, the giant cells (GCs), close to the vascular cylinder (Goverse et al., 2000; Gheysen and Fenoll, 2002). GCs are surrounded by a gall or knot, which gives RKN its vernacular name. Further parasitic nematode development occurs only after GCs are triggered. The female nematode increases its body volume considerably by drawing nutrients from the GCs, maintenance of these cells being dependent on nematode feeding. This suggests that the host root tissues can control RKN development, which may be specific to GCs.
The model nodulating legume Lotus japonicus (Handberg and Stougaard, 1992; Sato and Tabata, 2006) has been recently described as a powerful host to study RKN interactions (Lohar and Bird, 2003). Several kinds of L. japonicus mutations affect RKN parasitism. The mutant har1 is hyperinfected by both rhizobia (Wopereis et al., 2000) and RKNs (Lohar and Bird, 2003). It has been hypothesized that HAR1, which encodes a CLAVATA1-like receptor kinase with no shoot phenotype (Krusell et al., 2002; Nishimura et al., 2002), interacts with a putative nematode-encoded CLAVATA3-like peptide, ligand for the CLV1 receptor mimicking the endogenous plant ligand (Olsen and Skriver, 2003). Other aspects of the host early responses to RKNs were altered or abolished in the L. japonicus mutants nfr1 and nfr5, affected in Nod factor receptor genes, suggesting that RKNs produce a molecule with functional equivalence to Nod factors. Because the ability of RKNs to establish GCs and reproduce was markedly reduced in these mutant lines, it was proposed that RKNs have adapted part of the symbiont-response pathway to enhance their parasitic ability (Weerasinghe et al., 2005).
No systematic attempts to dissect genetically the infection processes induced by RKNs in host plants from other interacting microbes have been reported. However, some information about host genotypes affecting susceptibility or other interaction phenotypes to RKNs has been collated from other species, including model legumes (Roberts, 1992; Boiteux et al., 1999). In the case of root nodule symbiosis, this has led to the analysis of particular traits (Tirichine et al., 2000; De Mita et al., 2006). Ecotypes of the model plant Arabidopsis thaliana have been screened for phenotypic variation in their interactions with Meloidogyne hapla (Boiteux et al., 1999). However, Arabidopsis is generally a poor host for RKNs and does not establish symbiosis with rhizobia or mycorrhiza. Here, the potential of L. japonicus as a suitable source of genetic variability in the interactions with RKNs is investigated by assessing a collection of ecotypes (Kawaguchi et al., 2001) for relevant phenotypes. The screening protocol yielded plant variants that exhibited very different degrees of disease severity and/or nematode reproduction rates. Results allowed identification of novel sources of host variability to plant nematode parasitism that may help in the genetic characterization and identification of host pathways involved in nematode – and other microbes – interactions.
MATERIALS AND METHODS
Plant material and growth conditions
Seeds from 60 available ecotypes of Lotus japonicus (Regel) Larsen, randomly selected from the National BioResource Project collection, Japan, were scarified and disinfected by soaking in concentrated sulfuric acid for 20 min. The seeds were then washed five times with sterile water, left to soak for 3 h before sowing them on agar-MS medium at 4°C for 24 h, and transferred to a growth incubator with an 18/6 h day/night cycle and a 24/18°C day/night temperature regime for germination. Two-week-old seedlings were transferred to individual 10-cm plastic pots containing a mixture of sand and peat moss (1:1, v/v). Plants in the glasshouse were watered daily, with no artificial light supplied, and kept at a 24/18°C day/night temperature regime. Plants were inoculated with nematodes 3 weeks after potting, and galls and egg masses counted 6 weeks after inoculation. As relevant morphological features of the root systems may differ significantly between ecotypes, fresh weights of all roots were also measured and used to standardize both the galling and the egg mass counts for statistically meaningful comparisons between the two parameters.
Nematode culture and inoculations
A Meloidogyne incognita (Kofoid and White) line was established, maintained and bulked up from a single egg mass on susceptible tomato plants (Solanum lycopersicum ‘Tiny Tim’). This line originates from an M. incognita population that has been reared in our laboratory for over 10 years. To collect egg masses, soil was gently removed from infected roots, and 2–3-cm root pieces containing galls were placed in a Petri dish. A total of 4000 egg masses were picked with forceps using a stereomicroscope (250×), and stored at 4°C in 1·5-mL plastic tubes until further use. To produce infective J2, batches of 50–100 egg masses were transferred at a time to a sterile Petri dish containing a minimum volume of sterile distilled water. The gelatinous matrix was removed, and the released eggs were disinfected in a 0·2 g mL−1 sodium hypochlorite solution for 3 min. The contents of the Petri dish were collected onto a 30-μm mesh sieve slightly submerged in an evaporating basin containing tap water to wash out the sodium hypochlorite. The washing process was repeated three times. Three or four drops of 4 % (v/v) lactic acid were added to the sieve contents and left for 2 min. The eggs were then washed and poured into a Petri dish and two drops of 1 mg mL−1 malachite green and tomato root diffusate (1 : 1, v/v) were added. Eggs were incubated for 5–7 d at room temperature. After hatching, the J2 were poured and washed through a bank of sieves (100-, 53- and 30-μm mesh), and collected for inoculation. Pots were watered and allowed to drain overnight prior to nematode inoculation. The nematode inoculum was quantified and diluted with tap water to a density of 500 J2 mL−1. Two equidistant, 4-cm holes were made in the soil around the root systems, and 1 mL of the freshly hatched nematode suspension poured in each hole, which was then covered with soil. Pots were lightly watered during the first 7 d after inoculation.
Experimental design
As 60 ecotypes were to be tested simultaneously, the screening procedure was set up according to an alpha design (Patterson and Williams, 1976) in which block sizes were kept small (ten ecotypes per block) but the blocks were grouped together into sets of six within which each ecotype was replicated once. Two glasshouse compartments were used and three such sets of six blocks were set up in each compartment giving an overall replication of six plants per ecotype. Because not all plants could be inoculated on the same day, the design was set up sequentially, completing one block from each compartment (and from each set within each compartment in turn) on each of 18 consecutive days.
Staining and microscopy
Six weeks after nematode inoculation, root systems were soaked overnight and soil gently washed off the roots. Roots were stained for 15 min in 0·15 g L−1 phloxine B and subsequently rinsed. The excess water was gently removed from the roots with absorbent paper and galls and egg masses per root system were counted under a magnifying lens. Roots were stained for nematodes with acid fuchsin–lacto-glycerol, and photographs taken with a camera attached to a stereomicroscope (150–625×).
Statistical analysis
The numbers of galls and egg masses per gram root weight were transformed to logarithms to stabilize the variance (after adding a small offset of 0·5 to allow for zero counts) and analysed using the method of residual maximum likelihood with both the blocking structure and ecotypes as random effects. Thirty-five plants with missing data owing to premature death, and three ecotypes subsequently left with insufficient replication (MG-2, MG-90, MG-98), were excluded from the analysis. Hence the actual replication for the 57 included ecotypes ranged between three and six.
RESULTS
The complete data set from this experiment, including number of galls and egg masses, and root weights of all plants tested is provided in the Supplementary Information available online.
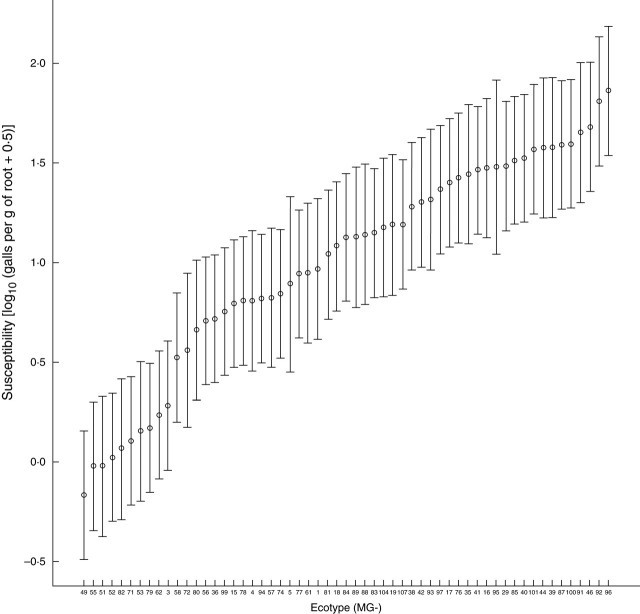
Susceptibilities of L. japonicus ecotypes to M. incognita, measured as the mean number of galls per gram root weight, range over a 100-fold scale (Fig. 1 and Supplementary Information). The most susceptible accession tested (MG-96) was severely affected by the nematode-induced disease, with between 164 and 674 galls in the roots. By contrast, ecotypes at the other end of the susceptibility spectrum, such as MG-49, showed virtually no sign of disease induced by M. incognita, with only 0–11 galls present in the root systems (Fig. 1; see also Fig. 3A).
Fig. 1.
Susceptibilities of Lotus japonicus ecotypes to the RKN Meloidogyne incognita. Mean logarithmically transformed numbers of galls per gram root weight are presented in increasing order and labelled on the horizontal axis by ecotype accession number (MG-). Bars represent 95% confidence intervals around the fitted means.
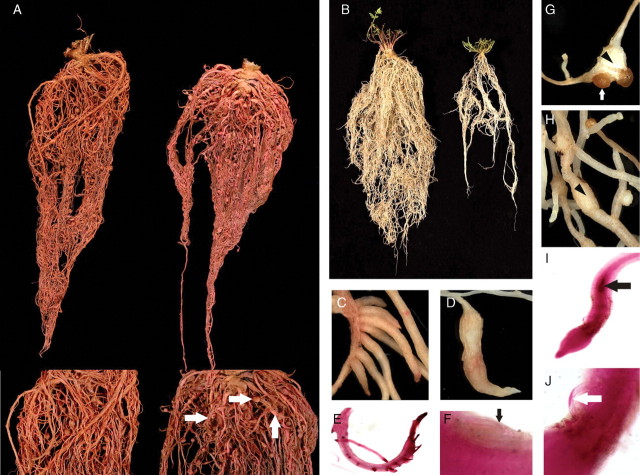
Fig. 3.
Responses of Lotus japonicus ecotypes to Meloidogyne incognita. (A) Infected root systems of a highly susceptible ecotype, MG-96 (right), and a resistant ecotype, MG-55 (left), and at higher magnification (lower panel) showing severe galling (arrows) in ecotype MG-96 and virtually no galls in ecotype MG-55. (B) Visible morphological differences between root systems of two L. japonicus ecotypes used in this study. (C–E) Lotus japonicus ecotypes showing abnormal root swellings (C, D) and necrotic tips and deformation of the lateral roots (E) after RKN infection. (F–H) Normal development of M. incognita nematodes in L. japonicus roots: (F) fourth stage juvenile of M. incognita inside root (arrow), and (G, H) galls (arrowheads) with egg masses (arrow) attached to mature females. (I, J) Necrosis (black arrow) in ‘pendulum-like’ swelling of the tip of lateral roots (I) associated with nematodes partially embedded in roots (J).
No bias to any particular susceptibility value among ecotypes was observed, and only relatively small, quantitative differences between accession susceptibilities were apparent when ecotypes were ordered by their susceptibility values (Fig. 1). There was no evidence that a saturation value for susceptibility (i.e. maximum count of nematode-induced galls an ecotype root can support) was reached among the ecotypes tested (Fig. 1).
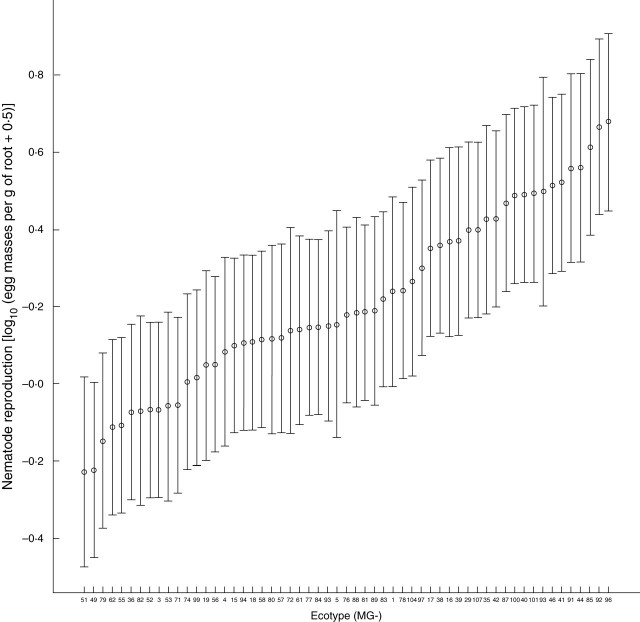
The effect that the different L. japonicus ecotypes had upon the reproduction rate of M. incognita was also measured (Supplementary Information and Fig. 2). This figure shows the same general features as Fig. 1: there was no apparent limit to the number of egg masses that an ecotype could support, and ecotypes did not show a bias in any particular nematode reproduction rate. The most and the least susceptible ecotypes to RKN challenge were also the best and the worst hosts for RKN reproduction, respectively (Figs 1 and 2).
Fig. 2.
Reproduction rate of Meloidogyne incognita in Lotus japonicus ecotypes. Mean logarithmically transformed numbers of egg masses per gram root weight are presented in increasing order and labelled on the horizontal axis by ecotype accession number (MG-). Bars represent 95% confidence intervals around the fitted means.
No evidence for a relationship between the collection sites of ecotypes and their phenotypes with nematode infections could be revealed. The ten most resistant ecotypes in Figs 1 and 2 were not collected close to each other, as seen from the prefecture names in the Legumebase website (see Supplementary Information and http://www.shigen.nig.ac.jp/legume/legumebase/index.jsp).
The logarithmically transformed numbers of galls and egg masses per gram root weight (Supplementary Information, and Figs 1 and 2) were highly correlated (r = 0·794, P < 0·001, d.f. = 314). Thus, over all replicates of the 57 ecotypes tested, there was a good correlation between the severity of the RKN-induced disease (susceptibility) and the rate at which RKNs reproduce in them. However, ecotypes MG-18, MG-93, MG-76 and MG-19 showed a relatively high discrepancy between these values: the ratio of mean egg masses to mean galls for these were 0·025, 0·028, 0·031 and 0·033, respectively.
Although host susceptibility to M. incognita and nematode reproduction rates were the interaction phenotypes targeted in the present study, they were not the only phenotypes found among the infected L. japonicus ecotypes. Distortions to the normal pattern of gall and root formation were observed in some ecotypes upon RKN challenge (Fig. 3A,G, H). Some ecotypes showed symptoms of necrosis or deformation of lateral roots upon RKN infection (Fig. 3E, I). In some cases, the necrosis was clearly associated with the presence of the nematode (Fig. 3I), whereas in others the necrosis extended to the newly formed lateral root tips, relatively distant from the nematode (Fig. 3E). Other ecotypes, for example MG-4 and MG-74, showed multiple lateral roots arranged in comb-like deformations of the infected roots (Fig. 3C), and roots of ecotypes MG-29 and MG-38 were distorted by RKN infection forming ‘chilli pepper’-like swellings (Fig. 3D).
DISCUSSION
Lotus japonicus has proved a very useful model plant in the genetic dissection of symbiotic interactions with nodulating bacteria (Wopereis et al., 2000; Radutoiu et al., 2003; Sandal et al., 2006), mycorrhizal fungi (Stracke et al., 2002; Kistner et al., 2005) and plant nematodes (Lohar and Bird, 2003). Recent studies have revealed that rhizobial symbiosis and RKN parasitism may involve shared pathways of transcriptional regulation (Koltai et al., 2001) and early host recognition (Weerasinghe et al., 2005). As a necessary first step to isolating and characterizing host genes involved in RKN parasitism, relevant interaction phenotypes between a collection of L. japonicus ecotypes and M. incognita were examined here.
Alhough asexual populations of RKNs are often represented as being evolutionarily inert (Trudgill and Blok, 2001; Castagnone-Sereno, 2006), molecular and laboratory studies have revealed an unexpected level of clonal diversity among populations within apomictic RKN species such as M. incognita (Castagnone-Sereno, 2006), which can rapidly adapt to new environmental constraints (e.g. host resistance), although with some fitness costs (Castagnone-Sereno et al., 1994). As genetic diversity within experimental M. incognita populations could affect the phenotype of the interactions with Lotus accessions, we performed our investigations using an M. incognita line bulked up from a single egg mass derived from a laboratory population (see Materials and Methods).
Molecular approaches have been employed to uncover putative nematode molecules used by RKNs to establish compatible interactions (Doyle and Lambert, 2002; Huang et al., 2003; Neveu et al., 2003). However, besides circumstantial evidence of host genes whose expressions are altered upon RKN infection, particularly in GCs (Gheysen and Fenoll, 2002; Jammes et al., 2005), direct genetic proof for involvement in susceptibility to RKNs is available only for a small number of plant genes (Lohar and Bird, 2003; Weerasinghe et al., 2005). All of these genes were first identified in Lotus for their altered phenotypes with interacting bacteria and fungal symbionts (Stracke et al., 2002; Radutoiu et al., 2003).
Much variability in interaction phenotypes (disease severity – galling – and nematode reproduction) was found within and between L. japonicus ecotypes with M. incognita. Variation within ecotypes could be due to segregation for the susceptibility traits evaluated. The quantitative differences in the galling observed among L. japonicus accessions ranged from high susceptibility to virtual lack of disease (Fig. 1). According to Trudgill (1991), resistance describes the effects of host genes that restrict or prevent nematode multiplication in a host species, and, as such, ecotypes in which RKN reproduction is severely impaired (such as MG-49 and MG-55) could be regarded as resistant. However, an associated hypersensitive response was not observed in these ecotypes. The lack of saturation points in the values of the interaction phenotypes tested (Figs 1 and 2) (i.e. maximum number of galls or egg masses per accession) suggests that other, as yet untested, L. japonicus ecotypes may show even higher degrees of nematode-induced disease than the accessions reported here. The high correlation shown between host susceptibilities and nematode reproduction rates among ecotypes reinforces these traits as indicators of the sequential biological processes occurring during compatible interactions between RKNs and their hosts. GCs, and surrounding galls, are triggered by the RKNs before they reproduce. However, RKN reproduction may be under the control of the same or different host gene(s) regulating host susceptibility to RKNs. Ecotypes showing a significant disparity between these values are candidates to contain an alteration in a host gene(s) uncoupling gall formation from subsequent RKN reproduction. Ecotypes MG-18, MG-19, MG-76 and MG-93 could be such candidates (Figs 1 and 2). Other abnormal interaction phenotypes such as developmental anomalies and necrosis (Fig. 3C–E, I, J) may also be under the genetic control of the host, and it will be of interest to elucidate the genetic basis behind these other phenotypes. It is tempting to hypothesize that Lotus may have evolved a mechanism(s) to dominate the parasitic relationship with RKNs, as found in Medicago and other legumes to control bacteroid differentiation (Mergaert et al., 2006).
Future work will look at the mechanisms and genetics of the relevant interaction phenotypes identified and evaluated in the present study with the goal of identifying, mapping, and isolating individual genes and/or quantitative trait loci conditioning these interactions. The ongoing programme to sequence the L. japonicus genome will be very helpful in advancing towards this aim. The extent of overlap and putative cross-talk between host transduction pathways involved in nematode parasitism and other (bacterial, mycorrhizal) Lotus symbiosis could also reveal interesting evolutionary aspects of the interactions between plants and a variety of micro-organisms.
SUPPLEMENTARY INFORMATION
A table of Supplementary Information is available online at http://aob.oxfordjournals.org/ giving the complete data set showing the number of galls and Meloidogyne incognita egg masses counted for each of the six replicate Lotus japonicus plants tested per ecotype. Each accession (MG-) is numbered after the code used by the National BioResource Project, Japan (http://www.shigen.nig.ac.jp/legume/legumebase/index.jsp). Other ecotype features such as the geographical location of collection and phenotypic characteristics of the ecotypes used in this study are available on its website. Also shown is the fresh weight of roots for all plants tested.
ACKNOWLEDGEMENTS
The National BioResource Project, Japan (http://www.nbrp.jp/index.jsp), kindly provided seeds of all L. japonicus accessions used in this study. Thanks to Graham Shephard (Rothamsted Research) for the preparation of Fig. 3. An anonymous reviewer is gratefully thanked for helpful comments. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council, UK.
LITERATURE CITED
- Bird DMcK. Signaling between nematodes and plants. Current Opinion in Plant Biology. 2004;7:372–376. doi: 10.1016/j.pbi.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Boiteux LS, Fonseca MEN, Simon PW. Host status and reaction of Arabidopsis thaliana ecotypes to infection by the northern root-knot nematode (Meloidogyne hapla) Plant Breeding. 1999;118:355–358. [Google Scholar]
- Cabrera Poch HL, Manzanilla López RH, Kanyuka K. Functionality of potato cyst nematode resistance gene Hero in leaves. Plant, Cell & Environment. 2006;29:1372–1378. doi: 10.1111/j.1365-3040.2006.01517.x. [DOI] [PubMed] [Google Scholar]
- Castagnone-Sereno P. Genetic variability and adaptive evolution in parthenogenetic root-knot nematodes. Heredity. 2006;96:282–289. doi: 10.1038/sj.hdy.6800794. [DOI] [PubMed] [Google Scholar]
- Castagnone-Sereno P, Vanlerberghe-Masutti F, Leroy F. Genetic polymorphism between and within Meloidogyne species detected with RAPD markers. Genome. 1994;37:904–909. doi: 10.1139/g94-129. [DOI] [PubMed] [Google Scholar]
- Davis EL, Hussey RS, Baum TJ, Bakker J, Schots A, Rosso MN, Abad P. Nematode parasitism genes. Annual Review of Phytopathology. 2000;38:365–396. doi: 10.1146/annurev.phyto.38.1.365. [DOI] [PubMed] [Google Scholar]
- De Mita S, Santoni S, Hochu I, Ronfort J, Bataillon T. Molecular evolution and positive selection of the symbiotic gene NORK in Medicago truncatula. Journal of Molecular Evolution. 2006;62:234–244. doi: 10.1007/s00239-004-0367-2. [DOI] [PubMed] [Google Scholar]
- Doyle EA, Lambert KN. Cloning and characterization of an esophageal-gland-specific pectate lyase from the root-knot nematode Meloidogyne javanica. Molecular Plant–Microbe Interactions. 2002;15:549–556. doi: 10.1094/MPMI.2002.15.6.549. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Fenoll C. Gene expression in nematode feeding sites. Annual Review of Phytopathology. 2002;40:191–219. doi: 10.1146/annurev.phyto.40.121201.093719. [DOI] [PubMed] [Google Scholar]
- Goverse A, de Almeida Engler J, Verhees J, van der Krol S, Helder J, Gheysen G. Cell cycle activation by plant parasitic nematodes. Plant Molecular Biology. 2000;43:747–761. doi: 10.1023/a:1006367126077. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant Journal. 1992;2:487–496. [Google Scholar]
- Huang G, Gao B, Maier T, Allen R, Davis EL, Baum TJ, Hussey RS. A profile of putative parasitism genes expressed in the esophageal gland cells of the root-knot nematode Meloidogyne incognita. Molecular Plant–Microbe Interactions. 2003;16:376–381. doi: 10.1094/MPMI.2003.16.5.376. [DOI] [PubMed] [Google Scholar]
- Jammes F, Lecomte P, de Almeida-Engler J, Bitton F, Martin-Magniette ML, Renous JP, et al. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant Journal. 2005;44:447–458. doi: 10.1111/j.1365-313X.2005.02532.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Motomura T, Imaizumi-Anraku H, Akao S, Kawasaki S. Providing the basis for genomics in Lotus japonicus: the accessions Miyakojima and Gifu are appropriate crossing partners for genetic analysis. Molecular Genetics and Genomics. 2001;266:157–166. doi: 10.1007/s004380100540. [DOI] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, et al. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. The Plant Cell. 2005;17:2217–2229. doi: 10.1105/tpc.105.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H, Dhandaydham M, Opperman C, Thomas J, Bird DMcK. Overlapping plant signal transduction pathways induced by a parasitic-nematode and a rhizobial endosymbiont. Molecular Plant–Microbe Interactions. 2001;14:1168–1177. doi: 10.1094/MPMI.2001.14.10.1168. [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420:422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- Lohar DP, Bird DMcK. Lotus japonicus: a new model to study root-parasitic nematodes. Plant and Cell Physiology. 2003;44:1176–1184. doi: 10.1093/pcp/pcg146. [DOI] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proceedings of the National Academy of Sciences of the USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu C, Jaubert S, Abad P, Castagnone-Sereno P. A set of genes differentially expressed between avirulent and virulent Meloidogyne incognita near-isogenic lines encode secreted proteins. Molecular Plant–Microbe Interactions. 2003;16:1077–1084. doi: 10.1094/MPMI.2003.16.12.1077. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, et al. Har1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Skriver K. Ligand mimicry? Plant-parasitic nematode polypeptide with similarity to CLAVATA3. Trends in Plant Sciences. 2003;8:55–57. doi: 10.1016/S1360-1385(03)00003-7. [DOI] [PubMed] [Google Scholar]
- Patterson HD, Williams ER. A new class of resolvable incomplete block designs. Biometrika. 1976;63:83–92. [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Roberts PA. Current status of the availability, development, and use of host plant resistance to nematodes. Journal of Nematology. 1992;24:213–227. [PMC free article] [PubMed] [Google Scholar]
- Sandal N, Petersen TR, Murray J, Umehara Y, Karas B, Yano K, et al. Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Molecular Plant–Microbe Interactions. 2006;19:80–91. doi: 10.1094/MPMI-19-0080. [DOI] [PubMed] [Google Scholar]
- Sasser JN, Freckman DW. A world perspective on nematology: the role of the society. In: Veech JA, Dickson DW, editors. Vistas on nematology. Hyattsville, MD, USA: Society of Nematology; 1987. pp. 7–14. [Google Scholar]
- Sato S, Tabata S. Lotus japonicus as a platform for legume research. Current Opinion in Plant Biology. 2006;9:128–132. doi: 10.1016/j.pbi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Tirichine L, de Billy F, Huguet T. Mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago truncatula. Plant Physiology. 2000;123:845–851. doi: 10.1104/pp.123.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudgill DL. Resistance to and tolerance of plant parasitic nematodes in plants. Annual Review of Phytopathology. 1991;29:167–192. [Google Scholar]
- Trudgill DL, Blok VC. Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annual Review of Phytopathology. 2001;39:53–77. doi: 10.1146/annurev.phyto.39.1.53. [DOI] [PubMed] [Google Scholar]
- Weerasinghe RR, Bird DMcK, Allen NS. Root-knot nematodes and bacterial Nod factors elicit common signal transduction events in Lotus japonicus. Proceedings of the National Academy of Sciences of the USA. 2005;102:3147–3152. doi: 10.1073/pnas.0407926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Gleason CA. Plant–nematode interactions. Current Opinion in Plant Biology. 2003;6:327–333. doi: 10.1016/s1369-5266(03)00059-1. [DOI] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, de Bruijn FJ, et al. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant Journal. 2000;23:97–114. doi: 10.1046/j.1365-313x.2000.00799.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.