Abstract
Eukaryotic origins of replication are selected by loading a head-to-head double hexamer of the Mcm2–7 replicative helicase around origin DNA. Cdt1 plays an essential but transient role during this event; however, its mechanism of action is unknown. Through analysis of Cdt1 mutations, we demonstrate that Cdt1 performs multiple functions during helicase loading. The C-terminus of Cdt1 binds Mcm2–7, and this interaction is required for efficient origin recruitment of both proteins. We show that origin recognition complex (ORC) and Cdc6 recruit multiple Cdt1 molecules to the origin during helicase loading, and disruption of this multi-Cdt1 intermediate prevents helicase loading. Although dispensable for loading Mcm2–7 double hexamers that are topologically linked to DNA, the essential N-terminal domain of Cdt1 is required to load Mcm2–7 complexes that are competent for association with the Cdc45 and GINS helicase-activating proteins and replication initiation. Our data support a model in which origin-bound ORC and Cdc6 recruit two Cdt1 molecules to initiate double-hexamer formation prior to helicase loading and demonstrate that Cdt1 influences the replication competence of loaded Mcm2–7 helicases.
Keywords: Cdc45, Cdt1, GINS, helicase, pre-replicative complex (pre-RC)
Introduction
Eukaryotic cells duplicate their genome in a timely fashion by initiating DNA replication from multiple genomic sites known as origins of replication. At each origin, DNA replication commences bi-directionally via a pair of replication forks. The establishment of bi-directional replication forks requires the DNA unwinding activity of the replicative DNA helicase, Mcm2–7 (Aparicio et al, 1997; Labib et al, 2000; Ying and Gautier, 2005; Ilves et al, 2010). In addition to this essential function, the loading of the Mcm2–7 helicase onto DNA (in G1 phase) and its subsequent activation (in S phase) are tightly regulated to ensure once-and-only-once initiation from eukaryotic origins of replication (Arias and Walter, 2007).
Helicase loading, also referred to as pre-replicative complex (pre-RC) formation or origin licensing, is nucleated by DNA-bound origin recognition complex (ORC). As cells enter G1, ORC bound to origins of replication recruits Cdc6 followed by Cdt1 and the Mcm2–7 helicase. These proteins load Mcm2–7 in an inactive form onto all potential origins of replication (Remus and Diffley, 2009). During this process, Cdt1 transiently interacts with the origin-bound ORC/Cdc6 complex but is released after Cdc6 ATP hydrolysis and Mcm2–7 loading (Randell et al, 2006). Notably, in vitro studies found that the Mcm2–7 helicase is loaded as a head-to-head double hexamer with dsDNA running through a central channel, but only hexameric Mcm2–7 complexes are observed in solution (Evrin et al, 2009; Remus et al, 2009; Gambus et al, 2011). These findings suggest that two Mcm2–7 hexamers are loaded in a coordinated process (Remus et al, 2009). The anti-parallel orientation of the Mcm2–7 hexamers within the double hexamer is proposed to be critical to establish bi-directional replication forks.
Because both the origin of replication (Bell, 1995) and ORC (Lee and Bell, 1997; Clarey et al, 2006; Chen et al, 2008) lack obvious symmetry, it is unclear how they direct the assembly of the symmetric Mcm2–7 double hexamer. One possibility is that two ORC molecules bind the origin in opposite orientations to coordinately load the head-to-head double hexamer. Another possibility is that one ORC molecule sequentially recruits and loads Mcm2–7 hexamers in opposite orientations. A third possibility is that a single ORC molecule directs the formation of the double hexamer by simultaneously recruiting and loading two Mcm2–7 molecules onto the origin DNA.
During late G1 and S phase, the activity of Dbf4-dependent Cdc7 kinase (DDK) and S-phase cyclin-dependent kinase (S-CDK) stimulate a subset of loaded Mcm2–7 double hexamers to initiate DNA unwinding and replisome assembly (Labib, 2010). In Saccharomyces cerevisiae, phosphorylation of the Mcm4 and Mcm6 subunits by DDK is required to alleviate the inhibitory function of the Mcm4 N-terminus (Randell et al, 2010; Sheu and Stillman, 2010). Beginning in late G1, DDK phosphorylation of Mcm2–7 is required for Cdc45 and Sld3 to associate with Mcm2–7 (Sheu and Stillman, 2006; Heller et al, 2011). Upon entry into S phase, phosphorylation of the replication initiation factors Sld2 and Sld3 by S-CDK is required to recruit a second helicase-activating protein, the GINS complex (Tanaka et al, 2007; Zegerman and Diffley, 2007; Muramatsu et al, 2010; Heller et al, 2011).
Although Cdt1 is essential for the loading of Mcm2–7 onto DNA, its function during the helicase-loading process is unknown. Studies of Xenopus Cdt1 identified the C-terminal two-thirds of the protein as required for helicase loading and the last 150 amino acids bound to a subset of the Mcm2–7 complex (Ferenbach et al, 2005). In addition, studies of mammalian Cdt1 have identified its C-terminus as mediating Mcm2–7 binding (Yanagi et al, 2002; Teer and Dutta, 2008; You and Masai, 2008; Jee et al, 2010). In S. cerevisiae, the only well-defined role of Cdt1 in helicase loading is its formation of a heptameric complex with Mcm2–7, which is necessary for the nuclear import of both proteins (Tanaka and Diffley, 2002). A role for budding yeast Cdt1 during the initial recruitment of Mcm2–7 to the origin is supported by direct interactions between Cdt1 and ORC (Semple et al, 2006; Asano et al, 2007; Chen et al, 2007; Chen and Bell, 2011). Cdt1 also could position the recruited helicase in the proper orientation for loading or assist in the opening or closing of the Mcm2–7 ring that is necessary to load the ring-shaped Mcm2–7 around the origin DNA.
In this study, we investigate Cdt1 function during helicase loading. We show that the formation of the Cdt1/Mcm2–7 complex drives the recruitment of both proteins to ORC- and Cdc6-bound origin DNA. We identify a multi-Cdt1 intermediate during helicase loading and show that a Cdt1 mutation that only allows recruitment of one Cdt1 to the origin prevents helicase loading. Intriguingly, we find that the essential N-terminus of Cdt1 is dispensable for Cdt1 to bind Mcm2–7, to be recruited to the origin, and to load Mcm2–7 double hexamers. Instead, Mcm2–7 complexes loaded by Cdt1 lacking its N-terminus fail to associate with the helicase-activating proteins Cdc45 and GINS. Our findings demonstrate that Cdt1 plays multiple roles during the helicase-loading process, facilitating the origin recruitment of a pair of Mcm2–7 complexes for subsequent loading and ensuring that loaded Mcm2–7 complexes are capable of replication initiation.
Results
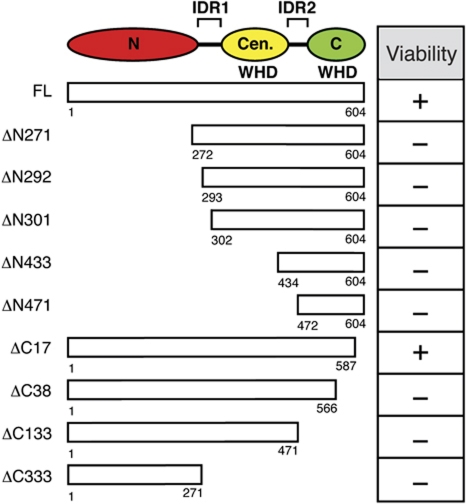
To investigate S. cerevisiae Cdt1 (unless otherwise noted, hereafter Cdt1 refers to the S. cerevisiae protein) function, we constructed a series of N- and C-terminal deletions based on structure-based profile–profile alignment (HHpred; Soding et al, 2005) and secondary structure prediction (Jpred3; Cole et al, 2008) tools. These analyses predicted three domains for Cdt1: an N-terminal domain (a.a. 11–272) as well as a central (a.a. 310–435) and C-terminal domain (a.a. 500–602), both of which are predicted to adopt a winged-helix domain (WHD) fold as observed for metazoan Cdt1 (Lee et al, 2004; Khayrutdinov et al, 2009; Jee et al, 2010). Inter-domain regions are predicted to separate the N-terminal from the central domain (IDR1) and the central from the C-terminal domain (IDR2) (Figure 1).
Figure 1.
In vivo complementation analysis of Cdt1-deletion mutants. (Top) Diagram of Cdt1 structural domains predicted by HHpred analysis. Cdt1 is predicted to contain three discrete domains (N-terminal, central, and C-terminal) and two inter-domain regions (IDR1 and IDR2). The central and C-terminal domains are predicted to fold into a WHD. (Left) Schematic representation of N- and C-terminal Cdt1-deletion constructs. End points of each construct are indicated. (Right) Complementation analysis of cdt1 mutants. Each cdt1 mutant was placed under its native promoter and tested for complementation of cell proliferation in a yeast strain lacking CDT1.
We first investigated the regions of Cdt1 that are required for its function in vivo. To this end, we assessed the ability of each of the Cdt1 mutations expressed from the endogenous Cdt1 promoter to complement a full deletion of the CDT1 gene. We observed that all three predicted domains of Cdt1 were indispensable in vivo. Only the smallest C-terminal deletion mutant (ΔC17 Cdt1) complemented a CDT1 deletion (Figure 1; Supplementary Figure S1A).
The N-terminal domain of human Cdt1 contains a nuclear localization signal (NLS) that is critical for its nuclear import and function (Nishitani et al, 2004). Consistent with Cdt1 nuclear localization being mediated through binding to Mcm2–7 (Tanaka and Diffley, 2002), we did not identify an NLS motif within the Cdt1-coding region. Nevertheless, we asked whether the addition of the SV-40 NLS to the N-terminal deletion mutants restored complementation. In all cases, this modification did not change the ability of the mutant to complement a CDT1 deletion (Supplementary Figure S1B).
Cdt1 origin recruitment requires IDR1 and the central domain
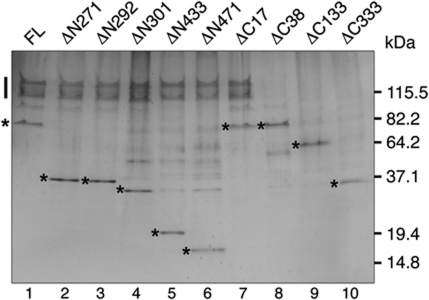
Nuclear accumulation of Cdt1 requires its interaction with the Mcm2–7 helicase, and neither protein is competent for nuclear entry alone (Tanaka and Diffley, 2002). Although the C-terminus of metazoan Cdt1 is critical for its interaction with Mcm2–7 (Yanagi et al, 2002; Ferenbach et al, 2005; Teer and Dutta, 2008; You and Masai, 2008), a Mcm2–7-binding site has not been identified in S. cerevisiae Cdt1. To identify this region in Cdt1, we generated yeast strains that overexpressed all six Mcm proteins and each Cdt1-deletion mutant fused to a C-terminal FLAG epitope. Cdt1 was immunoprecipitated from G1 phase-arrested extracts, and all immunoprecipitated proteins were examined by silver staining (Figure 2). Full-length Cdt1 and each N-terminal deletion co-precipitated approximately equimolar amounts of Mcm2–7. In contrast, only the smallest C-terminal truncation mutant (ΔC17) co-precipitated detectable Mcm2–7. These data demonstrate that the Mcm2–7-binding domain is contained within the C-terminal 133 residues (a.a. 472–604) of Cdt1.
Figure 2.
The C-terminal domain of Cdt1 binds Mcm2–7. FLAG immunoprecipitations of extracts derived from G1-arrested strains overexpressing all six Mcm proteins and the indicated FLAG-tagged Cdt1 constructs were analysed by 4–20% SDS–PAGE separation followed by silver staining to allow visualization of Mcm2–7 subunits and the Cdt1-deletion mutant proteins. The location of each Cdt1 protein is indicated with an * and, when present, a bar indicates the location of the Mcm2–7 proteins. The additional proteins between Cdt1 and Mcm2–7 in lanes 4–6 are contaminants that do not cross-react with Mcm2–7 antibodies.
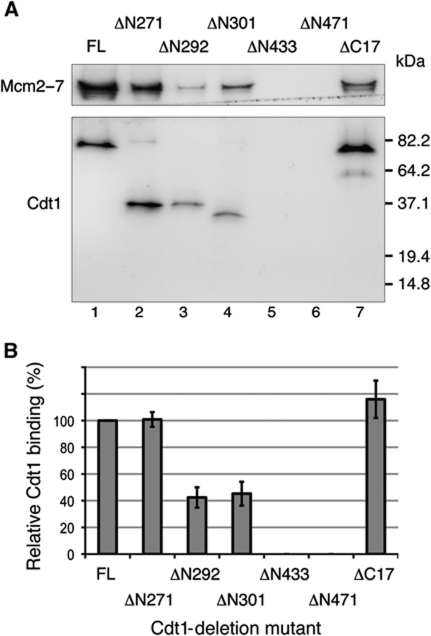
We used a reconstituted helicase-loading assay (Seki and Diffley, 2000; Evrin et al, 2009; Remus et al, 2009; Tsakraklides and Bell, 2010) to identify regions of Cdt1 that are required for the initial recruitment of the Cdt1 to the origin. To detect the origin recruitment of Cdt1, we added ATPγS to the assays to arrest the helicase-loading process prior to ATP hydrolysis by Cdc6. Under these conditions, Cdt1 and Mcm2–7 stably associate with origin DNA, but Mcm2–7 loading is inhibited (Randell et al, 2006). Deletion of the entire N-terminal domain (ΔN271) or the C-terminal 17 residues of Cdt1 did not affect Cdt1 origin recruitment (Figure 3A, compare lanes 1, 2, and 7). In contrast, deletion of the central domain of Cdt1 (ΔN433 and ΔN471) eliminated Cdt1 and Mcm2–7 recruitment (Figure 3A, lanes 5 and 6). Notably, two N-terminal deletions that removed the N-terminal domain and IDR1 (ΔN292 and ΔN301) consistently reduced Cdt1 origin association by approximately half (Figure 3B) and Mcm2–7 recruitment even more extensively. These data suggest that both IDR1 and the central domain contribute to origin recruitment of Cdt1 and Mcm2–7.
Figure 3.
IDR1 and the central domain of Cdt1 are required for full Cdt1 origin recruitment. (A) Reconstituted Cdt1 recruitment assays were performed on 4 pmol of 1039 bp ARS1 DNA using 12 pmol of purified ORC, Cdc6, and the indicated Cdt1/Mcm2–7 in the presence of ATPγS. Assembly was allowed to proceed for 20 min at 25 °C and origin-associated proteins were analysed by 4–20% SDS–PAGE separation followed by immunoblotting with anti-Mcm2–7 (UM174) and anti-FLAG (Sigma) antibodies to detect Cdt1-FLAG. Note that the use of a 4–20% SDS–PAGE gel results in the detection of only one (or two, lane 7) Mcm2–7 band(s) by immunoblot analysis (the polyclonal antibody does not recognize all Mcm2–7 subunits equally). (B) Quantification of Cdt1 origin association, expressed as a percentage relative to full-length Cdt1 origin recruitment. Error bars represent the standard deviation of three independent experiments.
Cdt1/Mcm2–7 complex formation is necessary for their recruitment to the origin
To test the Mcm2–7 loading function of the three C-terminal deletion mutants that were unable to bind the Mcm2–7 complex (Figure 2), we used an extract-based helicase-loading assay (Randell et al, 2006). We used immunodepletion to eliminate endogenous Cdt1 from a G1 phase-arrested helicase-loading extract (Randell et al, 2006). Immunodepletion of Cdt1 did not result in co-depletion of Mcm2–7 (Supplementary Figure S2A) due to excess Mcm2–7 in the extract. Consistent with previous studies (Randell et al, 2006), the Cdt1-depleted helicase-loading extract showed no residual Cdt1 origin recruitment in the presence of ATPγS (Supplementary Figure S2B, compare lanes 2 and 4) or Mcm2–7 loading in the presence of ATP (Supplementary Figure S2D, lanes 1 and 2). Addition of full-length purified Cdt1 to the depleted extract restored Cdt1 origin recruitment (Supplementary Figure S2C) and Mcm2–7 origin loading (Supplementary Figure S2D) to the Cdt1-depleted assembly extract. In contrast, Cdt1-deletion mutants compromised for Mcm2–7 binding (ΔC38, ΔC133, ΔC333) could not support either function (Supplementary Figure S2C and D). These data suggest that, in addition to possessing an intact IDR1 and central domain, Cdt1 must bind to Mcm2–7 to support helicase recruitment and loading.
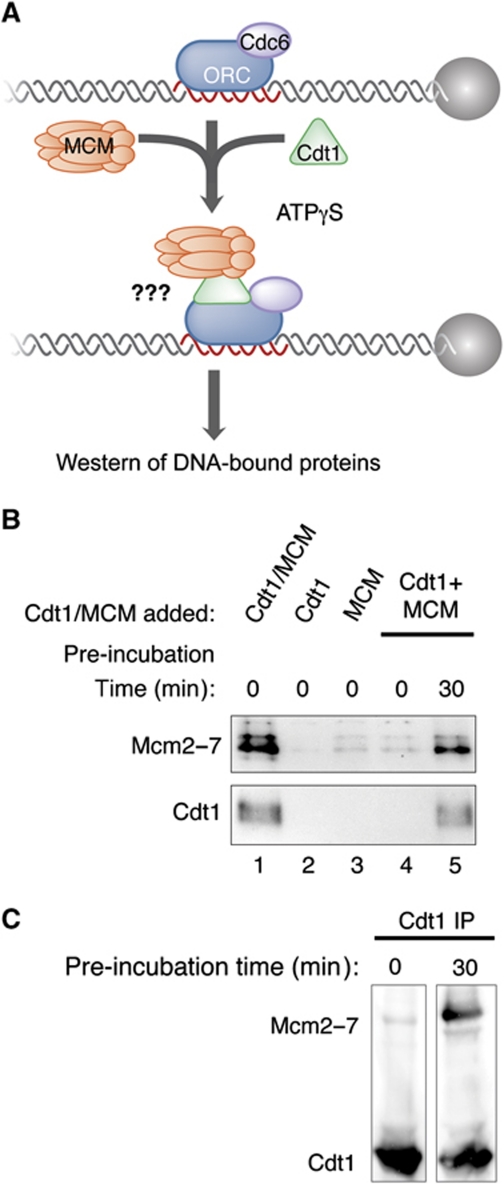
To test directly whether Cdt1/Mcm2–7 complex formation was critical for the recruitment of either component to the origin, we used the reconstituted Cdt1 origin recruitment assay to allow separate addition of Mcm2–7 and Cdt1 (Figure 4A). We found that neither Cdt1 nor Mcm2–7 showed robust origin association in the absence of the other protein (Figure 4B, lanes 2 and 3). Simultaneous addition of both Cdt1 and Mcm2–7 to the reaction did not enhance the levels of Cdt1 or Mcm2–7 recruitment (Figure 4B, lane 4). In contrast, if Cdt1 and Mcm2–7 were pre-incubated for 30 min before their addition to the helicase-loading assay, Cdt1 and Mcm2–7 recruitment was restored (Figure 4B, lane 5). Importantly, when Cdt1 is immunoprecipitated from Cdt1 and Mcm2–7 mixtures with or without a 30-min pre-incubation, the extent of Cdt1/Mcm2–7 complex formation correlated with their extent of origin recruitment (Figure 4C). Thus, formation of the Cdt1/Mcm2–7 complex is critical for the initial recruitment of either protein to the origin.
Figure 4.
Cdt1/Mcm2–7 complex formation is required for recruitment of either component to the origin. (A) Experimental outline for (B). ORC and Cdc6 were incubated with ARS1 DNA in the presence of ATPγS and supplemented with purified Cdt1/Mcm2–7 complex, Cdt1, Mcm2–7, or Cdt1 and Mcm2–7 that was or was not pre-incubated with each other in H/300 buffer for 30 min at 25 °C prior to their addition. Assays were performed using 1 pmol ARS1 DNA and 4 pmol of each pre-RC component. (B) Pre-incubation of Cdt1 and Mcm2–7 supports their origin recruitment. The experiment was performed as described in (A). DNA–protein complexes were isolated and analysed by immunoblot with anti-Mcm2–7 (UM174) and anti-Cdt1 (HM5352) antibodies. (C) Pre-incubation of Cdt1 and Mcm2–7 results in Cdt1/Mcm2–7 complex formation. In all, 8 pmol of purified Cdt1 and Mcm2–7 were incubated in H/300 buffer for 0 or 30 min at 25 °C. After the indicated pre-incubation time, half of the Cdt1 and Mcm2–7 pre-incubation mixture was incubated with anti-Cdt1 (HM5352) antisera coupled to beads for 20 min at 4 °C. The other half was utilized as described in (A). Immunoprecipitated fractions were collected following low-salt wash and analysed by immunoblot as in (B).
Multiple Cdt1 molecules assemble at the origin to load Mcm2–7
The origin recruitment studies demonstrate that Cdt1 plays a critical role in escorting the Mcm2–7 helicase to the origin. If IDR1 and the central domain of Cdt1 constitute one ORC/Cdc6-binding site, a single Mcm2–7 hexamer would be initially recruited to the origin. Alternatively, if IDR1 and the central domain of Cdt1 function as two independent ORC/Cdc6-binding sites, this would allow the initial recruitment of two Mcm2–7 hexamers to the origin. Consistent with the latter model, the Orc6 protein contains two essential, independent Cdt1-binding sites (Chen et al, 2007; Chen and Bell, 2011). The latter model also predicts that deletion of IDR1 would eliminate one of two Cdt1-binding sites, explaining the ∼50% reduction in Cdt1 origin recruitment observed for the ΔN292 and ΔN301 mutants (Figure 3B). To distinguish between these models, we asked if more than one Cdt1 molecule is recruited to the origin.
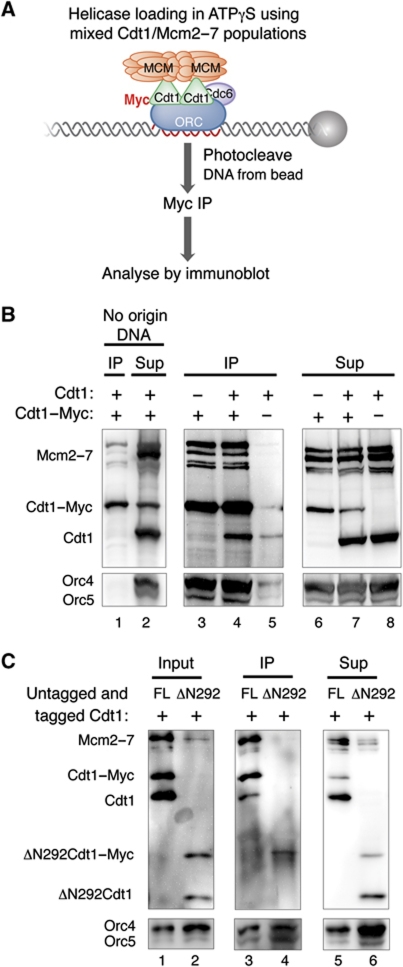
To determine whether one or multiple Cdt1 molecules assemble at the origin during helicase loading, we performed reconstituted Cdt1 recruitment assays with a mixture of untagged and Myc-tagged Cdt1 in complex with Mcm2–7. Myc-tagged Cdt1 bound Mcm2–7 (Supplementary Figure S3A), loaded Mcm2–7 (Supplementary Figure S3B), and was recruited to the origin DNA (Supplementary Figure S3C). Importantly, tagged and untagged Cdt1 were equally effective at mediating Cdt1 and Mcm2–7 origin recruitment (Supplementary Figure S3D). After performing the assembly reaction in the presence of ATPγS, we released DNA-bound intermediates from the beads and performed an immunoprecipitation using anti-Myc antibody (Figure 5A). Consistent with the formation of a multi-Cdt1 ATPγS-stabilized intermediate, untagged Cdt1 was present in the anti-Myc immunoprecipitate when both tagged and untagged Cdt1 were used in the recruitment assay (Figure 5B, lane 4). Importantly, the observed co-immunoprecipitation of untagged Cdt1 and ORC with Myc-tagged Cdt1 required the presence of origin DNA. In the absence of origin DNA, only Mcm proteins were associated with tagged Cdt1 (Figure 5B, lanes 1 and 2). These findings indicate that co-immunoprecipitation required formation of the ATPγS-stabilized intermediate on origin DNA and was not the result of protein–protein interactions off the DNA. We conclude that multiple molecules of Cdt1 are recruited to the origin during helicase loading.
Figure 5.
Multiple Cdt1 molecules associate with the origin to promote Mcm2–7 loading. (A) Experimental outline for (B). Fully reconstituted Cdt1 recruitment assays were performed using purified ORC, Cdc6, and Cdt1/Mcm2–7 in the presence of ATPγS. When tagged and untagged Cdt1/Mcm2–7 species were mixed, equimolar amounts of each species were used. Assembled complexes were washed, DNA and associated proteins were released from beads, and incubated with anti-Myc antibody coupled to beads. Immunoprecipitated fractions were separated from supernatant (Sup) fractions, and both fractions were TCA precipitated and analysed by immunoblotting with anti-Mcm2–7 (UM174), anti-Cdt1 (HM5352), and anti-ORC (1108) antibodies. (B) Helicase loading occurs via a multi-Cdt1 helicase-loading intermediate. The experiment was performed as outlined in (A). The ‘no-origin DNA’ control was performed with 8 pmol of all pre-RC components in the absence of ARS1 DNA. (C) IDR1 is essential for formation of a multi-Cdt1 intermediate. The experiment was performed as described in (A), except that tagged and untagged ΔN292 Cdt1/Mcm2–7 was substituted for full-length Cdt1/Mcm2–7 complexes in lanes 2, 4, and 6. All reactions contain equimolar amounts of tagged and untagged Cdt1 or ΔN292 Cdt1 as indicated.
The recruitment of multiple Cdt1 molecules to the origin could be achieved by two distinct means: (1) a single origin-bound ORC/Cdc6 complex interacting with multiple Cdt1 molecules or (2) one Cdt1 interacting with each of multiple ORC/Cdc6 complexes bound to a single origin. We distinguished between these models using two approaches. First, we used an analogous approach to that described above for Cdt1 to determine if multiple ORC molecules were associated with the origin DNA. In this case, we used ORC including either full-length Orc1-FLAG or untagged ΔN235 Orc1. This mutation removes the N-terminal BAH domain of Orc1 but is not required for Mcm2–7 recruitment (Supplementary Figure S4A, lane 3) and fully complements an ORC1 deletion (Bell et al, 1995). As expected for a single ORC bound to the origin, we found that only the tagged Orc1 subunit is detected after Mcm2–7 recruitment and anti-FLAG immunoprecipitation (Supplementary Figure S4A, lane 5). Although the ARS1 origin contains only one high-affinity ORC-binding site, the B2 element of ARS1 has the potential to accommodate a second ORC molecule in opposite orientation (Wilmes and Bell, 2002). To test if a second ORC molecule bound to B2 mediated the formation of a multi-Cdt1 helicase-loading intermediate, we performed the Cdt1 immunoprecipitation experiment described above using an ARS1 template lacking B2. As in the experiment using wild-type ARS1 template, untagged Cdt1 was co-precipitated with tagged Cdt1 (Supplementary Figure S4B). Together, these data indicate that a single ORC is associated with the origin DNA during the formation of the multi-Cdt1, helicase-loading intermediate.
If IDR1 and the central domain of Cdt1 function as two independent ORC/Cdc6-binding sites, then disrupting IDR1 should eliminate formation of the multi-Cdt1 intermediate and co-precipitation of untagged Cdt1. To test this possibility, we performed the same immunoprecipitation assay using wild-type ARS1 DNA template with tagged and untagged IDR1-deletion mutant ΔN292 Cdt1/Mcm2–7. In contrast to full-length Cdt1, we did not observe co-precipitation of untagged ΔN292 Cdt1 with Myc-tagged ΔN292 Cdt1 (Figure 5C, lane 4). This is not due to a lack of untagged ΔN292 Cdt1 in the sample prior to the immunoprecipitation. Both tagged and untagged ΔN292 Cdt1 are recruited to the origin DNA with equal efficiency (Figure 5C, lane 2; ‘input’ represents proteins recruited to the origin DNA prior to anti-Myc immunoprecipitation). Disruption of IDR1 also resulted in reduced Mcm2–7 origin recruitment (Figure 5C, compare lanes 1 and 2), suggesting that recruitment of multiple Cdt1 molecules enhances Mcm2–7 origin recruitment. Therefore, we suggest that Cdt1 possesses two distinct ORC/Cdc6-binding sites, one of which is impaired by disrupting IDR1.
Our previous studies showed that CDK phosphorylation inactivated the N-terminal Cdt1-binding site in Orc6 but had no effect on the C-terminal binding site (Chen and Bell, 2011). To address whether the Cdt1 and Mcm2–7 binding observed in the absence of IDR1 is sensitive to S-CDK (Clb5–Cdc28) phosphorylation of ORC, we compared Cdt1 and Mcm2–7 origin recruitment in reconstituted assays using either S-CDK-phosphorylated or unmodified ORC (Supplementary Figure S5). As observed previously, S-CDK phosphorylation of ORC resulted in an ∼50% reduction of wild-type Cdt1 recruitment (Supplementary Figure S5A). In contrast, when ΔN292 Cdt1 was used, Cdt1 and Mcm2–7 levels were reduced >10-fold (Supplementary Figure S5B). This finding suggests that ΔN292 Cdt1 retains the ability to associate with the N-terminal Orc6-binding site but has lost binding to the C-terminal binding site.
Mcm2–7 helicase loading requires normal levels of Cdt1 origin recruitment
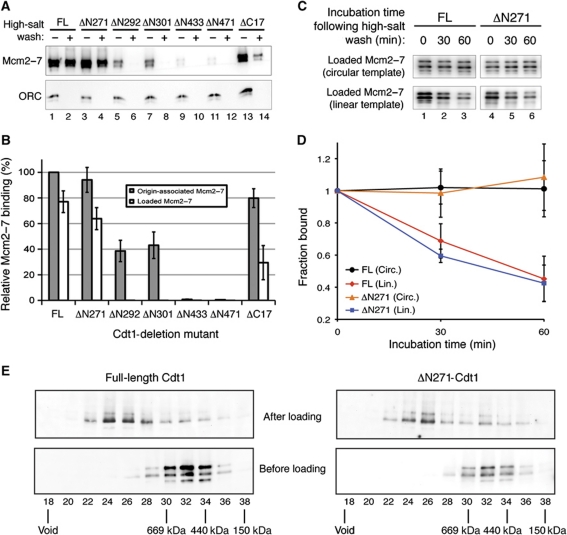
The recruitment of two Cdt1 molecules to the origin by a single ORC molecule suggests a mechanism for the coordinated loading of two Mcm2–7 hexamers (Remus et al, 2009). To address whether Cdt1 functions after the initial recruitment of Mcm2–7 hexamers to the origin, we tested all of the Cdt1 deletions that bound Mcm2–7 for the ability to load Mcm2–7 onto origin DNA (Figure 6A and B). These assays replaced ATPγS with ATP to support nucleotide hydrolysis and helicase loading, and high-salt washes distinguished loaded Mcm2–7 complexes from associated complexes (Donovan et al, 1997; Rowles et al, 1999; Bowers et al, 2004). Both the ΔN271 and ΔC17 Cdt1 mutants, which displayed no defect in their origin recruitment, supported Mcm2–7 loading (Figure 6A, lanes 4 and 14), although ΔC17 Cdt1 showed a partial defect (Figure 6A, compare lanes 2 and 14). In contrast, each of the Cdt1 mutations that was defective for the initial association of Cdt1/Mcm2–7 was also defective for Mcm2–7 loading. Consistent with their complete loss of origin recruitment, the two Cdt1-deletion mutants lacking the central domain (ΔN433 and ΔN471) showed no Mcm2–7 loading (Figure 6A, lanes 9–12). Intriguingly, the two Cdt1-deletion mutants that disrupt IDR1 (ΔN292 and ΔN301) and showed a two-fold reduction in their origin recruitment (Figure 3) exhibited a partial defect in Mcm2–7 recruitment but a complete defect in helicase loading (Figure 6A, lanes 5–8). This lack of correlation between the level of initial Cdt1 and Mcm2–7 origin recruitment and level of subsequent Mcm2–7 loading indicates that Cdt1 function during helicase loading is not limited to the initial recruitment of Mcm2–7 to the origin.
Figure 6.
The N-terminal domain of Cdt1 is not required to load topologically linked Mcm2–7 double hexamers. (A) Reconstituted helicase-loading assays were performed on 1 pmol ARS1 DNA using 4 pmol of ORC, Cdc6, and indicated Cdt1/Mcm2–7 in the presence of ATP. Assembled pre-RCs were washed with H buffer containing 300 mM KGlut (low-salt wash) or 500 mM NaCl (high-salt wash) to distinguish loaded Mcm2–7 complexes. DNA–protein complexes were analysed by 4–20% SDS–PAGE followed by immunoblotting with anti-Mcm2–7 (UM174) and anti-Orc4 (SB6; Chen et al, 2007) antibodies. (B) Quantification of Mcm2–7 origin association (low-salt wash) and loading (high-salt wash) for indicated Cdt1 proteins, expressed as a percentage relative to levels supported by full-length Cdt1. Error bars represent the standard deviation of three independent experiments. (C) Time course analysis of ΔN271 Cdt1-loaded Mcm2–7 and full-length Cdt1-loaded Mcm2–7 complexes on circular and linear DNA templates. Fully reconstituted helicase-loading assays were performed on 175 fmol pUC19 ARS1 wild-type circular DNA (top) using 2 pmol purified ORC, Cdc6, and indicated Cdt1/Mcm2–7 or 500 fmol 1 kb linear ARS1 DNA (bottom) using 1 pmol purified pre-RC components in the presence of ATP and H buffer containing 300 mM KGlut (H/300). Assembled pre-RCs were washed with H buffer containing 500 mM NaCl (high-salt wash) to isolate loaded Mcm2–7 complexes. The resulting loaded Mcm2–7 was incubated in H/300 for the indicated amounts of time at 25 °C. DNA–protein complexes were analysed by 8% SDS–PAGE followed by immunoblotting with anti-Mcm2–7 (UM174) antibody. (D) Quantification of Mcm2–7 western blot signals. Fraction bound reflects amount of Mcm2–7 at a given time point (30 or 60 min) relative to amount of Mcm2–7 with no incubation time (0 min) for a given DNA template and Cdt1 protein. Error bars represent the standard deviation of three independent experiments. (E) Gel filtration analysis of Mcm2–7 complexes before and after loading. (Top) Mcm2–7 loading assays were performed with Cdt1-depleted, G1-arrested whole-cell extracts complemented with the indicated Cdt1/Mcm2–7 complex. The resulting complexes were washed with high salt, released from the DNA by DNAse I digestion and separated on a Superose 6 gel filtration column. (Bottom) The indicated purified Cdt1/Mcm2–7 complexes (500–750 ng) were fractionated on a Superose 6 column. The void volume and peak fraction of protein standard elution are indicated.
Unlike all other lethal deletion mutations, the ΔN271 Cdt1 deletion mutant supported wild-type levels of Mcm2–7 loading as measured by salt-resistance (Figure 6B). To test more robustly if the Mcm2–7 complexes loaded by ΔN271 Cdt1 were properly loaded, we asked if these complexes were topologically linked to the DNA template and in the form of a double hexamer. We compared residence times of Mcm2–7 complexes loaded by ΔN271 Cdt1 onto either a circular or a linear ARS1 DNA template (Remus et al, 2009). Consistent with topological linkage, Mcm2–7 loaded by full-length or ΔN271 Cdt1 each showed the same reduced half-lives on the linear DNA templates (Figure 6C and D). We tested double-hexamer formation using gel filtration chromatography (Evrin et al, 2009). In contrast to free Mcm2–7 complexes, Mcm2–7 complexes assembled by either wild-type or ΔN271 Cdt1 primarily migrated as a double hexamer (Figure 6E).
Mcm2–7 helicases loaded by ΔN271 Cdt1 are defective for replication initiation
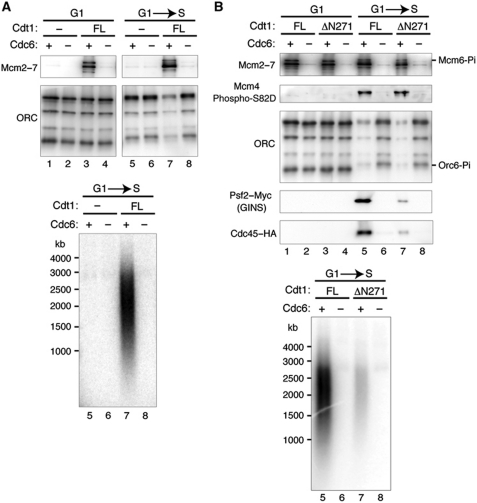
Our data indicate that the N-terminus of Cdt1 is dispensable for Mcm2–7 binding, origin recruitment, and loading of topologically linked, double-hexameric Mcm2–7; however, this mutation is unable to complement a CDT1 deletion (Figure 1). Therefore, we asked if the N-terminus is required for the downstream events of replication initiation using a recently developed assay for origin-dependent in vitro replication initiation that supports helicase activation, replisome assembly, and DNA replication initiation and elongation (Heller et al, 2011). This assay recapitulates the G1-to-S phase transition in a three-step process: (1) Mcm2–7 is loaded onto immobilized ARS1-containing DNA using a G1 phase-arrested extract; (2) loaded Mcm2–7 is treated with purified DDK; and (3) Mcm2–7 is activated by subsequent incubation with a S-phase extract.
To test the ability of ΔN271 Cdt1 to load replication-competent Mcm2–7, we immunodepleted endogenous Cdt1 from the G1 extract and added back purified Cdt1/Mcm2–7 complexes to support helicase loading before exposure to DDK and S-phase extract. Cdt1-depleted G1 extract was unable to support helicase loading (Figure 7A, top, lanes 1 and 2 and lanes 5 and 6) or replication elongation as analysed by alkaline gel electrophoresis of radiolabelled replication products (Figure 7A, bottom, lanes 5 and 6). Addition of full-length Cdt1/Mcm2–7 to the Cdt1-depleted G1 extract resulted in Cdc6-dependent Mcm2–7 loading and replication initiation (Figure 7A, top, lanes 3 and 4 and lanes 7 and 8; bottom, lanes 7 and 8). As expected, addition of ΔN271 Cdt1/Mcm2–7 to the G1 extract restored Cdc6-dependent Mcm2–7 loading at levels comparable to full-length Cdt1 (Figure 7B, lanes 1–4). Interestingly, when ΔN271 Cdt1-loaded Mcm2–7 complexes were exposed to S-phase extract, Mcm2–7 levels decreased modestly relative to that of full-length Cdt1-loaded Mcm2–7 (84% of full-length; Figure 7B, top, lanes 5–8) but replication initiation was severely impaired (12% of full-length; Figure 7B, bottom). The well-characterized targets of both CDK (Orc2 and Orc6) and DDK (Mcm4 and Mcm6) were equally phosphorylated in reactions containing full-length or ΔN271 Cdt1 following exposure to S-phase extract (Figure 7B, top, lanes 5–8; Supplementary Figure S6; the Mcm4-phospho-S82-D83 antibody detects a DDK target in Mcm4; Randell et al, 2010; ΔN271 Cdt1 signal was 94% of full-length Cdt1). Consistent with the DNA replication defects observed, Mcm2–7 complexes loaded with ΔN271 Cdt1 were severely impaired for the recruitment of the helicase-activating proteins GINS (Psf2, 10% of full-length) and Cdc45 (6% of full-length; Figure 7B, top, compare lanes 5 and 7). We also tested the possibility that ΔN271 Cdt1 inhibited Cdc45 and GINS binding because it was not released from the origin DNA during Mcm2–7 loading. In contrast to this possibility, we found that after assembly of Mcm2–7 onto origin DNA in the G1 extract, neither full-length nor ΔN271 Cdt1 was detectably associated with origin DNA (Supplementary Figure S7). Thus, although Cdt1 is no longer present at the origin when loaded Mcm2–7 complexes are activated, our data show that the N-terminus of Cdt1 is required to load Mcm2–7 complexes that are competent to interact with helicase-activating factors that drive the initiation of replication.
Figure 7.
Mcm2–7 helicases loaded by ΔN271 Cdt1 are not competent for replication initiation. (A) Full-length Cdt1 supports the loading of Mcm2–7 that are competent for replication. In vitro replication assays were performed on 175 fmol of bead-coupled pUC19 ARS1 wild-type DNA. An ORC-overexpressing whole-cell extract (ySK100) immunodepleted of endogenous Cdt1 was or was not supplied with purified full-length Cdt1/Mcm2–7 and Cdc6 during the helicase-loading reaction step (G1). DNA-bound proteins were subsequently treated with DDK and exposed to a cdc7–4 S-phase extract (yRH182) (G → S). Beads were washed and DNA–protein complexes were removed by boiling in Laemmli sample buffer for 5 min. (Top) Proteins were analysed by immunoblotting with anti-Mcm2–7 (UM174) and anti-ORC (1108) antibodies. (Bottom) Replication products from reactions 5–8 were analysed by alkaline gel electrophoresis and exposed to X-ray film. (B) In vitro replication assays were performed as in (A), except that the helicase-loading reactions were supplied with either full-length or ΔN271 Cdt1. (Top) Proteins were analysed by immunoblotting with anti-Mcm2–7 (UM174), anti-ORC (1108), anti-HA (12CA5; to detect Cdc45–HA), and anti-Myc (9E10; to detect Psf2–Myc, a subunit of the GINS complex) and Mcm4-phospho-S82-D83 (Randell et al, 2010) antibodies. HA-tagged Cdc45 and Myc-tagged Psf2 proteins are present in the yRH182 S-phase extract. (Bottom) Replication products from reactions 5–8 were analysed by alkaline gel electrophoresis and exposed to Phosphoimager screen.
Discussion
Loading of the Mcm2–7 replicative helicase is a highly regulated process that requires the coordinated action of ORC, Cdc6, and Cdt1. This event requires sequential ATP hydrolysis events by ORC and Cdc6, but the contribution of Cdt1 during the loading process has been less clear. Our findings identify the region of Cdt1 that binds to Mcm2–7 and show that this interaction is required for the initial recruitment of both proteins to ORC and Cdc6 bound at the origin. Mutations in Cdt1 that show a moderate defect in the initial recruitment of Cdt1 and Mcm2–7 but a complete defect in Mcm2–7 loading indicate that Cdt1 function is required for events after the initial recruitment of Mcm2–7. Based on our observation that multiple Cdt1 molecules are recruited to the origin during helicase loading, we propose that one of these functions is to facilitate formation of a multi-Mcm2–7 intermediate. Finally, our studies indicate that the N-terminus of Cdt1 is not required for formation of Mcm2–7 double hexamers that encircle origin DNA but instead is required to load Mcm2–7 that is competent for Cdc45 and GINS binding. Our findings reveal a multi-faceted role of Cdt1 during helicase loading and support a model in which the formation of a dimeric-Cdt1 helicase-loading intermediate is essential for helicase loading and replication initiation.
The requirement for formation of a Cdt1/Mcm2–7 complex to recruit either protein to the origin (Figure 4) imposes another layer of regulation during helicase loading. It is possible that Cdt1 and Mcm2–7 each have weak ORC/Cdc6-binding sites that are both required for tight binding. Alternatively, interaction between Mcm2–7 and Cdt1 may reveal binding sites that are otherwise obscured. The revealing of functional regions upon complex formation could also explain the requirement of Cdt1/Mcm2–7 complex formation for the nuclear import of both factors (Tanaka and Diffley, 2002). Because Cdt1 binds to Orc6 in solution (Chen et al, 2007), it is also possible that interaction of Cdt1/Mcm2–7 with ORC/Cdc6 is required to uncover these Cdt1-binding sites on Orc6. In each case, the requirement of prior complex formation would ensure that both Cdt1 and Mcm2–7 or ORC and Cdc6 are present prior to initiating helicase loading.
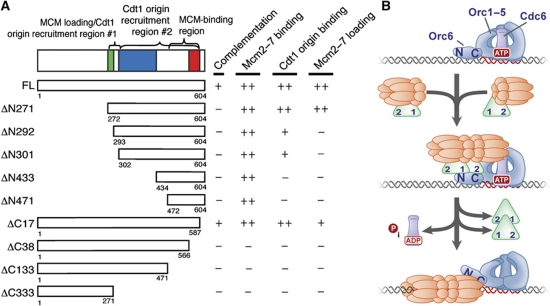
Although both the central and C-terminal domains of Cdt1 are predicted to adopt similar WHD folds (Lee et al, 2004; De Marco et al, 2009; Khayrutdinov et al, 2009; Jee et al, 2010), our studies reveal that the two domains are important for distinct functions during the helicase-loading process. The C-terminal domain of Cdt1 contains the Mcm2–7-binding site (a.a. 472–587) that mediates Cdt1/Mcm2–7 complex formation. We found that residues 566–587, predicted to contain β-strands 2 and 3 (Supplementary Figures S2 and S3) and Wing 1 of the C-terminal WHD, are essential for helicase binding (Figure 8A). This finding is consistent with studies of metazoan Cdt1 that implicate β-strand 2 in Mcm2–7 binding (Khayrutdinov et al, 2009). In contrast, the central domain of Cdt1 is not required for Mcm2–7 binding but is required for Cdt1 and Mcm2–7 origin recruitment and Mcm2–7 loading (Figure 8A), suggesting that this WHD mediates contacts with ORC or Cdc6 rather than Mcm2–7.
Figure 8.
Contribution of Cdt1 during the Mcm2–7 helicase-loading process. (A) Summary of Cdt1-deletion mutant analysis. (B) Proposed model of Mcm2–7 helicase loading via a multi-Cdt1 helicase-loading intermediate. A single origin-bound ORC/Cdc6 exposes two Cdt1-binding sites on Orc6 (labelled N and C) and recruits two Cdt1/Mcm2–7 molecules to the origin. The two Cdt1/Mcm2–7 molecules bind both Orc6-binding sites via its two non-identical origin recruitment regions (labelled 1 and 2). Based on the S-CDK inhibited binding of ΔN292 Cdt1 lacking IDR1, we propose that site 1 is the central WHD and site 2 is within IDR1. At this stage, the two Mcm2–7 hexamers are oppositely oriented with respect to one another and may form a double hexamer as shown. The origin recruitment of two Cdt1/Mcm2–7 complexes leads to ATP hydrolysis by Cdc6, resulting in the loading of the Mcm2–7 double hexamer and the disassociation of both Cdt1 molecules from the origin.
A multi-Cdt1 intermediate in helicase loading
Our data strongly suggest that the central domain and IDR1 of Cdt1 function as two independent origin recruitment sites. Disrupting IDR1 (ΔN292/301) consistently reduces Cdt1 origin recruitment by approximately half (Figure 3), and the same mutation reduces the number of Cdt1 molecules recruited to the origin to only one (Figure 5C). Interestingly, mutants that disrupt IDR1 show a larger reduction in Mcm2–7 origin recruitment than Cdt1 (Figures 3 and 6). Similarly, we observe no Mcm2–7 co-precipitation with Cdt1 lacking IDR1 in the presence of ATPγS (Figure 5C). These data suggest that positive interactions between the multiple Cdt1/Mcm2–7 complexes recruited to the origin stabilize Mcm2–7 association. One intriguing possibility is that Mcm2–7 double-hexamer formation is initiated at this earlier step in the helicase-loading process and the absence of a second Mcm2–7 complex negatively impacts the stable binding of the remaining complex.
Although it is possible that multiple ORC molecules with a single Cdt1-binding site facilitate the formation of a multi-Cdt1 helicase-loading intermediate at each origin of replication, our evidence does not support this model. First, unlike Cdt1, when we performed immunoprecipitation experiments with tagged and untagged ORC, we did not observe co-precipitation of the untagged protein (Supplementary Figure S4A). Second, the multi-Cdt1 intermediate can form in the absence of the B2 element of ARS1 (Supplementary Figure S4B), the second highest-affinity ORC-binding site at ARS1. Third, if the same Cdt1-binding site on two ORC molecules mediates the initial origin recruitment of Cdt1/Mcm2–7, then untagged ΔN292 Cdt1 bound to one ORC molecule would be expected to co-precipitate with tagged ΔN292 Cdt1 via an origin DNA-mediated interaction with the second ORC bound to tagged ΔN292 Cdt1. Instead, as expected if there were two distinct Cdt1 binding sites present in the ATPγS-dependent helicase-loading intermediate, we observe no detectable untagged ΔN292 Cdt1 in these experiments (Figure 5C). Finally, consistent with ORC recruiting multiple Cdt1 molecules, the Orc6 subunit of ORC (which is required to recruit Cdt1 to the origin) contains two unrelated and independent Cdt1-binding sites, both of which are essential for ORC function (Chen et al, 2007; Chen and Bell, 2011).
Based on our data, we propose that two Cdt1 molecules are recruited to the origin, each escorting a Mcm2–7 complex, to initiate Mcm2–7 double-hexamer loading during pre-RC formation (Figure 8B). In this model, an origin-bound ORC/Cdc6 complex recruits two Cdt1/Mcm2–7 molecules to the origin with Orc6 protein playing a primary role in these interactions. The origin recruitment of Cdt1 and Mcm2–7 is mediated by IDR1 and the central domain of Cdt1, which we envision act as two distinct Orc6-binding sites that are complementary to the two unrelated (N- and C-terminal) Cdt1-binding sites on Orc6. We propose that the two independent Cdt1–Orc6 interaction domains recruit and load Mcm2–7 helicases in opposite orientations to initiate head-to-head double-hexamer formation prior to helicase loading. Loading is completed by Cdc6 ATP hydrolysis, which also results in release of Cdt1 from the origin (Figure 8B).
Additional evidence supports the arrangement of ORC, Cdt1, and Mcm2–7 proteins illustrated in Figure 8B. Studies of the form of Mcm2–7 before and after helicase loading were unable to identify single-hexamer intermediates in Mcm2–7 loading (Remus et al, 2009). This suggests that both Mcm2–7 complexes are loaded simultaneously, which would require the simultaneous recruitment of two Mcm2–7 complexes. Previous crosslinking studies show that Orc6 is the only subunit that is exclusively crosslinked to DNA outside of the primary ORC DNA-binding site (ARS consensus sequence or ACS; Lee and Bell, 1997). Orc6 crosslinking occurs 40 bp from the ACS between the B1 and B2 elements of ARS1, the latter of which is implicated in modulating Mcm2–7 helicase loading (Zou and Stillman, 2000; Lipford and Bell, 2001). Given that one Mcm2–7 hexamer is thought to extend the length of 30 bp of DNA (Remus et al, 2009), this positioning is consistent with Orc6 interacting with two Cdt1/Mcm2–7 complexes extended along the length of the origin DNA adjacent to ORC. In this model, only one of the two Mcm2–7 complexes would be in proximity to ORC (Figure 8B). Because the Orc2–5 interaction region of Orc6 is located at its C-terminus (Chen et al, 2007), it is likely that the C-terminal Cdt1-binding site on Orc6 would be proximal to Orc2–5 and the N-terminal site would interact with the distal Cdt1/Mcm2–7 complex. If this arrangement is correct, then interfering with the more ORC-proximal Cdt1–Orc6 interaction would cause a greater disruption of Mcm2–7 association with ORC and the origin DNA than interfering with the ORC-distal site. Indeed, we observed 10–20% of wild-type Mcm2–7 recruitment by Cdt1 lacking IDR1 (which appears to interferes with the Cdt1 binding to the C-terminal Orc6-binding site; Figure 3A). In contrast, CDK phosphorylation of ORC (which interferes with the predicted distal, N-terminal Orc6–Cdt1 interaction site) results in only a 50% reduction of Mcm2–7 recruitment (Chen and Bell, 2011).
Disrupting the formation of a multi-Cdt1 helicase-loading intermediate inhibits Mcm2–7 loading. We demonstrate that loss of one putative ORC/Cdc6-binding site (ΔN292) on Cdt1 results in the origin association of only one Cdt1 species (Figure 5C). This ΔN292 Cdt1-deletion mutant also showed significantly lower levels of initial Mcm2–7 association and no helicase loading in the presence of ATP (Figure 6). Interestingly, a fusion of the C-terminal domain of Orc6 to Cdt1, in which there is one molecule of Cdt1 per ORC complex, also exhibits very inefficient helicase loading (Chen et al, 2007). We suspect the defect in Mcm2–7 loading by the Cdt1–ORC fusion could be due to an inability to recruit two Cdt1/Mcm2–7 complexes to the origin to initiate coordinated helicase loading. Notably, recent work has proposed that preventing the formation of a multi-Cdt1 helicase-loading intermediate is likely to be the primary mechanism by which CDK inhibits ORC function outside of G1 phase (Chen and Bell, 2011). By inactivating one of the two Cdt1-binding sites of Orc6, CDK causes a two-fold decrease in the amount of origin-associated Cdt1/Mcm2–7 complexes but results in more than a 10-fold reduction in Mcm2–7 loading (Chen and Bell, 2011). Combined with our current observations, these results support a model in which CDK regulates ORC function by preventing formation of a multi-Cdt1 helicase-loading intermediate and, therefore, subsequent helicase loading.
The N-terminus of Cdt1 is required to load activation-competent Mcm2–7
Intriguingly, we found that the essential N-terminal domain of Cdt1 is not required to assemble topologically linked Mcm2–7 double hexamers at the origin. Instead, we found that Mcm2–7 complexes loaded by ΔN271 Cdt1 are unable to recruit the Cdc45 and GINS helicase-activating proteins and are defective for replication initiation (Figure 7). Interestingly, this defect is not due to an inability of DDK to phosphorylate Mcm2–7 (Supplementary Figures S5 and 7B), providing strong evidence that DDK phosphorylation of Mcm2–7 is not sufficient to drive Cdc45 and GINS binding. Together, our data indicate that the N-terminus of Cdt1 is required to assemble Mcm2–7 that are competent for subsequent assembly of the active replicative helicase. Because Cdt1 is no longer present when loaded Mcm2–7 complexes associate with Cdc45 and GINS (Supplementary Figure S7), the inability for ΔN271 Cdt1-loaded Mcm2–7 to recruit Cdc45 and GINS must be the result of a defect that is maintained in the absence of the mutant Cdt1. This is in contrast to studies of Xenopus Cdt1 that found the Cdt1 N-terminus was not required for in vitro Mcm2–7 loading or replication in Xenopus egg extracts (Ferenbach et al, 2005). It remains possible that this domain of Xenopus Cdt1 could be required at other times in development in vivo.
Why are the Mcm2–7 complexes assembled by ΔN271 Cdt1 impaired for the recruitment of helicase-activating factors? These Mcm2–7 complexes show no difference in DDK phosphorylation (Supplementary Figures S5 and 7C), suggesting that Mcm2–7 loaded by both forms of Cdt1 are in a conformation that facilitates DDK binding and phosphorylation (Francis et al, 2009). It is possible that the N-terminus of Cdt1 is required to disrupt an interaction between ORC and Mcm2–7 that interferes with subsequent Cdc45 and GINS binding. If so, this interaction does not prevent the subsequent release of ORC observed at the end of the replication reaction (Figure 7B). Alternatively, Cdc45 and GINS binding could be compromised due to a change in the discontinuity between the Mcm2 and Mcm5 subunits of ΔN271 Cdt1-loaded Mcm2–7 complexes (Bochman and Schwacha, 2008; Costa et al, 2011). If the conformation of the Mcm2–5 ‘gate’ is compromised in a ΔN271 Cdt1-loaded Mcm2–7 complex, the gap is not drastic enough to also affect its affinity for DNA (Figure 6). Another possibility is that the individual hexamers within the head-to-head Mcm2–7 double hexamer are improperly oriented relative to one another. This model demands that the initial loading and DDK phosphorylation of Mcm2–7 is indifferent to inter-hexamer subunit orientation, whereas the recruitment of helicase-activating factors to the double hexamer is sensitive to the orientation of Mcm2–7.
Our studies demonstrate that Cdt1 plays multiple roles during the helicase-loading process and provide a simple solution to explain how the symmetric replication-competent Mcm2–7 double hexamers are loaded onto asymmetric origins of replication. Nevertheless, many aspects of the Mcm2–7 loading process remain to be elucidated. Does coordinated Mcm2–7 loading involve concerted or sequential loading of origin-recruited helicases? Are multiple ATP hydrolysis events by Cdc6 required to load the Mcm2–7 double hexamer? What is the stoichiometry of Cdc6 in the helicase-loading intermediates? How does the N-terminus of Cdt1 influence the conformation of the loaded Mcm2–7 helicase? The development of new assays that directly monitor the formation of the loaded Mcm2–7 double hexamer with better time resolution will be required to answer these questions.
Materials and methods
Strain construction
Strains were prepared using standard laboratory methods. Yeast strains used in this study are listed in Supplementary Table S1.
Complementation analysis
Each Cdt1-deletion mutant was placed under the control of the endogenous Cdt1 promoter in a Leu-marked test plasmid (pRS405) and integrated into the genome of yTJT73. Complementation was scored by the ability of the integrated Cdt1 test plasmid to support growth of yTJT73 in the presence of FOA, which selects against the presence of a plasmid including wild-type CDT1.
Protein purification
Purified ORC was prepared from yeast strain ySK100 as described previously (Tsakraklides and Bell, 2010). Cdc6 was purified from Escherichia coli as described previously (Randell et al, 2006). Cdt1/Mcm2–7 and Cdt1 proteins were prepared as described previously (Tsakraklides and Bell, 2010), except that Cdt1 instead of Mcm4 was tagged with the FLAG epitope and yeast cultures were arrested in G1 phase with α-factor for 3 h before galactose induction for 4.5 h. Extract was incubated with FLAG resin for 12 h at 4 °C. For all preparations, Complete protease inhibitors (Roche) were added to cell lysis buffers prior to extract preparation according to the manufacturer's instructions.
Cdt1 recruitment and helicase-loading assays
Reconstituted helicase-loading assays for Cdt1 that was capable of Mcm2–7 binding were performed as previously described (Tsakraklides and Bell, 2010). Cdt1 recruitment assays for Cdt1 that was capable of Mcm2–7 binding were performed as described above for the helicase-loading assay, except that ATP was replaced with ATPγS and 4 pmol of 1039 bp ARS1 DNA and 12 pmol purified ORC, Cdc6, and Cdt1/Mcm2–7 were used per reaction. Helicase-loading and Cdt1 recruitment assays for Cdt1 that was unable to bind Mcm2–7 were performed as described previously (Randell et al, 2006).
Co-immunoprecipitation assays
Cdt1-containing helicase-loading intermediates were assembled from purified proteins as described above (Cdt1 recruitment assay for Cdt1 capable of Mcm2–7 binding), except that 8 pmol of 1039 bp ARS1 (wild-type or B2−) DNA and 32 pmol of purified ORC, Cdc6, and Cdt1/Mcm2–7 were used per reaction. When tagged and untagged Cdt1/Mcm2–7 species were mixed, 16 pmol of tagged Cdt1/Mcm2–7 and 16 pmol of untagged Cdt1/Mcm2–7 were used. Assembled complexes were washed, DNA–protein complexes were released from beads by UV irradiation, and complexes were incubated with anti-Myc antibody coupled to GammaBind G Sepharose beads for 1.5 h at 4 °C in H/300 buffer supplemented with 3 mM ATPγS. Immunoprecipitated fractions were separated from supernatant fractions by three successive washes with H buffer (50 mM HEPES (pH 7.6); 5 mM Mg acetate; 1 mM EDTA; 1 mM EGTA; 10% glycerol; 0.01% NP-40) containing 300 mM potassium glutamate at 4 °C. Immunoprecipitated and supernatant fractions were TCA precipitated and analysed by immunoblotting with anti-Mcm2–7 (UM174; Chen and Bell, 2011), anti-Cdt1 (HM5352; Randell et al, 2006), and anti-ORC (1108; Bowers et al, 2004) antibodies. ORC co-immunoprecipitation was performed similarly except anti-FLAG antibodies were used for immunoprecipitation and the FLAG epitope on Cdt1 was removed after purification by site-specific protease cleavage.
Gel filtration analysis of loaded Mcm2–7
Assays were performed according to Evrin et al (2009). Briefly, helicase-loading assays were performed using 2 pmol of bead-coupled ARS1 DNA and G1-extract depleted for Cdt1 and complemented with the indicated Mcm2–7/Cdt1 complex. Assembled complexes were salt extracted (Randell et al, 2006) and resuspended in 15 μl of H buffer containing 300 mM potassium glutamate, 50 μM zinc acetate, 5 mM calcium chloride, 10 mM magnesium acetate, and 1 U DNase I. The resuspended beads were incubated at 25 °C for 6 min. The released Mcm2–7 complexes were loaded on a 2.4-ml Superose 6 column (GE Healthcare) equilibrated in H buffer with 300 mM potassium glutamate. The following protein standards were used to calibrate the column: Blue dextran 2000 (void volume); thyroglobulin (669 kDa); apoferritin (440 kDa); aldolase (158 kDa).
In vitro replication assays
Replication assays were performed as described in Heller et al (2011). Briefly, 3.7 kb pUC19 ARS1 wild-type containing plasmids were biotinylated and coupled to streptavidin-coated magnetic beads. Each replication reaction was performed using 175 fmol of bead-coupled pUC19 ARS1. Helicase loading was performed in a 40-μl reaction volume using standard in vitro helicase-loading conditions (Bowers et al, 2004) with Cdt1-depleted, G1-arrested ySK100 whole-cell extract supplied with 1 pmol purified full-length or ΔN271 Cdt1/Mcm2–7 complexes and Cdc6. Endogenous Cdt1 was immunodepleted from ySK100 whole-cell extract as previously described (Randell et al, 2006). After removal of G1-arrested extract, beads containing DNA–protein complexes were treated with DDK. DDK phosphorylation reactions were performed as previously described (Francis et al, 2009), except that reactions contained 225 mM KGlut, 3.5 mM MgOAc, 5% glycerol, 1 mM spermine, 3 mM DTT, and 1 mM ATP. After removal of DDK and buffer, DDK-treated beads were exposed to replication conditions. Each replication reaction was performed in a 40-μl reaction volume containing 25 mM HEPES-KOH (pH 7.6), 20 mM creatine phosphate, 2 mM DTT, 0.04 mg/ml creatine kinase, 225 mM KGlut, 12 mM MgOAc, 3 mM ATP, 200 μM GTP, 200 μM CTP, 200 μM UTP, 40 μM dNTPs, 10 μCi (40 μM) α-32PdCTP, and 750 μg cdc7–4 S-phase whole-cell extract (yRH182; Heller et al, 2011) at 25 °C for 150 min. Beads were washed and DNA–protein complexes were removed by boiling in Laemmli sample buffer for 5 min. Proteins were analysed by immunoblotting with anti-Mcm2–7 (UM174), anti-ORC (1108), anti-HA (12CA5), and anti-Myc (9E10) antibodies. Replicated DNA samples were run on a 0.8% agarose gel at 6 V/cm for 3 h in running buffer containing 2 mM EDTA and 30 mM NaOH. The gel was dried and exposed to X-ray film or Phosphoimager screen.
Supplementary Material
Acknowledgments
We thank R Sinapius and M Himmelright for providing purified ORC and Cdc6, M Miller for assistance with strain construction of yTJT73, S Kang and SH Chan for providing ARS1 pUC19 template, H Blitzblau and S Chen for critical reading of the manuscript, and the Bell Lab for helpful discussions and constructive comments. This work was supported by a grant from the National Institutes of Health to SPB (GM52339). SPB is an employee of the Howard Hughes Medical Institute.
Author contributions: TJT and SPB designed all of the experiments, analysed the data, and wrote the paper. TJT performed all of the experiments except the gel filtration analysis, which was performed by SPB.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Asano T, Makise M, Takehara M, Mizushima T (2007) Interaction between ORC and Cdt1p of Saccharomyces cerevisiae. FEMS Yeast Res 7: 1256–1262 [DOI] [PubMed] [Google Scholar]
- Bell SP (1995) Eukaryotic replicators and associated protein complexes. Curr Opin Genet Dev 5: 162–167 [DOI] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B (1995) The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83: 563–568 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A (2008) The Mcm2–7 complex has in vitro helicase activity. Mol Cell 31: 287–293 [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP (2004) ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell 16: 967–978 [DOI] [PubMed] [Google Scholar]
- Chen S, Bell SP (2011) CDK prevents Mcm2–7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev 25: 363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP (2007) Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev 21: 2897–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H (2008) The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 105: 10326–10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M (2006) Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol 13: 684–690 [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36 (Web Server issue): W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM (2011) The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18: 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, Lygerou Z, Medema RH, Blow JJ, Svergun DI, Taraviras S, Perrakis A (2009) Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing. Proc Natl Acad Sci USA 106: 19807–19812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA 94: 5611–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C (2009) A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA 106: 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenbach A, Li A, Brito-Martins M, Blow JJ (2005) Functional domains of the Xenopus replication licensing factor Cdt1. Nucleic Acids Res 33: 316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP (2009) Incorporation into the prereplicative complex activates the Mcm2–7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev 23: 643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Khoudoli GA, Jones RC, Blow JJ (2011) MCM2–7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem 286: 11855–11864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP (2011) Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR (2010) Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee J, Mizuno T, Kamada K, Tochio H, Chiba Y, Yanagi K, Yasuda G, Hiroaki H, Hanaoka F, Shirakawa M (2010) Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins. J Biol Chem 285: 15931–15940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayrutdinov BI, Bae WJ, Yun YM, Lee JH, Tsuyama T, Kim JJ, Hwang E, Ryu KS, Cheong HK, Cheong C, Ko JS, Enomoto T, Karplus PA, Guntert P, Tada S, Jeon YH, Cho Y (2009) Structure of the Cdt1 C-terminal domain: conservation of the winged helix fold in replication licensing factors. Protein Sci 18: 2252–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF (2000) Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Lee C, Hong B, Choi JM, Kim Y, Watanabe S, Ishimi Y, Enomoto T, Tada S, Cho Y (2004) Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature 430: 913–917 [DOI] [PubMed] [Google Scholar]
- Lee DG, Bell SP (1997) Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol 17: 7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Bell SP (2001) Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell 7: 21–30 [DOI] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H (2010) CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon), and GINS in budding yeast. Genes Dev 24: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T (2004) Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J Biol Chem 279: 30807–30816 [DOI] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP (2006) Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell 21: 29–39 [DOI] [PubMed] [Google Scholar]
- Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP (2010) Mec1 is one of multiple kinases that prime the Mcm2–7 helicase for phosphorylation by Cdc7. Mol Cell 40: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF (2009) Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Diffley JF (2009) Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol 21: 771–777 [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ (1999) Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci 112(Part 12): 2011–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Diffley JF (2000) Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci USA 97: 14115–14120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple JW, Da-Silva LF, Jervis EJ, Ah-Kee J, Al-Attar H, Kummer L, Heikkila JJ, Pasero P, Duncker BP (2006) An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J 25: 5150–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B (2010) The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33 (Web Server issue): W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Diffley JF (2002) Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2–7 during G1 phase. Nat Cell Biol 4: 198–207 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Teer JK, Dutta A (2008) Human Cdt1 lacking the evolutionarily conserved region that interacts with MCM2–7 is capable of inducing re-replication. J Biol Chem 283: 6817–6825 [DOI] [PubMed] [Google Scholar]
- Tsakraklides V, Bell SP (2010) Dynamics of pre-replicative complex assembly. J Biol Chem 285: 9437–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bell SP (2002) The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc Natl Acad Sci USA 99: 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi K, Mizuno T, You Z, Hanaoka F (2002) Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J Biol Chem 277: 40871–40880 [DOI] [PubMed] [Google Scholar]
- Ying CY, Gautier J (2005) The ATPase activity of MCM2–7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J 24: 4334–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Masai H (2008) Cdt1 forms a complex with the minichromosome maintenance protein (MCM) and activates its helicase activity. J Biol Chem 283: 24469–24477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol 20: 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.