Abstract
The causes and consequences of rapid radiations are major unresolved issues in evolutionary biology. This is in part because phylogeny estimation is confounded by processes such as stochastic lineage sorting and hybridization. Because these processes are expected to be heterogeneous across the genome, comparison among marker classes may provide a means of disentangling these elements. Here we use introns from nuclear-encoded reproductive protein genes expected to be resistant to introgression to estimate the phylogeny of the western chipmunks (Tamias: subgenus: Neotamias), a rapid radiation that has experienced introgressive hybridization of mitochondrial DNA (mtDNA). We analyze the nuclear loci using coalescent-based species-tree estimation methods and concatenation to estimate a species tree and we use parametric bootstraps and coalescent simulations to differentiate between phylogenetic error, coalescent stochasticity and introgressive hybridization. Results indicate that the mtDNA gene tree reflects several introgression events that have occurred between taxa of varying levels of divergence and at different time points in the tree. T. panamintinus and T. speciosus appear to be fixed for ancient mitochondrial introgressions from T. minimus. A southern Rocky Mountains clade appears well sorted (i.e., species are largely monophyletic) at multiple nuclear loci, while five of six taxa are nonmonophyletic based on cytochrome b. Our simulations reject phylogenetic error and coalescent stochasticity as causes. The results represent an advance in our understanding of the processes at work during the radiation of Tamias and suggest that sampling reproductive-protein genes may be a viable strategy for phylogeny estimation of rapid radiations in which reproductive isolation is incomplete. However, a genome-scale survey that can statistically compare heterogeneity of genealogical process at many more loci will be necessary to test this conclusion.
Keywords: Coalescent simulation, gene tree, hybridization, introgression, lineage sorting, multilocus, reproductive proteins, Sciuridae, species tree, Tamias
The study of rapid radiations is of central importance to evolutionary biology because much of the diversity of life has been generated in great bursts of speciation (Dobzhansky 1951; Schluter 2000; Alfaro et al. 2009). Understanding the ecological and genetic processes at work during these radiations is therefore of great interest. In order to elucidate those processes most clearly, however, an understanding of phylogeny is necessary. Unfortunately, it is precisely during rapid sequences of speciation events that the inference of phylogeny becomes most difficult. The primary reason for this is that when lineages diverge in rapid succession, individual gene genealogies may not reflect the actual phylogeny of the organisms under study. There are two main sources of this gene tree discordance (exclusive of phylogenetic error): stochasticity in the coalescent process and interspecific gene flow. If there is a series of rapid speciation events, the expected time to coalescence of alleles in an ancestral population ( generations; Nordborg 2001) may be greater than the time between divergence events, and the shorter the time between speciation events, the more likely gene trees will differ from the actual speciation history simply due to coalescent stochasticity (Pamilo and Nei 1988). In fact, for some combinations of topology and speciation times, the most probable gene trees are discordant with the species tree (Degnan and Rosenberg 2006). The second major source of discordance, introgressive hybridization, has been a topic of controversy (e.g., Dowling and Secor 1997). Although it has historically been viewed as unimportant and uncommon in the zoological literature (e.g., Fisher 1958; Mayr 1963), a rapidly increasing number of cases demonstrate its near ubiquity (e.g., Schwenk et al. 2008). Furthermore, many examples involve introgression among nonsister taxa (e.g., Good et al. 2003, Good et al. 2008; Buckley et al. 2006; Linnen and Farrell 2008; Maureira-Butler et al. 2008; Bossu and Near 2009). Indeed, the increasing frequency of such observations has led to the emerging divergence-with-gene-flow model of speciation (Machado et al. 2002; Llopart et al. 2005), in which the genetic factors that underlie speciation may continue to accumulate despite incomplete reproductive isolation.
Currently, the most common approach to molecular phylogenetic estimation from multiple loci is concatenation into a supermatrix that is then subject to analysis (e.g., Ward et al. 2010). This approach is typically justified by the assertion that by combining data, the vagaries of mutational variance and other sources of error should be overcome by the underlying phylogenetic signal, producing a robust estimate of the species tree (e.g., Rokas et al. 2003, Gatesy and Baker 2005). It has recently been demonstrated, however, that under the conditions expected during a rapid radiation, stochasticity in the coalescent can produce enough gene tree discordance that concatenation becomes positively misleading (Kubatko and Degnan 2007). If, by contrast, coalescent stochasticity is taken into account in these circumstances, reliable species-tree estimation may be possible (Maddison and Knowles 2006, Edwards et al. 2007, Knowles and Carstens 2007 ). Several methods are now available for the estimation of species trees that account for gene tree discordance under the assumption that it is due only to coalescent stochasticity and mutational variance. Among them are *BEAST (Heled and Drummond 2010), Bayesian estimation of species trees (BEST; Edwards et al. 2007; Liu and Pearl 2007), species tree estimation using maximum likelihood (STEM; Kubatko et al. 2009) and minimization of deep coalescences (MDC; Maddison 1997; Maddison and Knowles 2006). Each method can estimate moderately large species trees with multiple haplotypes per species.
These emerging methods represent an important advance in phylogenetic estimation, particularly for rapid radiations (Edwards 2008); however, they do not account for hybridization. As introgressive hybridization appears to be relatively common (e.g., Schwenk et al. 2008), it becomes important because the gene tree discordance it causes can often be very difficult to distinguish from that caused by stochasticity in the coalescent (Holder et al. 2001). It is not clear how introgressive hybridization will affect these methods, but there is some indication that if it occurs between nonsister taxa and is not accounted for, it may render species-tree estimation more difficult (Eckert and Carstens 2008).
Maddison (1997) described a phylogeny as a “cloudogram,” composed of a statistical distribution of gene trees produced by various evolutionary processes, including selection, gene flow, stochastic lineage sorting, and gene duplication and extinction. It is becoming clear that those processes are heterogeneous across the genome (Funk and Omland 2003; Payseur et al. 2004; Carling and Brumfield 2009). This is especially critical in the case of introgression. If introgression is heterogeneous in a predictable way across the genome (e.g., Harrison 1990; Rieseberg et al. 1999; Wu 2001), we may be able to select classes of markers and compare patterns of variation among them that allow us to illuminate various aspects of the biology of diversification, including phylogenetic history and reticulate evolution (e.g., Carling and Brumfield 2009; Maroja et al. 2009; Petit and Excoffier 2009). Here we take this approach to phylogeny estimation in a well-studied group of rodents, the diverse chipmunks of western North America (Tamias), comparing patterns at three reproductive protein genes, one anonymous locus, and one mitochondrial gene.
Study System
Chipmunks (Sciuridae: Tamias) exemplify many of the challenges associated with studies of rapid radiations, and despite much attention, the relationships among them remain poorly understood. There are 25 described species in Tamias, and most of the diversity (23 species) occurs in the monophyletic western North American clade, subgenus Neotamias (e.g., Levenson et al. 1985; Piaggio and Spicer 2001). The morphology of the baculum (the os penis or male genital bone) is a key taxonomic character in Tamias (e.g., White 1953; Sutton 1995); it occupies a distal position in the penis and exhibits little variation within species but strong discontinuities among species (White 1953; Patterson and Thaeler 1982; Sutton and Patterson 2000). Because these discontinuities often occur along some type of physical feature (e.g., rivers) or ecological boundaries (e.g., ecotones) and because there are usually no intermediate morphologies at these boundaries, bacular discontinuities in Tamias have been interpreted as reliable indicators of reproductive isolation and species limits (e.g., Sutton and Patterson 2000).
Phylogenetic hypotheses from chromosomal rearrangements, allozymes, and mitochondrial DNA (mtDNA) all conflict at some level (Nadler et al. 1977, Levenson et al. 1985; Piaggio and Spicer 2001). The published mtDNA phylogeny suffers from poor support at many branches and there are several instances in which species are not monophyletic. Additionally, studies of two northern Rockies species, T. amoenus and T. ruficaudus, have revealed a complex and temporally structured series of mtDNA introgression events (Good et al. 2003, Good et al. 2008; Hird and Sullivan 2009; Reid et al. 2010). These species span the deepest divergence in the mtDNA genealogy, suggesting that reproductive isolation may be widely incomplete in the genus, or at least decoupled from the process of lineage divergence. Taken together, the situation suggests the possibility that coalescent stochasticity and introgressive hybridization may be confounding our understanding of the phylogenetic relationships within the Tamias radiation.
Here we explore the use of reproductive-protein genes, which are expected to be both resistant to introgression (e.g., Rieseberg et al. 1999, Maroja et al. 2009), and to evolve rapidly (e.g., Swanson et al. 2003), to estimate the phylogeny of a rapid mammalian radiation and test hypotheses about introgressive hybridization among its species. We examine patterns of variation at multiple nuclear loci using coalescent-based species-tree estimation methods and concatenation to estimate a species tree, use parametric bootstraps to assess the effects of mutational variance and coalescent simulations to differentiate between coalescent stochasticity and introgressive hybridization.
MATERIALS AND METHODS
Sampling
Given the problems associated with species identification, introgression and nonmonophyly in Tamias, we incorporated intraspecific diversity through widespread geographic sampling within taxa. This approach should indicate the extent of nonmonophyly and, should introgressed alleles remain geographically structured, aid in the identification of instances of nonmonophyly due to introgression (e.g., Good and Sullivan 2001). Samples were obtained through fieldwork by us or from, when possible, vouchered specimens deposited in natural history museums (Table A1). Protocols for field collection and handling of chipmunks followed the guidelines of the American Society of Mammalogists Animal Care and Use Committee (Gannon and Sikes 2007) and were approved by the Animal Care and Use Committee of California State Polytechnic University, Pomona and the University of Idaho. Our focus is on inferring relationships among the monophyletic western North American chipmunks (subgenus Neotamias), so we used T. sibiricus and T. striatus, belonging to the subgenera Eutamias and Tamias, respectively, as outgroup taxa (Thorington and Hoffmann 2005).
For this study, we employed five loci: the mitochondrial gene cytochrome b (Cytb) and four nuclear loci: an anonymous locus (Anon), and introns from the genes zonadhesin, acrosin, and zona pellucida protein 2 (ZAN, ACR, and Zp2, respectively). The anonymous locus is noncoding, whereas the three gene fragments are exon-primed intron sequences. Anon, ZAN and Zp2 were developed in Tamias for this study by aligning sequences from Mus, Rattus and Spermophilus, and placing primers in conserved exon regions. Zp2 has previously been used in a phylogenetic context in rodents (Turner and Hoekstra 2006). We focused on these nuclear regions because three of them are found in reproductive-protein genes, which may be resistant to introgression (e.g., Rieseberg et al. 1999; Maroja et al. 2009) and are expected to be rapidly evolving (e.g., Swanson et al. 2003). ACR is a protease in the sperm acrosome that lyses the zonapellucida and facilitates the penetration of the sperm by the egg; ZAN is a transmembrane sperm protein that functions in the binding of sperm the zonapellucida; Zp2 is an egg protein in the zona pellucida that participates in sperm binding (Wasserman et al. 2001).
Extraction, Amplification, and Sequencing
Genomic DNA was extracted from tissues frozen or preserved in either ethanol or lysis buffer using QIAGEN DNeasy extraction kits (QIAGEN, Valencia, CA). Fragments were amplified via polymerase chain reaction (primers in Table S1; available from http://www.sysbio.oxfordjournals.org); we amplified Cytb, ZAN, Anon, and Zp2 using roughly the same thermal profile, differing only in annealing temperature (: 30 s at 94°C, 30 s at TA and 60 s at 72°C for 38 cycles. Annealing temperatures were 50°C (Cytb), 52°C (ZAN), 53°C (Anon), and 51°C (Zp2). We amplified ACR using a “touchdown” procedure following Good et al. (2008). PCR products were purified using QIAquick PCR purification kits (Qiagen, Valencia, CA) and then sequenced in both directions on the ABI PRISM 3100 using BigDye Terminator chemistry v. 3.1 (ABI, Foster City, CA). Chromatograms were aligned and edited using CodonCode ALIGNER (CodonCode Corp., Dedham, MA). In order to identify heterozygous sites, we examined regions of low-quality sequence (as identified by ALIGNER) and inspected chromatograms by eye. We considered double peaks in which the lower peak was at least 60% of the intensity of the higher to be heterozygous (if surrounding sequence was high quality) and coded them according to standard nucleotide ambiguity codes.
Haplotype Resolution
We used a combination of statistical and experimental methods to infer gametic phase of sequences with more than one heterozygous site. First, we used Phase (Stephens et al. 2001; Stephen and Donnelly 2003), a Bayesian method for inference of haplotypes from population data that implement an approximate coalescent prior on haplotype frequencies. Thus, higher prior probability is placed on the inference of haplotypes similar to those already present in the population. We ran Phase twice on the full data set for each locus for 500 iterations, changing the block size for the partition–ligation procedure for each run (from 10 to 50 bp). For a subset of haplotypes that we were unable to resolve statistically (those that contained two or more sites resolved with <90% posterior probability), we used Topo TA cloning kits (Invitrogen, Carlsbad, CA) to determine haplotypes experimentally. We prepared cloning reactions according to the manufacturer's instructions and sequenced 5–10 clones from each reaction. We observed chimeric clone sequences and thus sequenced enough clones to determine a majority-rule haplotype pair for each individual. These were incorporated into a second set of Phase runs as known haplotypes in an attempt to increase resolution. After the final Phase runs, heterozygous sites that we did not verify with cloning and that were inferred with less than 90% posterior probability were recoded as ambiguous.
Phylogenetic Analyses
We conducted phylogenetic analyses on each locus individually using maximum-likelihood (ML) and Bayesian methods. Redundant sequences were removed from each alignment with MacClade (D.R. Maddison and W.P. Maddison 2003). Models of sequence evolution were selected using PAUP* 4.0b (Swofford 2000) in combination with DT-ModSel (Minin et al. 2003), which employs a decision-theoretic criterion that selects the simplest model that is expected to perform well. Using the identified models, we performed ML searches in GARLI (Zwickl 2006); we optimized model parameters on a neighbor-joining tree in PAUP*, fixed them in GARLI, and ran the algorithm set to terminate automatically when 200,000 generations passed with no increase in log likelihood of the best tree. We ran each data set at least five times (or until the same best likelihood score was returned twice), starting from random topologies, to ensure that the tree search was not trapped in local optima. Bootstrap support was also evaluated using GARLI, with the termination condition set to 35,000 generations and we performed 200 replicates.
We also performed Bayesian phylogenetic analyses using MrBayes 3.1 (Huelsenbeck and Ronquist 2001, Ronquist and Huelsenbeck 2003). When unavailable models were selected, we chose the next most parameterized model available in MrBayes. For Cytb, we also conducted both partitioned and unpartitioned Bayesian analyses. In the partitioned analysis, we assigned first and second codon positions to one partition and third codon positions to a second partition. Preliminary analyses indicated that the default prior probabilities placed on branch lengths by MrBayes were overwhelming branch-length signal in all nuclear loci both individually and when concatenated. We came to this conclusion because observed tree lengths were approximately equal to the product of the per branch branch length prior and the number of branches in the tree, 2 orders of magnitude greater than ML estimates. Therefore, after experimenting with a range of priors, we used the unconstrained exponential branch length prior with a mean of 0.002 for all final analyses, which resulted in branch lengths that more closely followed ML estimates. It should be noted that bipartition posterior probabilities (i.e., split frequencies) did not appear to be affected by this branch-length estimation issue. This is the approach recommended by Brown et al. (2010). All MrBayes analyses consisted of two independent runs from different starting points and employed Metropolis-coupled Markov chain Monte Carlo analyses. Because all MrBayes analyses were conducted on a multiprocessor cluster, we increased the number of Markov chains per run from the default 4 to 8 and used a heating parameter of 0.1. We assessed convergence of the Markov chains using the standard deviation of split frequencies, which is calculated at regular intervals by MrBayes. We assumed that runs had converged when this value was less than 0.01. We further examined parameter estimates in Tracer (Rambaut and Drummond 2007) to assure that these had reached stable values.
Concatenated Analysis
We then concatenated the nuclear data in a single analysis using methods similar to those described above. The concatenated data set was generated by arbitrarily joining haplotypes from each locus for each individual, resulting in two concatenated sequences per individual. Only individuals with complete data were included in the concatenated analysis. We performed two separate analyses of the concatenated data set: an unpartitioned analysis using a model identified by DT-ModSel (Table S2) and a partitioned analysis in which each locus was assigned a separate model. We did not explore partitioning sites within genes because of the relatively low levels of divergence. In the partitioned analysis, gaps were coded as binary characters and consolidated in a fifth data partition assigned the Mk model. We conducted these analyses in both MrBayes and the partitioned version of GARLI (0.97). We used the Akaike information criterion (Akaike 1974) with the likelihoods from GARLI to select the partitioning scheme.
Species-Tree Estimation
Because the single-gene analyses indicated a lack of species monophyly within loci and statistically supported conflict among loci, we also estimated the phylogeny using methods that account for stochasticity in the coalescent process: MDC (Than and Nakhleh 2009, Maddison and Knowles 2006) as implemented in Phylonet (v. 2.1; Than and Nakhleh 2009), STEM (Kubatko et al. 2009) and *BEAST (Heled and Drummond 2010). The Phylonet implementation of MDC takes previously estimated gene tree topologies as input and finds the tree that minimizes the number of extra lineages across loci. We used the unrooted genealogies option. STEM takes previously estimated ML gene trees and an estimate of (to be applied to the whole tree) as input, and finds the ML species tree analytically using an estimator similar to the GLASS tree of Mossel and Roch (2008) or the maximum tree of Liu and Pearl (2007). *BEAST is a Bayesian method that takes multilocus sequence data as input and uses Markov chain Monte Carlo to sample from the posterior distribution of species trees. It accounts for uncertainty in gene trees as well as the species tree. We used substitution models chosen by DT-Modsel or the next most general model, and uncorrelated relaxed clock models. We ran *BEAST for 200 million generations, sampling every 20,000, at which point effective sample size values for all parameters were above 200 and examination of split frequencies across the Markov chain in AWTY (Wilgenbusch et al. 2004) indicated they had stabilized. Species-tree estimation included redundant sequences excluded for previous phylogenetic analyses. All methods of species-tree estimation used here assume the following. (i) There is no recombination within loci and free recombination between loci. (ii) Loci conform to a neutral coalescent model (i.e., there is no selection and loci conform to a molecular clock). (iii) There is no migration between taxa under consideration. (iv) Population (or species) membership is known. (v) There is no substructure within designated populations or species. STEM and MDC additionally assume that gene trees are estimated without error.
Therefore, before the initiating analyses, we explored some of these assumptions. We tested for recombination using the “difference of the sum of squares” method in TOPALi (v2; Milne et al. 2009). We tested for deviations from neutrality by calculating Tajima's D for each locus for species with over four sequences using DnaSP (v. 5.10; Librado and Rozas 2009) and by conducting pairwise Hudson–Kreitman–Aguadé tests (HKA; Hudson et al. 1987) using the program HKA (Hey 2004). Because we have a priori reason to expect some of the incongruence at Cytb is due to introgression, we excluded it from these analyses. Upon examination of nuclear gene trees, we note that analysis of the Zp2 locus yielded a gene tree with many partitions having very low branch support values. We therefore excluded it from our STEM and MDC analyses because of the much higher probability that it will violate the assumption that gene trees are estimated without error. We conducted *BEAST analyses with and without Zp2 in order to compare results across methods. We assigned each individual to species following bacular morphology, pelage characteristics, and distributional information when available. It was clear from multilocus genotypes and locality information that three museum specimens (Table A1; MSB84515, MSB90785, and MVZ199281) were potentially misidentified or have been subject to taxonomic revisions not reflected in current museum records (e.g., MVZ199281, T. rufus) and we assigned these specimens in analyses to their proper taxon. We also conducted species-tree analyses with these three specimens excluded. In other cases, we noted spatially defined populations that exhibited highly divergent genealogical patterns from the taxon to which they were assigned (specifically T. amoenus cratericus, and T. minimus grisescens). For coalescent-based species-tree estimation, we assigned them to new taxa assuming that it would be better to erroneously split a single true species than lump two nonsister taxa. STEM requires the user to input a value for θ, which it assumes is constant across all branches in the tree. We used the program LAMARC (v 2.0; Kuhner 2006), a coalescent genealogy sampler that estimates parameters (including θ from sequence data, and conducted several independent runs on nuclear data from several species with high intraspecific sampling in order to obtain a range of estimates of θ.
Assessment of Hybridization
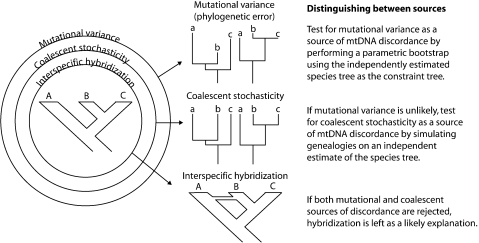
The Cytb genealogy is strongly discordant with the genealogies estimated for the three nuclear loci. This discordance could have three main sources: mutational variance, coalescent variance, and hybridization (Fig. 1). The first results in phylogenetic error (when the estimated tree differs from the true tree due to sampling error), whereas the latter two generate truly discordant gene trees. If we can reject the first two, introgressive hybridization is left as the likely source of discordance. In order to test the hypothesis that mutational variance is the source of phylogenetic incongruence, we performed parametric bootstrapping (e.g., Goldman et al. 2000, Sullivan et al. 2000). Briefly, we conducted ML searches with the Cytb genealogy constrained to match the estimated ML species tree from STEM. We then calculated the difference in likelihood scores between the constrained and unconstrained ML trees as our test statistic and generated a null distribution for this test statistic by simulation. One thousand DNA sequence data sets were simulated on the constrained topology with characteristics that matched the empirical data (using Seq-Gen; Rambaut and Grassly 1997). We then conducted ML searches that were both unconstrained and constrained to match the species tree, and calculated test statistics for each replicate.
Figure 1.
Testing hypotheses of lineage sorting and introgression in Tamias.
We used coalescent simulations similar to those used by Buckley et al. (2006) to test the hypothesis that coalescent stochasticity is causing the observed incongruence. We treated the STEM species-tree estimate as the model tree and used MESQUITE (Maddison and Maddison 2009) to simulate genealogies on it to match our sampling under two tree depths, with 10,000 trees for each depth. One depth was estimated from nuclear data, and another was 4× the depth, corresponding to an expected reduction in effective population size () for the mitochondrial genome. Simulated gene tree distributions also account for the number of samples for each data type (e.g., half the number for mtDNA). We then counted the number of deep coalescences required to fit each gene tree to the species tree and compared the values for empirical gene trees with the simulated distributions. If the number of deep coalescences for a given locus is greater than 95% of the simulated genealogies, we can reject the hypothesis that coalescent stochasticity is a source of discordance. These tests can be thought of as “global” tests of this hypothesis because no specific hybridization events are evaluated. If discordance is most apparent in one part of the tree, however, the test can be repeated using just that clade. We additionally tested specific “local” hypotheses (Table 2) by asking how many times a given phylogenetic relationship occurred between two species in the simulated data sets. For example, in the mtDNA tree, T. speciosus is nested within T. minimus, so we filtered the simulated data sets for a partition that reflects that relationship, treating its frequency as its probability given the model.
TABLE 2.
Summary of hypotheses of hybridization in Tamias
| Hypothesis | Time | Context | Test type | Outcome | P value |
| Global test of phylogenetic error | n/a | n/a | Parametric bootstrap | Reject null | <0.0066 |
| Global test of coalescent variance | n/a | n/a | Deep coalescences | Reject null | <0.001 |
| Southern Rocky Mountains clade | Recent | Within one clade | Deep coalescences | Reject null | <0.001 |
| minimus→panamintinus | Ancient | Nonsister taxa | Coalescent simulation | Reject null | <0.001 |
| minimus→speciosus | Ancient | Nonsister taxa | Coalescent simulation | Reject null | <0.001 |
| minimus→panamintinus/speciosus | Ancient | Nonsister taxa | Coalescent simulation | Reject null | <0.001 |
| minimus→T. a. cratericus | Ancient | Nonsister taxa | Coalescent simulation | Reject null | <0.001 |
| amoenus→T. m. grisescens | Ancient | Unresolved | Coalescent simulation | Fail to reject | 0.213 |
| ruficaudus→amoenus | Ancient and recent | Nonsister taxa | n/a | See Good et al (2008) | |
| T. ruficaudus ruficaudus→T. r. simulans | Recent | Sister taxa | n/a | See .Hird and Sullivan (2009) |
Notes:→indicates the direction of hypothesized introgression. Time column indicates whether or not putatively introgressed haplotypes are shared across species (recent) or divergent from putative donor taxon (ancient). Context column indicates phylogenetic context of hybridizing taxa. Deep coalescences indicate that we created a null distribution of deep coalescence scores to compare with the empirical Cytb genealogy. Coalescent simulation indicates that we filtered simulated gene-tree distributions to calculate the frequency of partitions present in Cytb that were hypothesized to be the result of hybridization.
In order to obtain appropriate empirical estimates of tree depth, we assumed a 3-year generation time, a divergence in the late Miocene (6 Ma; based on fossil data, Sutton 2002) and the largest θ estimated in LAMARC. Choosing the largest plausible θ is conservative, as it should make simulated gene tree discordance more likely, therefore making the null model harder to reject.
RESULTS
Sampling, Haplotype Resolution, and Patternsof Variation
We sampled 22 out of 25 named species of Tamias, with at least 2 samples from 21 of those, and up to 29 individuals in the case of the widespread species, T. minimus. In total, we collected 117 full and 165 partial genotypes (Table A1, GenBank accession numbers JN042160-JN043256, TreeBASE study TB2:S11722). Some adjacent exon sequence was also amplified with the intron loci. ACR contained 138 bp of coding sequence, ZAN contained none, and Zp2 contained 132 bp. Phase initially failed to resolve many haplotypes with high confidence, however; comparison of Phase-inferred haplotypes with subcloning results indicated the accuracy of Phase was high where it did make an assignment. For subclone-verified haplotypes, 96% of sites that Phase predicted with >90% posterior probability were correct. For sites predicted with <90% posterior probability, the method appeared to do little better than chance (61% correct). In total, 45 sites contained unresolved ambiguities after phasing. Nucleotide variation by locus is summarized in Table 1. Tests of recombination with DSS using the default sliding window size of 100 bp found no evidence of recombination within any of the nuclear loci. Tests of neutrality using Tajima's D yielded two significant deviations; in T. panamintinus and T. siskiyou at the Anon locus. HKA tests yielded no significant deviations; therefore, we do not regard these data as strongly deviating from neutrality.
Table 1.
Summary of nucleotide variation
| Locus | Length | Gene region* | n | nHaps | #PI | #var |
| Cytb | 1119 bp | ∼Entire gene | 168 | 147 | 360 | 413 |
| Anon | 751 bp | Noncoding | 139 | 134 | 99 | 138 |
| ACR | 1094 bp | ex1–ex2 | 149 | 75 | 107 | 147 |
| ZAN | 858 bp | ex25–ex27 | 141 | 111 | 112 | 144 |
| Zp2 | 851 bp | ex11–ex12 | 143 | 124 | 86 | 121 |
| Concatenated | 3584 bp | — | 117 | 198 | 429 | 521 |
| With gaps | 3616 | — | — | — | 461 | 553 |
n = number of individuals; nhaps = number of unique haplotypes; #PI = number of parsimony-informative sites; #var = number of variable sites.
All flanking exon sequences are partial.
Single-Locus Phylogenetic Analyses
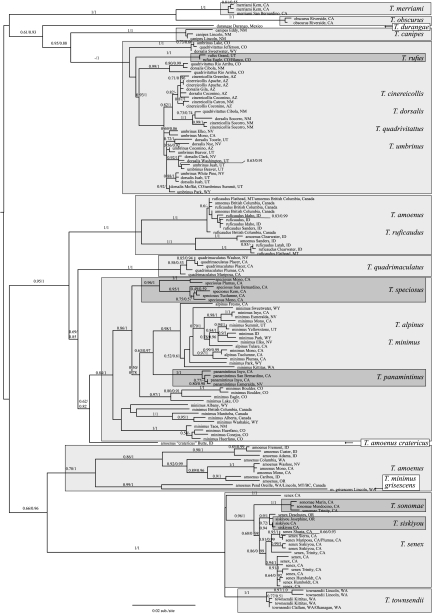
Models selected by DT-ModSel are listed in Table S2. Overall, topologies for nuclear loci were robust to the use of phased haplotype data versus unphased diploid sequences, model selection, and analysis method. The topology and branch support for Cytb, by contrast, were very sensitive to analysis method, model employed, partitioning scheme, and priors (in Bayesian analysis). We repeatedly observed what seem to be spurious topologies using both GARLI and MrBayes; in these, the outgroup was placed within T. minimus, leaving the rest of the tree nested within that species. Across runs that returned this topology, support values varied strongly. Examination of the posterior distribution of trees in multiple runs revealed that outgroup placement varied widely, reflecting significant uncertainty in this part of the tree. We therefore repeated our analyses with the outgroups pruned from the mtDNA (Fig. 2).
Figure 2.
ML tree, midpoint rooted, estimated from Cytb. Values on branches are ML bootstraps followed by Bayesian posterior probabilities. Species are indicated by shaded boxes, followed by country/state and province/county locales.
During the course of the single-locus analyses, we uncovered two populations that appeared to be substantially divergent from the taxa to which they have been historically assigned. One population corresponds to the subspecies T. minimus grisescens (Howell 1925) from the channeled scablands of central Washington. The other corresponds to samples identified as T. amoenus cratericus (Blossom 1937) from south-central Idaho (type locality, Craters of the Moon National Monument). In subsequent species-tree analyses, we treated these as independent lineages (named “T. m. grisescens” and “T. a. cratericus”).
Interlocus Conflict in Nuclear Loci
Three of our nuclear loci returned relatively well-supported topologies (Fig. 3a–c): ACR (Figs. 3a and S1), Anon (Figs. 3b and S2) and ZAN (Figs. 3c and S3). Zp2 (Figs. 3 and S4), although characterized by similar levels of variation, yielded trees with overall very low support values and many polyphyletic taxa. The three well-supported topologies conflict substantially with each other, containing mutually incompatible, yet statistically supported partitions (Bayesian posterior probability >0.95 or ML bootstrap >0.70; Fig. 3). In general, the only relationships corroborated by all three lociare characterized by shallow divergence such that the taxa in question are usually not reciprocally monophyletic in at least one locus (such as the southern Rocky Mountains group T. cinereicollis, T. canipes, T. dorsalis, T. canipes, T. rufus, T.quadrivitattus, and the Pacific Northwest group T. townsendii, T. senex, and T. siskiyou). The source of this conflict is not immediately evident as there is very little haplotype sharing, and with the exception of Zp2, nonmonophyly is only observed between taxa that are consistently closely associated in the gene trees.
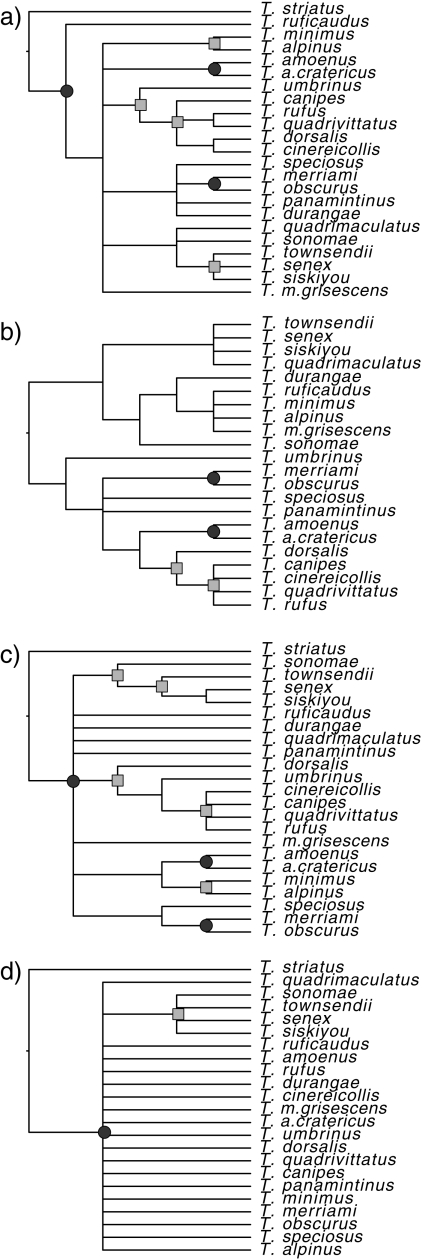
Figure 3.
Simplified nuclear gene-tree estimates. All species collapsed to a single tip. Only branches with either 0.7 bootstrap proportion or 0.95 posterior probability are retained. Partitions occurring in three gene trees marked with a circle, and two gene trees marked with a square. Tips contained in polytomies may be monophyletic or paraphyletic species (see supplementary figures).
Concatenated Analysis
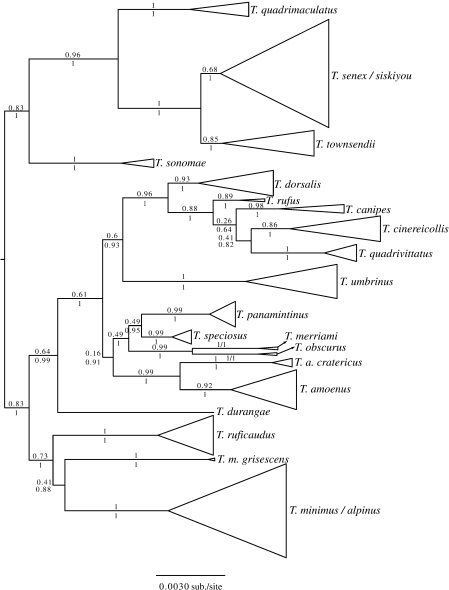
We could not amplify the Anon locus for either outgroup taxon, so they were excluded from the concatenated analysis (except for a Bayesian analysis following the above methods in which root placement was highly uncertain) and trees in figures are midpoint rooted. The AIC favored the partitioned model. We mapped the ML bootstrap and Bayesian posterior probabilities to the ML tree using SumTrees (Sukumaran and Holder 2010; Figs. and S5). Despite apparent conflict between loci, the analysis of the concatenated nuclear markers yields a tree that is strongly supported at many branches (Fig. 4). Additionally, with the exception of two species pairs (T. senex/T. siskiyou and T.minimus/T. alpinus), every species is recovered as monophyletic, most with strong support. Tamias senex and T. siskiyou are paraphyletic, and T. senex sequences tend to be nested within T. siskiyou. Tamias alpinus, is a monophyletic group nested within T. minimus and is characterized by strongly divergent ACR sequences in all three samples. This is consistent with expectations for a peripheral isolate of a widespread species. Branches with low support include relationships among three closely related southern Rocky Mountain species (T. cinereicollis, T. canipes, and T. quadrivittatus), and branches supporting relationships between previously proposed species groups (Levenson et al. 1985; Banbury and Spicer 2007) at the base of the tree. Finally, the Mexican endemic, T. durangae, is placed inconsistently across three well-supported nuclear genealogies, and in the concatenated analysis appears sister to a major clade, but this branch received little support.
Figure 4.
Phylogeny estimate from the concatenated nuclear data. The topology is the ML tree from GARLI and is midpoint rooted. Support values are ML bootstraps (above branches) and Bayesian posterior probabilities (below branches). Samples within species are collapsed for clarity. The full tree is in Figure S5).
Nuclear/mtDNA Conflict
The nuclear loci and Cytb phylogenies disagree substantially (Figs. 2 and 3); there are five instances of conflict that appear to be the result of interspecific introgressive hybridization. The first has been previously documented. Some individuals assigned to T. amoenus based on bacular morphology nest within T. ruficaudus in the Cytb phylogeny (Fig. 2; Good and Sullivan 2001, Good et al. 2003, Good et al. 2008, Demboski and Sullivan 2003). With the exception of Zp2, in these nuclear loci both taxa are distantly related. Zp2 indicates a potentially close relationship, but given the generally poor support in that locus, we have excluded it from further analyses. Second, T. speciosus and T. panamintinus are closely related to each other but distantly related to T. minimus at nuclear loci (Figs. 3, and S1–S3). Both, however, nest as monophyletic groups within T. minimus at Cytb (Fig. 2). Third, the group of southern Rocky Mountains species (T. umbrinus, T. dorsalis, T. rufus, T. quadrivittatus, T. cinereicollis, and T. canipes) appears relatively well sorted and monophyletic with nuclear markers but is characterized by low divergence and rampant nonmonophyly at Cytb. Fourth, the divergent T. minimus grisescens lineage is nested within T. amoenus at Cytb. Fifth, the divergent T. amoenus cratericus lineage appears sister to T. minimus at Cytb, but not at nuclear loci. Because mtDNA has a higher mutation rate and lower effective population size, it is expected to sort more quickly than nuclear DNA, making these conflicts between mitochondrial and nuclear phylogenies suggestive of multiple introgression events. These hypotheses are summarized in Table 2 and are tested using coalescent simulations below.
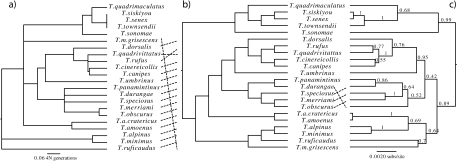
Species-Tree Estimates
Because of the strong incongruence among markers, we used three methods of phylogenetic estimation that account for incongruence assuming that it is due to incomplete lineage sorting. Estimates of θ per site from LAMARC used in these analyses ranged from 0.000796 (T. quadrivitattus) to 0.012119 (T. minimus; Table S3). The phylogeny estimates based on three nuclear loci from MDC (Fig. 5a) and STEM (Fig. 5b) and *BEAST (Fig. 5c) are broadly congruent with each other and with the nuclear concatenated tree (Fig. 4). *BEAST results using all four nuclear loci are extremely similar (Fig. S6). MDC and *BEAST differ only in the placement of T. durangae and T. amoenus. The Pacific Northwest group containing T. siskiyou, T. senex, T. townsendii, T. sonomae and T. quadrimaculatus, a southern Rocky Mountains group consisting of T. umbrinus, T. dorsalis, T. rufus, T. quadrivittatus, T. canipes and T. cinereicollis, and a group including T.durangae, T. speciosus, T. panamintinus, T. obscurus, and T. merriami(which are distributed across central/southern California, western Nevada and Baja Mexico) are corroborated by all three methods. These groupings are in general agreement with the concatenated estimate with the exception of T. durangae, the placement of which has very little support. Analyses with the three misidentified or revised specimens deleted yielded similar results. We also analyzed the data including Cytb. Its inclusion drastically altered the topology, forcing taxa that have undergone recent introgression (T. ruficaudus and T. amoenus) to be sister taxa, and having variable effects on those that have experienced putatively ancient introgression (Fig. S7).
Figure 5.
Species-tree estimates derived from STEM (a), MDC(b), and the maximum clade credibility tree from *BEAST (c) using three nuclear genes, ACR, Anon, and ZAN. Support values on the MDC and STEM trees are *BEAST posterior probabilities mapped onto the trees using SumTrees.
Assessment of Introgressive Hybridization
In order to assess more objectively the hypothesis that discordance between the mtDNA and the nuclear species-tree estimates is due to introgressive hybridization, we took a combination of simulation approaches. Our parametric bootstrap tests (a.k.a., SOWH tests) strongly rejected () the null hypothesis that the discordance between Cytb and the ML species tree is due solely to phylogenetic error.
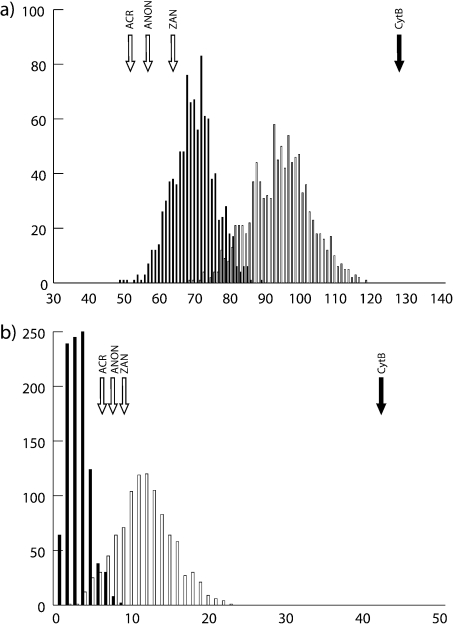
For coalescent simulations, we used the ML species tree estimated by STEM and simulated a tree depth of 8.6 generations (34.4 for mtDNA) by assuming a late Miocene divergence, a 3-year generation time and the largest θ estimated in LAMARC (that of T. minimus, 0.01). We generated distributions of deep coalescence scores for gene trees simulated at both the “nuclear” and “mitochondrial” tree depths for comparison (Fig. 5). Our first coalescent simulation strongly rejected the null hypothesis that the Cytb gene tree differed from the ML species tree as a result of coalescent stochasticity(; Fig. 6). Because the southern Rocky Mountains clade contains rampant nonmonophyly with respect to the mtDNA gene tree, we repeated this test solely on that group; we also rejected the null hypothesis (; Fig. 6). Using the simulated null distribution of gene trees, we also tested specific hypotheses of hybridization, detailed in Table 2. We filtered the gene-tree distribution for the following relationships that correspond to the mtDNA tree: that T. panamintinus, T. speciosus, or both group with T. minimus; that the T. amoenus cratericus lineage groups with T. minimus; and that the central Washington scablands lineage (T. minimus grisescens) groups with T. amoenus. The only case in which the null hypothesis of incomplete lineage sorting was not rejected was that of the association between T. amoenus and the T. minimus grisescens lineage.
Figure 6.
Null distributions for number of deep coalescences expected absent introgression and empirical test statistics for the full tree (a) and for the southern Rocky Mountains clade (b). Distributions and test statistics for nuclear DNA with a total species tree depth of 8.6 N generations are denoted by clear bars and arrows, those for mtDNA, with a total tree depth of 34.4 N generations, indicated by black bars and arrows.
DISCUSSION
The results presented here exemplify many of the challenges facing phylogenetic estimation in recent rapid radiations. Most notably, multiple unlinked gene genealogies conflict with one another at many statistically supported branches. This conflict is apparent both between mitochondrial and nuclear loci, and among nuclear loci. Nevertheless, the analysis of this multigene data set in a coalescent framework has provided a better understanding both of the phylogeny and of the population processes at work during diversification of the western North America species of chipmunks (subgenus Neotamias). There are some methodological issues with our analyses, however, that bear discussing.
Marker Selection
Three of the nuclear markers used in this study are introns from genes that code for proteins involved in fertilization reactions that are likely to have experienced some mode of selection at some point in their histories (Swanson et al. 2003, Turner and Hoekstra 2006), although a signal of selection was not detectable in this study. Though the use of genes thought to be under selection is not standard practice in phylogenetics, we selected these genes for two primary reasons. First, preliminary assessment of nuclear loci successfully used in higher-level phylogenetic analysis of rodents (Acp-5, C-myc, and Rag-1) suggested they would not be sufficiently variable for resolving relationships within Tamias (Good et al. 2008). Genes involved in reproduction, however, are frequently found to be rapidly evolving (Vacquier 1998; Swanson et al. 2003; Good and Nachman 2005) and thus may be a valuable source of phylogenetic information (Gatesy and Swanson 2007), especially for recent radiations. An important caveat, however, is that selection impacts the shape of gene genealogies, influencing the estimates of effective size and thus the probabilities of species trees. It is unclear what influence including genes affected by heterogeneous nonneutral evolutionary processes might have on species-tree inference. Second, because the chosen genes are directly involved in interactions between gametes during fertilization, they may be likely to acquire substitutions associated with reproductive isolation, making them less likely to introgress and more likely to reflect the “true” species history (e.g., Rieseberg et al. 1999, Maroja et al. 2009). In our study, two introns associated with these genes proved to be a useful source of phylogenetic information. We did not detect the effects of selection using tests that examine patterns of polymorphism, but because we used noncoding DNA we cannot assess amino acid substitution processes. We are also currently unable to assess the hypotheses of reduced introgression or elevated substitution at these loci, but with the rapid development of comparative genomics approaches we may be able to do so in the near future. Regardless, in an era of increasing genomic resources, researchers will have a large array of options in the markers they choose to employ, and both biological insight and efficiency of analysis may be achieved through the active selection of and comparison among different classes of markers.
Species-Tree Estimation
In this study, we employed three methods of species-tree estimation that account for gene tree incongruence: MDC, STEM, and *BEAST. These methods produced trees whose topologies are largely congruent with each other and also with our concatenated tree. It is clear from our analyses, however, that work remains to be done to increase the resolution of the species tree. Bayesian posterior probabilities of clades on the species trees are low and there is some conflict, particularly with respect to T. amoenus and T. durangae. There are several factors that complicate species-tree estimation. Most important to our study is the identification of multiple examples of introgressive hybridization in western North America chipmunks. As first indicated by previous research that documented hybridization between T. ruficaudus and T. amoenus (Good et al. 2003, Good et al. 2008), and expanded upon here, there have been many instances of interspecific mtDNA introgression in Tamias. In most of the cases examined here, much of this introgression has occurred between nonsister taxa; however, neither of the species-tree estimation methods used here account for gene tree discordance due to hybridization. Additionally, the robustness of these methods to the violation of this assumption is unclear (but see Eckert and Carstens 2008). We attempted to reduce the effects of this violation by comparing analyses excluding mtDNA from our species-tree analyses with those with the mtDNA included. When recent introgression between nonsister taxa results in shared mtDNA haplotypes (such as between T. ruficaudus and T. amoenus and between T. dorsalis and T. umbrinus), the effect was dramatic. The taxa spanning the deepest node in the species-tree estimates from nuclear data alone (T. amoenus and T. ruficaudus) are placed as sister taxa when the mtDNA are included. Conversely, for T. speciosus and T. panamintinus (putative recipients of an ancient introgression from T. minimus), their divergence-time estimate, but not topological placement, was altered in STEM, but in *BEAST they were placed as sister to T. minimus, T. alpinus. Although we see little clear evidence of hybridization in our nuclear loci, we cannot say with confidence that ancient introgression events have not caused some of the nuclear gene tree discordance at deeper nodes, and this may have biased our estimates in unpredictable ways.
An important assumption of STEM and MDC is that gene trees are estimated without error; failure to take into account uncertainty in topology (STEM and MDC) and branch lengths (STEM) caused by mutational variance may have strongly distorting effects on species-tree estimation. Indeed, though we excluded our most poorly supported gene tree (Zp2), there were still branches in the other gene trees with low support values and zero-length branches in the ML topologies. When zero-length branches occur between haplotypes or clades from different species (an issue expected to stem from the difficulty of finding sufficiently variable DNA regions in rapidly diverging groups), they may strongly affect STEM's estimates of topology and branch lengths. This may be evident in our ML species-tree estimate (Fig. 5), particularly in the case of the senex–siskiyou–townsendii grouping, where a visual inspection of nuclear gene trees would seem to indicate an obvious resolution of ((senex,siskiyou)townsendii), a result found with the *BEAST estimate. However, the ML species-tree estimate indicates a polytomy with zero-length branches. Estimates of nodal support in ML species-tree estimates (e.g., from nonparametric bootstrap replicates or Bayesian posterior distributions) should incorporate phylogenetic uncertainty in the gene-tree estimates, but it is unclear what the effects of polytomies stemming from a paucity of variation will be.
Assessment of Introgressive Hybridization
Several recent phylogenetic studies have presented cases in which there is very strong gene-tree discordance attributable to introgressive hybridization (Buckley et al. 2006; Linnen and Farrell 2008; Maureira-Butler et al. 2008; Bossu and Near 2009; Spinks and Shaffer 2009) and have addressed hypotheses about the source of the discordance in different ways. In their study of Neodiprion, Linnen and Farrell (2008) fit their data to the isolation-with-migration model (Hey and Nielsen 2004) in a pairwise fashion to estimate migration rates for all possible pairs of taxa in their tree. In further work on the genus, Linnen and Farrell (2008) used Bayesian tests of monophyly under the null hypotheses that if introgression is absent, more species should be recovered as monophyletic at mitochondrial loci than nuclear loci. Spinks and Shaffer (2009) used fossil calibrated divergence-date estimation to show a discrepancy between divergence times at different loci.
Here, we generally follow Buckley et al. (2006) in conducting coalescent simulations based on an estimate of the species tree to test both global hypotheses of gene-tree/species-tree fit (using counts of deep coalescences as our metric) as well as specific hypotheses of introgression. Our analyses indicated for both the full phylogeny and the southern Rocky Mountains clade that Cytb contained many more deep coalescent events than would be expected under a model of lineage divergence without migration (i.e., with no introgression). Additionally, four out of five tests of the frequency of relationships observed in the mtDNA tree indicated those relationships were unlikely to be observed under our model. This approach, however, comes with the caveat that it is conditioned on the validity of the model (species tree and effective population sizes) that we used to simulate the null distributions. Although we have discussed above potential issues with the topology and relative branch lengths of the species tree, there is also the issue of the absolute tree depth to be used in the coalescent simulation. We attempted to be conservative in coming to this estimate. Although this analysis ignores many sources of uncertainty, most of our assumptions are conservative in that they tend toward a short tree depth; this should make it harder to reject the null hypotheses that coalescent stochasticity is sufficient to explain the incongruence. This argument is partially born out by the observation that the nuclear loci generally have fewer deep coalescences than would be expected by chance under our model, indicating that we may have assumed an unrealistically short tree. This may, however, also be a result of the overall lower resolution of the empirical nuclear gene trees. When counting deep coalescences, polytomies are resolved in such a way as to minimize them, whereas the simulated gene trees are fully resolved, giving them more opportunities to conflict with the species tree. We have also assumed stable effective sizes and no population subdivision within species. Changing effective population sizes may either increase or decrease the rate of coalescence within populations, biasing our null distributions in unknown ways. Population structure may increase the time to coalescence within a species (Nordborg 2001), making gene tree discordance more likely. Finally, an important issue in this analysis is the original assignment of individuals to species. Multilocus genotypes seemed to indicate that three specimens were misidentified or have been the subject of taxonomic revisions (indicated by asterisks in Table A1). In these cases, we treated them as such and reassigned them. Exclusion of these specimens had no effect on species-tree estimation. Their inclusion as originally identified in the species-tree analyses would likely have caused drastic changes in the topology and had unpredictable effects on our hypothesis tests. This points to an area where current methods need further development. Although methods for simultaneous species delimitation and species-tree estimation are becoming available (O'Meara 2010, Yang and Rannala 2010), their models still do not account for introgression.
Causes and Consequences of Introgressive Hybridization
Although introgressive hybridization is becoming increasingly well documented in animals, we still have a poor understanding of its causes and consequences. In terms of causation, neutral hypotheses posit that incomplete reproductive isolation and genetic drift can result in the introduction and spread of a mitochondrial type (Wirtz 1999, Chan and Levin 2005). These explanations often invoke reproductive isolating factors such as Haldane's (1922) rule or mechanical isolation to explain the frequently asymmetric nature of introgression. Chan and Levin (2005) modeled mate choice and hybridization at species range edges and found that mtDNA could introgress rapidly and unidirectionally at a contact zone when one species is rare, the other is common, and females are highly selective. In this case, females of the rare species eventually accept heterospecific matings, whereas rare males cannot find mates. This facilitates the introgression of the rare species mtDNA into the common species. Alternatively, selection may cause widespread introgression. In a study of a hybrid zone between Manacu svitellinus and Manacu scandei in Central America, Stein and Uy (2006) found that a divergent cline in plumage coloration could be explained by female preference for a heterospecific trait. In the case of mtDNA, recent work has found evidence for pervasive nonneutral evolution (Bazin et al. 2006), and specific patterns of regional mtDNA variation have been suggested to be a result of altitudinal or climatic adaptation (Cheviron and Brumfield 2009, Ruiz-Pesini et al. 2004).
In Tamias, a number of studies have documented hybridization between T. amoenus and T. ruficaudus and between subspecies of T. ruficaudus (Good and Sullivan 2001, Good et al. 2003, Good et al. 2008, Hird and Sullivan 2009, Hird et al. 2010, Reid et al. 2010) at distinct contact zones. The predominant pattern is one of unidirectional mtDNA introgression with evidence for either very little (Reid et al. 2010), or substantial (Hird and Sullivan 2009) nuclear gene flow accompanying it. Given the current information, it is difficult to distinguish between neutral and selective hypotheses for these introgression events. Many of the species in the southern Rocky Mountain clade partition into different ecological niches characterized by differing habitats and elevations (e.g., Bergstrom 1992, Brown 1971), forming a multitude of discrete contact zones throughout their respective ranges. The widespread distribution of these contact zones throughout the southern Rockies may be a driver in the high level of introgression apparent in our analyses. Further understanding of the actual mechanisms will require a broader survey of nuclear markers that can identify the rates and level of heterogeneity of nuclear introgression and analyses of the signature of selection in both patterns of polymorphism and amino acid substitution.
SUPPLEMENTARY MATERIAL
Supplementary material, including data files and/or online-only appendices, can be found at http://www.sysbio.oxfordjournals.org/.
FUNDING
Funding for this work was provided by National Science Foundation (DEB-0717426 to J.S., DEB-0716200 to J.D.), the Denver Museum of Nature & Science, and National Institute of Health (P20 RR16448 to The Institute for Bioinformatics and Evolutionary Studies).
Supplementary Material
Acknowledgments
We would like to acknowledge the contributions of K. Bell, D. Nagorsen, K. Reiss, K. Ellis, and J. Good for collection of specimens and for identification of bacular morphology. The following museums provided tissue samples: Royal British Columbia Museum, Denver Museum of Nature & Science, Museum of Southwestern Biology, Field Museum of Natural History, Humboldt State University Vertebrate Museum, and Museum of Vertebrate Zoology. B. Carstens, M. Koopman, J. McVay, D. Tryant, S. Nuismer, M. Webster, J. Cloud, S. Hird, R. Debry, E. Jockusch and two anonymous reviewers provided valuable comments on the manuscript.
TABLE A1.
Specimen information
| Name | Sample | Species | New locality | Source |
| n3136 | MSB43429 (NK3136) | townsendii | WA: Kittitas Co. | MSB |
| n3253 | MSB43547 (NK3253) | townsendii | WA: Clallam Co. | MSB |
| n8531 | MSB58069 (NK8531) | townsendii | WA: Okanogan Co. | MSB |
| jeb279 | UWBM79489 (JEB279) | townsendii | WA: Lincoln Co. | UWBM |
| jeb283 | UWBM79493 (JEB283 | townsendii | WA: Lincoln Co. | UWBM |
| meh013 | UWBM78741 (MEH013) | townsendii | WA: Kittitas Co. | UWBM |
| m152777 | MVZ152777 | sonomae | CA: Marin Co. | MVZ |
| kzr33 | KZR33 | sonomae | CA: Mendocino Co. | KZR |
| kzr89 | KZR89 | sonomae | CA: Trinity Co. | KZR |
| kzr59 | KZR59 | sonomae | CA: Trinity Co. | KZR |
| w2920 | W2920 | siskiyou | Near HSU | HSUVM |
| w2924 | W2924 | siskiyou | Near HSU | HSUVM |
| w2925 | W2925 | siskiyou | Near HSU | HSUVM |
| kzr66 | KZR66 | siskiyou | OR: Josephine Co. | KZR |
| kzr67 | KZR67 | siskiyou | OR: Josephine Co. | KZR |
| jrd075 | JRD075 | senex | OR: Deschutes Co. | SL |
| m196677 | MVZ196677 | senex | CA: Shasta Co. | MVZ |
| m207216 | MVZ207216 | senex | CA: Mariposa Co. | MVZ |
| m208612 | MVZ208612 | senex | CA: Sierra Co. | MVZ |
| k5032 | K5032 | senex | CA: Plumas Co. | SL |
| k4919 | K4919 | senex | CA: Plumas Co. | SL |
| w2921 | W2921 | senex | Near Humboldt State University (HSU) | HSUVM |
| w2922 | W2922 | senex | Near HSU | HSUVM |
| w2923 | W2923 | senex | Near HSU | HSUVM |
| w2926 | W2926 | senex | Near HSU | HSUVM |
| w2927 | W2927 | senex | Near HSU | HSUVM |
| w2928 | W2928 | senex | Near HSU | HSUVM |
| w2929 | W2929 | senex | Near HSU | HSUVM |
| kzr2 | KZR2 | senex | CA: Siskiyou Co. | KZR |
| kzr5 | KZR5 | senex | CA: Siskiyou Co. | KZR |
| kzr39 | KZR39 | senex | CA: Humboldt Co. | KZR |
| kzr40 | KZR40 | senex | CA: Humboldt Co. | KZR |
| kzr60 | KZR60 | senex | CA: Trinity Co. | KZR |
| kzr110 | KZR110 | senex | CA: Trinity Co. | KZR |
| d185 | ZM.11203 (DZTM185) | rufus | CO: Eagle Co. | DMNS |
| n56221 | MSB76520 (NK56221) | rufus | CO: Rio Blanco Co. | MSB |
| m199281 | MVZ199281* | rufus (quadrivittatus) | UT: Grand Co. | MVZ |
| d156 | ZM.11091 (DZTM156) | canipes | NM: Lincoln Co. | DMNS |
| d158 | ZM.11093 (DZTM158) | canipes | NM: Eddy Co. | DMNS |
| d160 | ZM.11095 (DZTM160) | canipes | NM: Lincoln Co. | DMNS |
| d151 | ZM.11086 (DZTM151 | cinereicollis | NM: Socorro Co. | DMNS |
| d152 | ZM.11087 (DZTM152) | cinereicollis | NM: Socorro Co. | DMNS |
| d223 | ZM.11110 (DZTM223) | cinereicollis | NM: Catron Co. | DMNS |
| d226 | ZM.11111 (DZTM226) | cinereicollis | AZ: Greenlee Co. | DMNS |
| d229 | ZM.11114 (DZTM229) | cinereicollis | AZ: Apache Co. | DMNS |
| d230 | ZM.11115 (DZTM230) | cinereicollis | AZ: Apache Co. | DMNS |
| d231 | ZM.11116 (DZTM231) | cinereicollis | AZ: Coconino Co. | DMNS |
| d237 | ZM.11378 (DZTM237) | cinereicollis | AZ: Coconino Co. | DMNS |
| m197180 | MVZ197180 | dorsalis | AZ: Coconino Co. | MVZ |
| d154 | ZM.11089 (DZTM154) | dorsalis | NM: Socorro Co. | DMNS |
| d201 | ZM.11393 (DZTM201) | dorsalis | CO: Moffat Co. | DMNS |
| d217 | ZM.11133 (DZTM217) | dorsalis | NM: Cibola Co. | DMNS |
| d236 | ZM.11121 (DZTM236) | dorsalis | AZ: Gila Co. | DMNS |
| d250 | ZM.11144 (DZTM250) | dorsalis | WY: Sweetwater Co. | DMNS |
| h6491 | HSU6491 | dorsalis | UT: Juab Co. | HSU |
| n55375 | MSB76753 (NK55375) | dorsalis | UT: Washington Co. | MSB |
| n55222 | MSB76872 (NK55222) | dorsalis | UT: Beaver Co. | MSB |
| n43680 | MSB86620 (NK43680) | dorsalis | UT: Tooele Co. | MSB |
| h5705 | HSU5705 | dorsalis | NV: Clark Co. | HSUVM |
| h6543 | HSU6543 | dorsalis | NV: Nye Co. | HSUVM |
| h5504 | HSU5504 | umbrinus | NV: Elko Co. | HSUVM |
| h6070 | HSU6070 | umbrinus | UT: Beaver Co. | HSUVM |
| h6474 | HSU6474 | umbrinus | UT: Juab Co. | HSUVM |
| n55419 | MSB76754 (NK55419) | umbrinus | UT: Beaver Co. | MSB |
| d163 | ZM.11188 (DZTM163) | umbrinus | CO: Lake Co. | DMNS |
| d238 | ZM.11379 (DZTM238) | umbrinus | AZ: Coconino Co. | DMNS |
| d251 | ZM.11160 (DZTM251) | umbrinus | UT: Summit Co. | DMNS |
| d256 | ZM.11165 (DZTM256) | umbrinus | UT: Summit Co. | DMNS |
| d267 | ZM.11147 (DZTM267) | umbrinus | WY: Park Co. | DMNS |
| h5779 | HSU5779 | umbrinus | CA: Mono Co. | HSUVM |
| h6239 | HSU6239 | umbrinus | NV: White Pine Co. | HSUVM |
| d60 | ZM.11031 (DZTM60) | quadrivittatus | CO: Jefferson Co. | DMNS |
| d70 | ZM.11024 (DZTM70) | quadrivittatus | CO: Jefferson Co. | DMNS |
| d85 | ZM.11078 (DZTM85) | quadrivittatus | NM: Rio Arriba Co. | DMNS |
| d87 | ZM.11085 (DZTM87) | quadrivittatus | NM: Rio Arriba Co. | DMNS |
| d176 | ZM.11096 (DZTM176) | quadrivittatus | CO: Saguache Co. | DMNS |
| d218 | ZM.11134 (DZTM218) | quadrivittatus | NM: Cibola Co. | DMNS |
| m201430 | MVZ201430 | alpinus | CA: Tuolumne Co. | MVZ |
| m206400 | MVZ206400 | alpinus | CA: Tulare Co. | MVZ |
| m206395 | MVZ206395 | alpinus | CA: Fresno Co. | MVZ |
| jrd107 | JRD107 | quadrimaculatus | NV: Washoe Co. | SL |
| m201431 | MVZ201431 | quadrimaculatus | CA: Mariposa Co. | MVZ |
| k7133 | K7133 | quadrimaculatus | CA: Plumas Co. | KZR |
| kzr90 | KZR90 | quadrimaculatus | CA: Placer Co. | KZR |
| kzr91 | KZR91 | quadrimaculatus | CA: Placer Co. | KZR |
| kwe61 | KWE061 | merriami | CA: Kern Co. | DMNS |
| kwe4 | KWE004 | merriami | CA: Kern Co. | DMNS |
| kwe21 | KWE021 | merriami | CA: San Bernardino Co. | DMNS |
| kwe51 | KWE051 | obscurus | CA: Riverside Co. | DMNS |
| kwe49 | KWE049 | obscurus | CA: Riverside Co. | DMNS |
| n69922 | MSB84515 (NK69922)a | speciosus (senex) | CA: Tuolumne Co. | MSB |
| n73186 | MSB90785a | speciosus (umbrinus) | CA: Mono Co. | MSB |
| k4216 | K4216 | speciosus | CA: Plumas Co. | SL |
| jrd288 | JRD288 | speciosus | CA: Mono Co. | DMNS |
| kwe13 | KWE013 | speciosus | CA: San Bernardino Co. | DMNS |
| kwe3 | KWE003 | speciosus | CA: Kern Co. | DMNS |
| m196353 | MVZ196353 | panamintinus | CA: San Bernardino Co. | MVZ |
| m199507 | MVZ199507 | panamintinus | CA: Inyo Co. | MVZ |
| m216375 | MVZ216375 | panamintinus | CA: Inyo Co. | MVZ |
| jrd176 | JRD176 | panamintinus | CA: Inyo Co. | DMNS |
| jrd280 | ZM.11505 | panamintinus | NV: Esmeralda Co. | DMNS |
| jeb900 | UWBM77758 | minimus | WA: Lincoln Co. | UWBM |
| jeb910 | UWBM77768 | minimus | WA: Lincoln Co. | UWBM |
| jr2438 | UWBM78465 | minimus | WA: Kittitas Co. | UWBM |
| m197184 | MVZ197184 | minimus | NV: Elko Co. | MVZ |
| m202384 | MVZ202384 | minimus | CA: Mono Co. | MVZ |
| m216306 | MVZ216306 | minimus | CA: Inyo Co. | MVZ |
| m217078 | MVZ217078 | minimus | CA: Plumas Co. | MVZ |
| n7876 | MSB53280 | minimus | Canada: Manitoba | MSB |
| n2113 | MSB55759 | minimus | Canada: Alberta | MSB |
| n55538 | MSB77073 | minimus | UT: Summit Co. | MSB |
| n147828 | MSB15209 | minimus | ID: Butte Co. | MSB |
| jrd281 | ZM.11504 | minimus | NV: Esmeralda Co. | DMNS |
| jrd326 | JRD326 | minimus | CA: Mono Co. | DMNS |
| d6 | ZM.10995 (DZTM6) | minimus | CO: Huerfano Co. | DMNS |
| d13 | ZM.10996 (DZTM13) | minimus | CO: Huerfano Co. | DMNS |
| d76 | ZM.11027 (DZTM76) | minimus | CO: Boulder Co. | DMNS |
| d77 | ZM.11026 (DZTM77) | minimus | CO: Boulder Co. | DMNS |
| d90 | ZM.11082 (DZTM90) | minimus | NM: Taos Co. | DMNS |
| d161 | ZM.11186 (DZTM161) | minimus | CO: Lake Co. | DMNS |
| d184 | ZM.11202 (DZTM184) | minimus | CO: Eagle Co. | DMNS |
| d206 | ZM.11122 DZTM206) | minimus | CO: Conejos Co. | DMNS |
| d275 | ZM.11155 (DZTM275) | minimus | WY: Washakie Co. | DMNS |
| r19761 | RBCM19761 | minimus | Canada: British Columbia | RBCM |
| jrd41 | JRD041 | minimus | WY: Park Co. | SL |
| d261 | ZM.11170 (DZTM261) | minimus | UT: Summit Co. | DMNS |
| d242 | ZM.11180 (DZTM242) | minimus | WY: Albany Co. | DMNS |
| h5463 | HSU5463 | minimus | NV: Nye Co. | DMNS |
| d247 | ZM.11141 (DZTM247) | minimus | WY: Sweetwater Co. | DMNS |
| d265 | ZM.11145 (DZTM265) | minimus | WY: Park Co. | DMNS |
| smh08 | SMH008 | ruficaudus | ID: Kootenai Co. | SL |
| jrd022 | JRD022 | ruficaudus | ID: Idaho Co. | SL |
| jmg113 | JMG113 | ruficaudus | MT: Flathead Co. | SL |
| jms209 | JMS209 | ruficaudus | WA: Pend Oreille Co. | SL |
| jmg161 | JMG161 | ruficaudus | MT: Sanders Co. | SL |
| jmg139 | JMG139 | ruficaudus | MT: Flathead Co. | SL |
| jmg077 | JMG077 | ruficaudus | ID: Idaho Co. | SL |
| jms257 | JMS257 | ruficaudus | ID: Clearwater Co. | SL |
| jms127 | JMS127 | ruficaudus | ID: Latah Co. | SL |
| nmr040 | NMR040 | ruficaudus | MT: Flathead Co. | SL |
| r19920 | RBCM19920 | ruficaudus | Canada: British Columbia | RBCM |
| jms265 | JMS265 | amoenus | ID: Adams Co. | SL |
| jrd120 | JRD120 | amoenus | ID: Butte Co. | SL |
| jrd309 | JRD309 | amoenus | CA: Mono Co. | SL |
| jrd108 | JRD108 | amoenus | ID: Custer Co. | SL |
| nmr31 | NMR031 | amoenus | Canada: British Columbia | SL |
| r19921 | RBCM19921 | amoenus | Canada: British Columbia | RBCM |
| jms198 | JMS198 | amoenus | MT: Lincoln Co. | SL |
| jrd128 | JRD128 | amoenus | ID: Fremont Co. | SL |
| jrd063 | JRD063 | amoenus | WA: Columbia C. | SL |
| jrd126 | JRD126 | amoenus | ID: Caribou Co. | SL |
| jrd104 | JRD104 | amoenus | NV: Washoe Co. | SL |
| jms207 | JMS207 | amoenus | WA: Pend Oreille Co. | SL |
| jmg005 | JMG005 | amoenus | OR: Harney Co. | SL |
| jmg160 | JMG160 | amoenus | MT: Sanders Co. | SL |
| jmg057 | JMG057 | amoenus | ID: Clearwater Co. | SL |
| r19771 | RBCM19771 | amoenus | Canada: British Columbia | RBCM |
| r19886 | RBCM19886 | amoenus | Canada: British Columbia | RBCM |
| kzr10 | KZR10 | amoenus | CA: Mono Co. | KZR |
| kzr11 | KZR11 | amoenus | CA: Mono Co. | KZR |
| uvm217 | UVM217 | durangae | Mexico: Durango | UVM |
| avb01 | UWBM77286 | sibiricus | Russia: PrimorskyKray | UWBM |
| can003 | CAN003 | striatus | SL | |
| cgg003 | CGG003 | striatus | SL |
DMNS (ZM) = Denver Museum of Nature & Science; HSUVM = Humboldt State University Vertebrate Museum; KZR = sample on loan from Karen Z. Reiss; MSB = Museum of Southwestern Biology; MVZ = Berkeley Museum of Vertebrate Zoology; RBMC = Royal British Columbia Museum; SL = indicates sample resides in laboratory of Jack Sullivan; UWBM = University of Washington Burke Museum; UVM = Zadock Thompson Natural History Collection, University of Vermont.
These specimen records were determined by multilocus genotype or locality to be misidentified or not reflective of taxonomic revisions and reassigned to this taxon.
References
- Akaike H. A new look at statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- Alfaro ME, Santini F, Brock CD, Alamillo H, Dornburg A, Carnevale G, Rabosky D, Harmon LJ. Vol. 106. Proc. Natl. Acad. Sci. U.S.A; 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates; pp. 13410–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banbury JL, Spicer GS. Molecular systematics of chipmunks (Neotamias) inferred by mitochondrial control region sequences. J. Mammal. Evol. 2007;14:149–162. [Google Scholar]
- Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- Bergstrom BJ. Parapatry and encounter competition between chipmunk (Tamias) species and the hypothesized role of parasitism. Am. Midl. Nat. 1992;128:168–179. [Google Scholar]
- Blossom PM. Description of a race of chipmunk from south central Idaho. Occas. Papers Mus. Zool. 1937;366:1–3. [Google Scholar]
- Bossu CM, Near TJ. Gene trees reveal repeated instances of introgression in orangethroat darters (Percidae: Etheostoma) Syst. Biol. 2009;58:114–129. doi: 10.1093/sysbio/syp014. [DOI] [PubMed] [Google Scholar]
- Brown JH. Mechanisms of competitive exclusion between two species of chipmunks. Ecology. 1971;52:305–311. [Google Scholar]
- Buckley TR, Cordeiro M, Marshall DC, Simon C. Differentiating between hypotheses of lineage sorting and introgression in New Zealand alpine cicadas (Maoricicada Dugdale) Syst. Biol. 2006;55:411–425. doi: 10.1080/10635150600697283. [DOI] [PubMed] [Google Scholar]
- Brown JM, Hedtke SM, Lemmon AR, Lemmon EM. When trees grow too long: investigating the causes of highly inaccurate Bayesian branch-length estimates. Syst. Biol. 2010;59:145–161. doi: 10.1093/sysbio/syp081. [DOI] [PubMed] [Google Scholar]
- Carling MD, Brumfield RT. Speciation in Passerina buntings: introgression patterns of sex-linked loci identify a candidate region for reproductive isolation. Mol. Ecol. 2009;18:834–847. doi: 10.1111/j.1365-294X.2008.04038.x. [DOI] [PubMed] [Google Scholar]
- Chan K, Levin S. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution. 2005;59:720–729. [PubMed] [Google Scholar]
- Cheviron ZA, Brumfield RT. Migration-selection balance and local adaptation of mitochondrial haplotypes in rufous-collared sparrows (Zonotrichia capensis) along an elevational gradient. Evolution. 2009;63:1593–1605. doi: 10.1111/j.1558-5646.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:762–768. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demboski JR, Sullivan J. Extensive mtDNA variation within the yellow-pine chipmunk, Tamias amoenus (Rodentia: Sciuridae), and phylogeographic inferences for northwest North America. Mol. Phylogent. Evol. 2003;26:389–408. doi: 10.1016/s1055-7903(02)00363-9. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. 3rd ed. New York: Columbia University Press; 1951. [Google Scholar]
- Dowling TE, Secor CL. The role of hybridization in the evolutionary diversification of animals. Ann. Rev. Ecol. Syst. 1997;28:593–619. [Google Scholar]
- Eckert AJ, Carstens BC. Does gene flow destroy phylogenetic signal? The performance of three methods for estimating species phylogenies in the presence of gene flow. Mol. Phylogent. Evol. 2008;49:834–842. doi: 10.1016/j.ympev.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2008;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Liu L, Pearl DK. Vol. 104. Proc. Natl. Acad. Sci. U.S.A; 2007. High resolution species trees without concatenation; pp. 5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. New York: Dover; 1958. [Google Scholar]
- Funk DJ, Omland KE. Species-level paraphyly and polyphyly: frequency, causes, consequences, with insight from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003;34:397–423. [Google Scholar]
- Gannon W, Sikes RS. Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J. Mammal. 2007;88:809–823. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatesy J, Baker RH. Hidden likelihood support in genomic data: can forty-five wrongs make a right. 2005;54:483–492. doi: 10.1080/10635150590945368. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Swanson WJ. Adaptive evolution and phylogenetic utility of ACR (Acrosin), a rapidly evolving mammalian fertilization gene. J. Mammal. 2007;88:32–42. [Google Scholar]
- Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- Good JM, Demboski JR, Nagorsen DW, Sullivan J. Phylogeography and introgressive hybridization: Chipmunks (genus: Tamias) in the northern Rocky Mountains. Evolution. 2003;57:1900–1916. doi: 10.1111/j.0014-3820.2003.tb00597.x. [DOI] [PubMed] [Google Scholar]
- Good JM, Hird S, Reid N, Demboski JR, Steppan SJ, Martin-Nims TR, Sullivan J. Ancient hybridization and mitochondrial capture between two species of chipmunks. Mol. Ecol. 2008;17:1313–1327. doi: 10.1111/j.1365-294X.2007.03640.x. [DOI] [PubMed] [Google Scholar]
- Good JM, Nachman MW. Rates of protein evolution are positively correlated with timing of expression during mouse spermatogenesis. Mol. Bio. Evol. 2005;22:1044–1052. doi: 10.1093/molbev/msi087. [DOI] [PubMed] [Google Scholar]
- Good JM, Sullivan J. Phylogeography of the red-tailed chipmunk (Tamias ruficaudus), a northern Rocky Mountain endemic. Mol. Ecol. 2001;10:2683–2695. doi: 10.1046/j.0962-1083.2001.01397.x. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 1922;12:101–109. [Google Scholar]
- Harrison RG. Hybrid zones: windows on evolutionary process. Oxford Surv. Evol. Biol. 1990;7:69–128. [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. HKA—a computer program for tests of natural selection. [Internet] Available from. 2004 http://lifesci.rutgers.edu/∼heylab/ProgramsandData/Programs/HKA/HKA_Documentation.htm. [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird S, Reid N, Demboski J, Sullivan J. Introgression at differentially aged hybrid zones in red-tailed chipmunks. Genetica. 2010;138:869–883. doi: 10.1007/s10709-010-9470-z. [DOI] [PubMed] [Google Scholar]
- Hird S, Sullivan J. Assessing gene flow across a hybrid zone in red-tailed chipmunks (Tamias ruficaudus) Mol. Ecol. 2009;18:3097–3109. doi: 10.1111/j.1365-294X.2009.04196.x. [DOI] [PubMed] [Google Scholar]
- Holder MT, Anderson JA, Holloway AK. Difficulties in detecting hybridization. Syst. Biol. 2001;50:978–982. doi: 10.1080/106351501753462911. [DOI] [PubMed] [Google Scholar]
- Howell AH. Preliminary descriptions of five new chipmunks from North America. J. Mammal. 1925;6:51–54. [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J, Ronquist F. MRBAYES: Bayesian inference of phylogenetic tress. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Knowles LL, Carstens BC. Delimiting species without monophyletic gene trees. Syst. Biol. 2007;56:887–895. doi: 10.1080/10635150701701091. [DOI] [PubMed] [Google Scholar]
- Kubatko LS, Carstens BC, Knowles LL. STEM: species-tree estimation using maximum-likelihood for gene trees under coalescence. Bioinformatics. 2009;25:971–973. doi: 10.1093/bioinformatics/btp079. [DOI] [PubMed] [Google Scholar]
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Kuhner MK. LAMARC 2.0: Maximum likelihood and Bayesian estimation of population parameters. Bioinformatics Appl. Note. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Levenson H, Hoffmann RS, Nadler CF, Deutsch L, Freeman SD. Systematics of the holarctic chipmunks (Tamias) J. Mammal. 1985;66:219–242. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Farrell BD. Phylogenetic analysis of nuclear and mitochondrial genes reveals evolutionary relationships and mitochondrial introgression in the sertifer species group of the genus Neodiprion (Hymenoptera: Diprionidae) Mol. Phylogent. Evol. 2008;48:240–257. doi: 10.1016/j.ympev.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Liu L, Pearl DK. Species trees from gene trees: Reconstructing Bayesian posterior distributions from a species phylogeny using estimated gene tree distributions. Syst. Biol. 2007;56:504–514. doi: 10.1080/10635150701429982. [DOI] [PubMed] [Google Scholar]
- Llopart A, Lachaise D, Coyne JA. Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea. Genetics. 2005;171:197–210. doi: 10.1534/genetics.104.033597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- Maddison WP. Gene trees in species trees. Syst. Biol. 1997;46:523–536. [Google Scholar]
- Maddison WP, Knowles L. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 2006;55:21–30. doi: 10.1080/10635150500354928. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade. Sunderland (MA): Sinauer& Associates. 2003 [Google Scholar]
- Maddison DR, Maddison WP. Mesquite: a modular system for evolutionary analysis, version 2.71 [Internet] 2009 Available from: http://mesquiteproject.org/mesquite/mesquite.html. [Google Scholar]
- Maroja LS, Andres JA, Harrison RG. Genealogical discordance and patterns of introgression and selection across a cricket hybrid zone. Evolution. 2009;63:2999–3015. doi: 10.1111/j.1558-5646.2009.00767.x. [DOI] [PubMed] [Google Scholar]
- Maureira-Butler IJ, Pfeil BE, Muangprom A, Osborn TC, Doyle JJ. The reticulate history of medicago (Fabaceae) Syst. Biol. 2008;57:466–482. doi: 10.1080/10635150802172168. [DOI] [PubMed] [Google Scholar]
- Mayr E. Cambridge (MA): Harvard University Press; 1963. Animal Species and Evolution. [Google Scholar]
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 2009;25:126–127. doi: 10.1093/bioinformatics/btn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 2003;52:674–683. doi: 10.1080/10635150390235494. [DOI] [PubMed] [Google Scholar]
- Mossel E, Roch S. Incomplete lineage sorting: consistent phylogeny estimation from multiple loci. IEEE Comput. Biol. Bioinform. 2008;7:166–171. doi: 10.1109/TCBB.2008.66. [DOI] [PubMed] [Google Scholar]
- Nadler CF, Hoffmann RS, Honacki JH, Pozin D. Chromosomal evolution in chipmunks, with a special emphasis on A and B karyotypes of the subgenus Neotamias. Am. Midl. Nat. 1977;98:343–353. [Google Scholar]
- Nordborg M. Coalescent theory. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of statistical genetics. Chichester (UK): Wiley; 2001. pp. 179–212. [Google Scholar]
- O'Meara BC. New heuristic methods for joint species delimitation and species tree inference. Syst. Biol. 2010;59:59–73. doi: 10.1093/sysbio/syp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P, Nei M. Relationships between gene trees and species trees. Mol. Biol. Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Patterson BD, Thaeler CS., Jr The mammalian baculum: hypotheses on the nature of bacular variability. J. Mammal. 1982;63:1–15. [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol. Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Piaggio AJ, Spicer GS. Molecular phylogeny of the chipmunks inferred from mitochondrial cytochrome b and cytochrome oxidase II gene sequences. Mol. Phylogent. Evol. 2001;20:335–350. doi: 10.1006/mpev.2001.0975. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4 [Internet] 2007 Available from: http://beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Reid N, Hird S, Schulte-Hostedde A, Sullivan J. Examination of nuclear loci across a zone of mitochondrial introgression between Tamias ruficaudus and Tamias amoenus. J. Mammal. 2010;91:1389–1400. [Google Scholar]
- Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- Schluter D. Oxford (UK): Oxford University Press; 2000. The Ecology of Adaptive Radiation. [Google Scholar]
- Schwenk K, Brede N, Streit B. Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philos. Trans. R. Soc. Lond., Ser. B. 2008;363:2805–2811. doi: 10.1098/rstb.2008.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinks PQ, Shaffer HB. Conflicting mitochondrial and nuclear phylogenies for the widly disjunct Emys (Testudines:Emydidae) species complex and what they tell us about biogeography and hybridization. Syst. Biol. 2009;58(1):1–20. doi: 10.1093/sysbio/syp005. [DOI] [PubMed] [Google Scholar]
- Stein AC, Uy JAC. Unidirectional spread of a secondary sexual trait across an avian hybrid zone. Evolution. 2006;60:1476–1485. [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. DendroPy: a python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- Sullivan J, Arellano EA, Rogers DS. Comparative phylogeography of Mesoamerican highland rodents: concerted versus independent responses to past climatic fluctuations. Am. Nat. 2000;155:755–768. doi: 10.1086/303362. [DOI] [PubMed] [Google Scholar]
- Sutton DA. Tamias amoenus. Mamm. Species. 1992;390:1–8. [Google Scholar]
- Sutton DA. Problems of taxonomy and distribution in fourspecies of chipmunks. J. Mammal. 1995;76:843–850. [Google Scholar]
- Sutton DA, Patterson BD. Geographic variation of the western chipmunks Tamiassenex and T. siskiyou, with two new subspecies from California. J. Mammal. 2000;81:299–316. [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2000 PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sunderland (MA): Sinauer and Associates. [Google Scholar]
- Than C, Nakhleh L. Species tree inference by minimizing deep coalescences. PLoS Comput. Biol. 2009 doi: 10.1371/journal.pcbi.1000501. 5:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Hoekstra HE. Adaptive evolution of fertilization proteins within a genus: variation in Zp2 and Zp3 in deer mice (Peromyscus) Mol. Biol. Evol. 2006;23:1656–1669. doi: 10.1093/molbev/msl035. [DOI] [PubMed] [Google Scholar]
- Vacquier VD. Evolution of gamete recognition proteins. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. [DOI] [PubMed] [Google Scholar]
- Ward PS, Brady SG, Fisher BL, Schultz TR. Phylogeny and biogeography of Dolichoderine ants: effects of data partitioning and relict taxa on historical inference. Syst. Biol. 2010;59:342–362. doi: 10.1093/sysbio/syq012. [DOI] [PubMed] [Google Scholar]
- Wasserman PM, Jovine L, Litscher ES. A profile of fertilization in mammals. Nat. Cell Biol. 2001;3:E59–E64. doi: 10.1038/35055178. [DOI] [PubMed] [Google Scholar]
- White JA. Vol. 5. Univ. Kansas Publ. Mus. Nat. Hist; 1953. The baculum in the chipmunks of western North America; pp. 611–631. [Google Scholar]
- Wilgenbusch JC, Warren DL, Swofford DL. AWTY: a system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference [Internet] 2004 doi: 10.1093/bioinformatics/btm388. Available from: http://ceb.csit.fsu.edu/awty. [DOI] [PubMed] [Google Scholar]
- Thorington RW, Jr, Hoffmann RS. Family Sciuridae. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd edn. Baltimore (MD): John Hopkins University Press; 2005. pp. 754–818. [Google Scholar]
- Wirtz P. Mother species-father species: unidirectional hybridization in animals with female choice. Anim. Behav. 1999;58:1–12. doi: 10.1006/anbe.1999.1144. [DOI] [PubMed] [Google Scholar]
- Wu CI. The genic view of the process of speciation. J. Evol. Biol. 2001;14:851–865. [Google Scholar]
- Yang Z, Rannala B. Vol. 107. Proc. Natl. Acad. Sci. U.S.A; 2010. Bayesian species delimitation using multilocus data; pp. 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Austin (TX): University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [dissertation] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.