Abstract
Caenorhabditis elegans let-7, a founding member of the microRNA family, is predicted to bind to six sites in the 3′UTR of the mRNA of its target gene, lin-41, to down-regulate LIN-41. Here, we demonstrate that wild-type let-7 microRNA binds in vitro to RNA from the lin-41 3′UTR. This interaction is dependent on two conserved let-7 complementary sites (LCSs). A 27-nucleotide sequence between the LCSs is also necessary for down-regulation in vivo. LCS mutations compensatory to the lesion in let-7(n2853) can partially restore lin-41 3′UTR function in a let-7(n2853) background, providing the first experimental evidence for an animal miRNA binding directly to its validated target in vivo.
Keywords: stRNA, microRNA, heterochronic, developmental timing, 3′UTR
Genes that regulate the timing of Caenorhabditis elegans post-embryonic development are called heterochronic genes. Several heterochronic genes are regulated post-transcriptionally through elements in their 3′UTR. This regulation is mediated by small temporal RNAs (stRNAs), like lin-4 and let-7, expressed at the first larval stage (L1) and L3 stage, respectively (Wightman et al. 1993; Feinbaum and Ambros 1999; Reinhart et al. 2000; Johnson et al. 2003). stRNAs are predicted to interact with partially complementary sequences in the 3′UTRs of their target genes and down-regulate gene expression through an unknown mechanism; however, the direct binding of an stRNA to its validated target has not been demonstrated experimentally in vivo.
lin-4 is thought to act by antisense base pairing to seven complementary sites in the lin-14 3′UTR to down-regulate lin-14 post-transcriptionally (Lee et al. 1993; Wightman et al. 1993). The 3′UTR of lin-14 is sufficient for temporal down-regulation; experiments using a lacZ reporter gene fused to the lin-14 3′UTR show stage-specific down-regulation (Wightman et al. 1993). The seven complementary sites in the lin-14 3′UTR are also conserved in Caenorhabditis briggsae, a closely related Caenorhabditis species, and are deleted in lin-14 gain-of-function mutants, demonstrating that these sites are crucial to normal lin-14 regulation (Wightman et al. 1993; Ha et al. 1996). Binding of lin-4 to lin-14 is thought to block protein synthesis or accumulation after the initiation of translation (Olsen and Ambros 1999).
let-7 controls the larval-to-adult (L/A) transition and let-7 mutants display retarded terminal differentiation of seam cells (Reinhart et al. 2000). lin-41 mutants display precocious terminal differentiation of these cells at the L4 stage (Slack et al. 2000). As judged by LIN-41/GFP fusion, LIN-41 appears to be down-regulated in the seam cells during the L4 stage, coinciding with the up-regulation of let-7 RNA (Slack et al. 2000). As in the case with lin-14, the lin-41 3′UTR placed behind a heterologous promoter is sufficient to down-regulate a reporter gene. This down-regulation requires let-7 and a region in the lin-41 3′UTR with sites complementary to let-7.
let-7 and lin-41 are conserved from C. elegans to mammals, and LCSs are found in Drosophila and zebrafish lin-41 3′UTRs (Pasquinelli et al. 2000; Slack et al. 2000). This conservation suggests that the mechanism of gene regulation is also present in higher eukaryotes. let-7 has targets in addition to lin-41, namely, hbl-1, which has eight predicted LCSs in its 3′UTR (Lin et al. 2003).
lin-4 and let-7 are the founding members of a larger family of microRNAs (miRNAs). MiRNAs are genomically encoded, untranslated RNA molecules of ∼20-25 nucleotides (nt), transcribed as precursors that can form a hairpin loop (Ambros et al. 2003). For example, the let-7 miRNA precursor RNA is ∼70 nt and is processed into a mature 22-nt molecule by the RNAse III nuclease Dicer (dcr-1 in C. elegans; Bernstein et al. 2001; Grishok et al. 2001).
Numerous groups have reported the cloning of >100 miRNAs using HeLa cell culture, Drosophila (Lagos-Quintana et al. 2001; Mourelatos et al. 2002), C. elegans (Lau et al. 2001; Lee and Ambros 2001; Lim et al. 2003), plants (Reinhart et al. 2002), and embryonic stem cells (Houbaviy et al. 2003). Like let-7, (Pasquinelli et al. 2000), some of the recently discovered miRNAs have been shown to be developmentally regulated and are highly conserved in Drosophila and humans (Lau et al. 2001; Mourelatos et al. 2002). The function of some of these novel miRNAs has now been deciphered. In Drosophila, bantam encodes a miRNA involved in cell proliferation and cell death (Brennecke et al. 2003), and mir-14 plays a role in cell death and fat metabolism (Xu et al. 2003). Loss of mir-15/16 leads to chronic lymphocytic leukemia (CLL) in humans (Calin et al. 2002). It is now apparent that a novel mechanism of gene regulation involving untranslated RNA molecules, first described with lin-4 and let-7, may be a general and ancient one.
To investigate the mechanism(s) by which animal miRNAs regulate their targets, we have dissected the interaction between the let-7 miRNA and one of its target genes, lin-41. We have determined that the sequences both necessary and sufficient for lin-41 down-regulation map to three elements in the lin-41 3′UTR, two of which are complementary to let-7. We also show that let-7 binds both in vitro and in vivo to RNA from the lin-41 3′UTR containing these two conserved LCSs, presenting the first evidence for a direct interaction between an animal miRNA and imperfect complementary sites in a validated miRNA target.
Results and Discussion
Multiple conserved LCSs in the lin-41 3′UTR
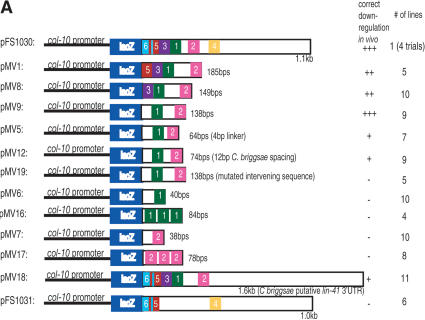
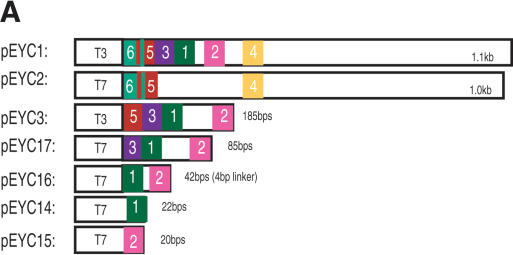
The 1.1-kb lin-41 3′UTR is sufficient to down-regulate a lacZ reporter gene driven by the col-10 promoter in a let-7-dependent manner, during late larval development in C. elegans (Reinhart et al. 2000). Additionally, an 85-bp deletion of the lin-41 3′UTR compromises temporal down-regulation of the reporter (Reinhart et al. 2000). Within this 85-bp region, are at least two sequences complementary to the let-7 miRNA, except for small bulged regions within the predicted duplexes. We found four additional putative LCSs, sites 3-6, in the lin-41 3′UTR with a lower predicted binding affinity to let-7 than have the previously reported sites (LCS 1-LCS 2, Supplemental Fig. S1a,b). Of the six predicted LCSs in the 3′UTR of lin-41 (Supplemental Fig. S1b), three are removed (LCS 1-LCS 3) in the 85-bp deletion (pFS1031, Fig. 1A).
Figure 1.
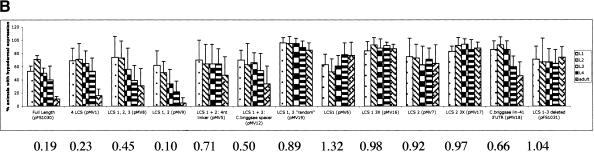
LCSs are necessary and sufficient for proper down-regulation in vivo. (A) Schematic of in vivo LCS constructs. All share a common col-10 promoter fused to the Escherichia coli lacZ reporter gene. Each construct contains various LCSs from the Caenorhabditis elegans lin-41 3′UTR inserted into the unc-54 3′UTR. pMV18 contains Caenorhabditis briggsae lin-41 DNA. (+++) 0%-12% of animals with hypodermal expression at the adult stage); (++) 13%-32%); (+) 33%-60%); (-) >60%. (B) In vivo lacZ expression analysis. We analyzed multiple lines for all constructs and averaged the data for each construct. Error bars indicate standard deviation between lines/trials where indicated. See Supplemental materials for number of lines/animals scored. Numbers below graph refer to ratio of hypodermal lacZ expression between adult vs. L1-L3 animals.
LCS 1 and LCS 2 are conserved in C. briggsae
At the protein level, C. elegans lin-41 is well conserved in C. briggsae (Stein et al. 2003). The NHL domain (Slack and Ruvkun 1998; Slack et al. 2000) of LIN-41 is 95% conserved between species as determined by BLAST analysis. In addition, sequences in the 3′UTR of C. briggsae lin-41 are also conserved, and the C. briggsae lin-41 3′UTR was able to partially down-regulate a reporter gene during C. elegans development (see below).
To determine which of the six 3′UTR LCS sequences may be important for lin-41 function, we performed a sequence alignment between the C. elegans and C. briggsae lin-41 3′UTRs (Supplemental Fig. S2). LCS 1 and LCS 2 are 100% conserved, suggesting they are important for LIN-41 down-regulation. LCS 3-LCS 6, in contrast, are poorly conserved, suggesting that they are less relevant for lin-41 function.
We investigated the ability of the C. briggsae lin-41 3′UTR to down-regulate a reporter gene in C. elegans by fusing 1.5kb of sequence downstream of the C. briggsae lin-41 stop codon to our lacZ reporter (Fig. 1A, pMV18). lacZ expression was down-regulated over time in vivo; 46% of adult animals expressed the reporter gene as compared with ∼90% of early larval animals. However, this down-regulation was less effective than the C. elegans lin-41 3′UTR (Fig. 1B, pFS1030), in which only 11% of adult animals showed expression compared with ∼80% of early larval animals. This demonstrates that sequences in the C. briggsae lin-41 3′UTR are capable of providing partial lin-41 down-regulation.
Two conserved LCSs are sufficient for LIN-41 down-regulation in vivo
We determined the minimal sequence in the lin-41 3′UTR that is necessary and sufficient for proper lin-41 down-regulation. Like the entire 3′UTR (pFS1030; Reinhart et al. 2000), LCS 1, LCS 2, LCS 3, and LCS 5together (pMV1) were found to be sufficient for proper reporter gene down-regulation (Fig. 1B). Further, only LCS 1 and LCS 2 together (pMV9) could direct proper down-regulation of the reporter; 5% of adult animals expressed the reporter gene. Interestingly, neither LCS 1 (pMV6) nor LCS 2 (pMV7) alone was sufficient to effect down-regulation of the reporter gene. Increasing the number of bulged siRNA/miRNA-binding sites in the 3′UTR of a reporter gene increases the degree of repression (Doench et al. 2003). To test whether we could achieve reporter gene down-regulation with multiple copies of either LCS 1 or LCS 2, we concatemerized either LCS 1 (pMV16) or LCS 2 (pMV17) three times with a 4-nt linker between each LCS. This did not result in lin-41 down-regulation in vivo (Fig. 1B). These experiments indicate either that only both copies of LCS 1 and LCS 2 together are sufficient for lin-41 down-regulation, and/or that the intervening 27-nt sequence between LCS 1 and LCS 2 is also required. We distinguished between these below.
The intervening sequence between LCS 1 and LCS 2 is required for full down-regulation
To determine the significance of the 27-nt spacer sequence between LCS 1 and LCS 2, we investigated the contribution of the spacer sequence length and the specific spacer sequence itself. We found that LCS 1 and LCS 2 with an artificial 4-nt linker was insufficient for reporter down-regulation (pMV5, Fig. 1B). Similarly, replacing the 27-nt native sequence with the C. briggsae 12-nt spacer sequence (pMV12) also did not restore complete reporter down-regulation. These experiments demonstrate that the spacer sequence is important for 3′UTR down-regulation activity. A shortened sequence replacing the native 27-nt C. elegans spacer sequence may cause the let-7 molecules to encounter steric hindrance and interfere with their ability to effectively bind lin-41 in vivo and/or function properly. Alternatively, the intervening sequence may provide a binding site for additional necessary factors, either protein or RNA.
To test the importance of the specific sequence of the 27-nt spacer, we altered the sequence, but not length of the spacer between LCS 1 and LCS 2 (pMV9 vs. pMV19, Supplemental Materials). We found that 85% of adult animals carrying the altered 27-nt sequence in pMV19 showed lacZ expression compared with 5% of pMV9 containing adult animals with the native 27 nt (Fig. 1B). This demonstrates that the specific sequence between LCS 1 and LCS 2 is required for lin-41 down-regulation. We speculate that this sequence binds additional necessary factors not yet identified, but we cannot rule out that by mutating the 27-nt sequence we may have introduced alternative secondary structure that may inhibit the binding of let-7. Mfold predictions do not, however, suggest secondary structure in this region of the 3′UTR (data not shown; Mathews et al. 1999; Zuker et al. 1999).
Point mutations in the LCS compromise reporter gene regulation in wild-type animals in vivo
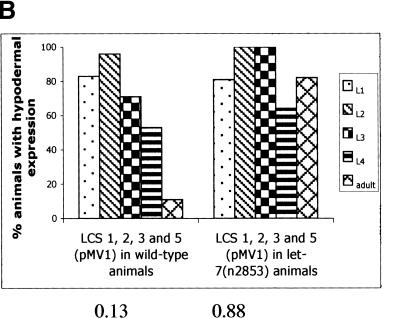
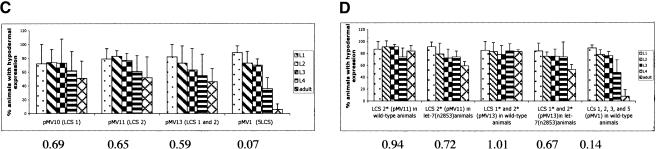
To understand better the in vivo interaction of the let-7 miRNA with its target sequences in the lin-41 3′UTR, we engineered point mutations in the LCSs. The let-7(n2853) lesion is a G to A point mutation at position 5 on the let-7 RNA (Reinhart et al. 2000), that is predicted to disrupt the base pairing between LCS and let-7 (Supplemental Fig. S1b; Fig. 2A). Consistent with this, let-7(n2853) animals carrying col-10-lacZ- LCS 1, LCS 2, LCS 3, and LCS 5(pMV1) failed to down-regulate the reporter gene at the adult stage, demonstrating that wild-type let-7 RNA is required for proper down-regulation of this reporter in vivo (Fig. 2B).
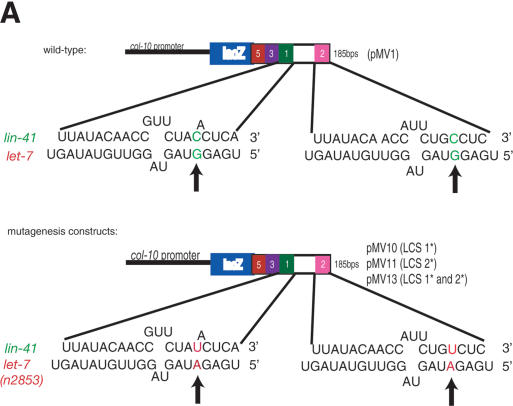
Figure 2.
Effect of point mutations in LCS 1 and LCS 2. (A) The sequence of the wild-type LCS 1 and LCS 2, the predicted base pairing with the let-7 RNA, and mutations compensatory to let-7(n2853) in the LCSs of the lin-41 3′UTR. Arrows indicate base nucleotide substitutions (C to U) made in LCS 1, LCS 2, or both, and predicted base pairing with the let-7(n2853) RNA. (B-D) See Supplemental materials for number of lines/animals scored. All data were analyzed as in Fig. 1B. Numbers below graph refer to ratio of hypodermal expression between adult vs. L1-L3 animals. (B) Down-regulation of col-10-lacZ-LCS 1, LCS 2, LCS 3, and LCS 5(pMV1) is abrogated in let-7(n2853) animals. (C) Effect of LCS 1* and LCS 2* mutations in a wild-type background. (D) A let-7(n2853) compensatory mutation in LCS 2 and LCS 1 partially restores down-regulation in a let-7(n2853) background. Lines that showed the most severe lack of down-regulation at the adult stage in a wild-type background were assayed.
We engineered nucleotide substitutions compensatory to the let-7(n2853) mutation in reporter constructs carrying the LCSs, changing the corresponding C to U in LCS 1 or LCS 2, or both LCS 1 and LCS 2 (Fig. 2A) of pMV1. The presence of a point mutation (designated *) in all three constructs in a wild-type background compromised down-regulation of the reporter gene at the adult stage in vivo, with between 40% and 50% of adults expressing the reporter (pMV10, pMV1011, and pMV1013, Fig. 2C). This compares with the <20% expression at the adult stage seen with the control wild-type plasmid, pMV1.
Thus, single point mutations in LCS 1 or LCS 2 that potentially disrupt let-7/lin-41 duplex formation resulted in decreased reporter down-regulation in wild-type animals (Fig. 2A,C). Although these data provide evidence that LCS 1 and LCS 2 are necessary for full LIN-41 down-regulation, surprisingly, we did not observe an additive effect when LCS 1 and LCS 2 were mutated concurrently; the expression pattern of pMV13 at the adult stage was comparable with pMV10 and pMV1011 at the same stage (Fig. 2C). There may be some other sequence(s) responsible for target down-regulation in the LCS 1* (pMV10) and LCS 2* (pMV11) constructs, a role perhaps played by the 27-nt spacer sequence between LCS 1 and LCS 2. These data, along with the deletion study (pFS1031, ΔLCS 1-LCS 3, Fig. 1B) indicate that both LCS 1 and LCS 2 and the specific 27-nt spacer sequence are necessary and sufficient for the regulation of our reporter, and by inference, lin-41 itself.
let-7 binds to specific complementary sequences from the lin-41 3′UTR in vivo
To establish in vivo experimental evidence for the direct binding of the let-7 RNA to the lin-41 3′UTR, we tested whether let-7(n2853) animals with reporter constructs with compensatory LCS* mutations could show reporter gene down-regulation at the adult stage. This was accomplished by crossing animals carrying mutant transgenic arrays carrying the LCS 2* (pMV11) or both LCS 1* and LCS 2* (pMV13) mutations into let-7(n2853) animals (Fig. 2A). Comparing lacZ expression patterns of LCS 2* (pMV11) in let-7(n2853) and wild-type adult animals, we saw 25% greater adult reporter gene down-regulation in let-7(n2853) animals; lacZ expression was seen in 84% of wild-type animals versus 59% of let-7(n2853) animals (Fig. 2D). These data indicate that enabling at least one mutant let-7 molecule to bind to one LCS in the lin-41 3′UTR and form a let-7 RNA (n2853)/lin-41* RNA duplex (at LCS 2) partially rescues down-regulation at the adult stage in vivo. We hypothesized that increasing the potential binding sites for mutated let-7 (LCS 1* and LCS 2*, pMV13) would increase the repression of lacZ expression at the adult stage. However, we observed only 30% greater (slightly higher than that of LCS 2* alone) adult reporter gene down-regulation in let-7(n2853) animals; lacZ expression was seen in 83% of wild-type animals (pMV13) versus 53% of let-7(n2853) animals carrying LCS 1* and LCS 2* (Fig. 2D). The demonstration that compensatory mutations in LCS 1 and LCS 2 partially restore reporter down-regulation in a let-7(n2853) mutant strongly suggests that let-7 binds directly to at least LCS 2.
A likely explanation as to why the compensatory mutations in LCS 1 and LCS 2 did not restore full down-regulation of lin-41 at the adult stage in a let-7(n2853) background is that let-7 RNA levels are reduced in let-7(n2853) animals (Reinhart et al. 2000). If let-7 levels are below a threshold for activity in a let-7(n2853) mutant, no amount of complementarity would completely rescue. Another possibility is that another LCS (LCS 3 and/or LCS 5, LCS 6, for example) may need to be mutated (*) to allow for better down-regulation of lin-41 at the adult stage in a let-7(n2853) background.
let-7 binds to RNA from the lin-41 3′UTR in vitro
We tested whether the let-7 RNA can bind to RNA from the lin-41 3′UTR in vitro. 32P-labeled in vitro-transcribed wild-type let-7 RNA and mutant let-7(n2853) RNA were incubated separately with three in vitro-transcribed versions of lin-41 3′UTR RNA, respectively, and the resulting complexes were resolved by native gel electrophoresis (Fig. 3B). The wild type let-7 binds to and retards the mobility of the full-length lin-41 3′UTR (pEYC1; Fig. 3A,B), as well as RNA with LCS 1, LCS 2, LCS 3, and LCS 5(pEYC3; Fig. 3A,B, lane 4), but not to a lin-41 3′UTR deleted for LCS 1-LCS 3 (pEYC2; Fig. 3A,B, lanes 2,3). let-7 also bound to the RNA sequence containing LCS 1, LCS 2, and LCS 3 (Fig. 3C, lane 6) but not to in vitro-transcribed RNA containing either LCS 1 or LCS 2 alone (Fig. 3c, lanes 2,3). However, we could detect binding of let-7 to a synthetic RNA that contained both LCS 1 and LCS 2 linked to each other by a 4-nt linker. These data showed that let-7 RNA can bind to lin-41 3′UTR RNA in vitro, and that a sequence containing LCS 1, LCS 2, and LCS 3 within a 185-nt region is necessary and sufficient for binding. The minimal sequence required for let-7 binding was LCS 1 and LCS 2 (Fig. 3C, lane 5), supporting the finding that the minimal sequence determined to be sufficient for lin-41 down-regulation in vivo contains LCS 1 and LCS 2 (see above). We observe multiple retarded complexes (small arrows, Fig. 3B, lane 4) that may represent multiple loading of let-7 molecules at various LCSs.
Figure 3.
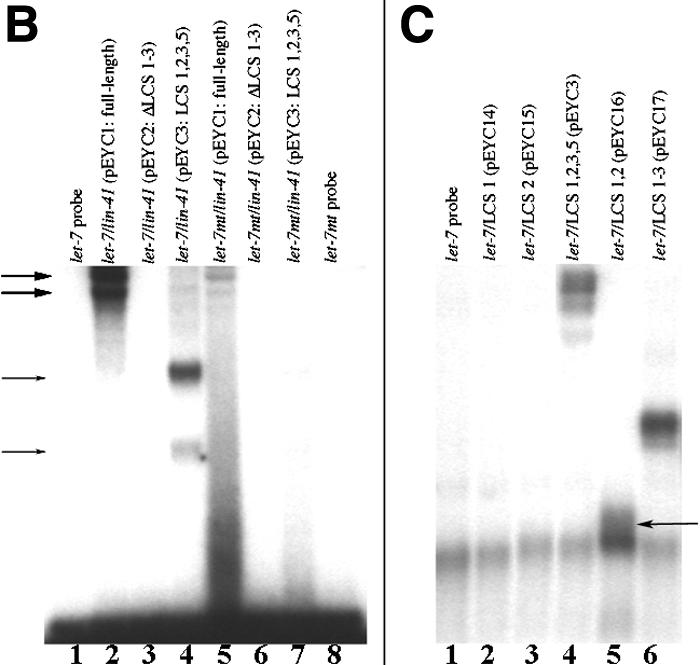
let-7 can bind to sequences from the lin-41 3′UTR in vitro. (A) Schematic of in vitro LCS constructs used to generate RNAs. (B) Wild-type let-7 RNA, but not let-7(n2853) mutant RNA can bind the full-length lin-41 3′UTR RNA. Lanes as indicated; retarded products indicated by two large arrows (lane 1), and two small arrows (lane 4). (C) LCS 1 and LCS 2 are sufficient to bind let-7 RNA in vitro. Lanes as indicated. The let-7 free probe has run off the gel and is not shown.
The mutant variant of let-7 RNA, carrying a single point mutation corresponding to let-7(n2853) shows lower in vitro-binding affinity than the wild-type let-7 RNA to the full-length lin-41 3′UTR RNA (Fig. 3B, lane 5) and to the RNA with LCS 1, LCS 2, LCS 3, and LCS 5 (Fig. 3B, lane 7). This suggests that the let-7(n2853) mutation destabilizes the let-7/lin-41 RNA duplex, consistent with the thermodynamic calculations of the effects of the G to A transition on duplex formation (data not shown; Supplemental Fig. S1b).
Conclusion
Our data define the minimal region responsible for lin-41 3′UTR down-regulation activity in vivo. This region contains two sequences complementary to let-7, LCS 1, and LCS 2, as well as a specific 27-nt spacer sequence. This work reveals that more than one miRNA complementary site is required for down-regulation activity. Perhaps let-7 miRNAs act in a cooperative fashion, stabilizing each other's interaction with the 3′UTR of their target gene, thereby increasing the degree of regulation imparted on their target gene. It has been shown in HeLa cell culture that increasing the number of miRNA/siRNA-binding sites increases the degree of repression of a target gene (Doench et al. 2003). By using in vivo transgenic arrays of a reporter gene and endogenously expressed let-7, we find that it is not enough to merely multimerize miRNA complementary sites. We predict that there are probably other factors, RNA or protein, that need to be present along with wild-type let-7 to ensure proper lin-41 repression at the adult stage. In the case of lin-41, this activity may reside in the LCS spacer sequence.
We altered the sequence of the 27-nt spacer and found that these changes severely affect lin-41 down-regulation activity. The spacer sequence may be important for secondary structure of the lin-41 3′UTR and/or for binding of additional required factors such as a specific RNA-binding protein and/or complex, or another miRNA. Conserved sequences flanking other miRNA-binding sites have been found in the hbl-1 3′UTR that are similar to NRE elements known to bind regulatory proteins such as Pumillio (Lin et al. 2003).
In this study, we demonstrate that our let-7 and lin-41 RNAs directly bind each other in vitro, and in vivo strongly suggesting that native let-7 and lin-41 bind each other. We have demonstrated for the first time that an animal miRNA binds to its target RNA in vivo through imperfect complementary sequences.
Materials and methods
Plasmid constructions, injections, and lacZ expression analysis
All plasmids with designation pMV#:DNA fragments were digested with SacII and NcoI and ligated into the unc-54 3′UTR of the B29 vector (Reinhart et al. 2000) cut with SacII and NcoI behind the Escherichia coli lacZ reporter gene. B29 contains the col-10 promoter fused to lacZ, and the unc-54 3′UTR, a 3′UTR thought to be absent of temporal regulatory elements (Wightman et al. 1993). Refer to published methods for pFS1030 and pFS1031 (Reinhart et al. 2000). For primer sequences and insert data, see Supplemental Table 1. Resulting plasmid sequences were verified by sequencing, injected into wild-type animals (5ng/μL) with injection marker pRF4 (rol-6) (80 ng/μL) to create extrachromosomal array lines. Refer to Supplemental materials for β-galactosidase assays. Control animals contained extrachromosomal array mgEx540 (pFS1030, 5ng/μL; Slack et al. 2000).
Site-directed mutagenesis and in vitro RNA binding
To make mutations in the lin-41 3′UTR LCS 1 and LCS 2 (designated 1* and 2*) plasmids pMV10, pMV11, and pMV13 were constructed as described in Supplemental materials. In vitro let-7/lin-41 duplex formation is described in Supplemental materials.
Acknowledgments
We thank members of the Slack Lab for experimental support and review of the manuscript, C. Guerrier-Takada (Yale University, New Haven, CT) for advice with in vitro binding assays, T-W. Lo (Yale University, New Haven, CT) for C. briggsae DNA, and M. Koelle (Yale University, New Haven, CT) for help with data analysis. This work was supported by an NIH grant to F.J.S (# GM 62594).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1165404.
References
- Ambros V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., et al. 2003. A uniform system for microRNA annotation. RNA 9: 277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 295-296. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25-36. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Dumitru, C.D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 99: 15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum R. and Ambros, V. 1999. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev. Biol. 210: 87-95. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C. 2001. Genes and mechanisms related to RNA interference regulate expression of small temporal RNAs that control C. elegans developmental timing. Cell 106: 23-34. [DOI] [PubMed] [Google Scholar]
- Ha I., Wightman, B., and Ruvkun, G. 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 10: 3041-3050. [DOI] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific MicroRNAs. Dev. Cell 5: 351-358. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Lin, S.Y., and Slack, F.J. 2003. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 259: 364-379. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862-864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843-854. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003. The microRNAs of Caenorhabditis elegans. Genes & Dev. 17: 991-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Johnson, S.M., Abraham, M., Vella, M.C., Pasquinelli, A., Gamberi, C., Gottlieb, E., and Slack, F.J. 2003. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639-650. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911-940. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671-680. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A., Reinhart, B., Slack, F., Maller, B., and Ruvkun, G. 2000. Conservation across animal phylogeny of the sequence and temporal regulation of the 21 nucleotide C. elegans let-7 heterochronic regulatory RNA. Nature 408: 86-89. [DOI] [PubMed] [Google Scholar]
- Reinhart B., Slack, F., Basson, M., Pasquinelli, A., Bettinger, J., Rougvie, A., Horvitz, R., and Ruvkun, G. 2000. The 21 nucleotide let-7 RNA regulates C. elegans developmental timing. Nature 403: 901-906. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. and Ruvkun, G. 1998. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 23: 474-475. [DOI] [PubMed] [Google Scholar]
- Slack F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the lin-29 transcription factor. Mol. Cell 5: 659-669. [DOI] [PubMed] [Google Scholar]
- Stein L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855-862. [DOI] [PubMed] [Google Scholar]
- Xu P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790-795. [DOI] [PubMed] [Google Scholar]
- Zuker M., Mathews, D.H., and Turner, D.H. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide in RNA biochemistry and biotechnology. Kluwer Academic Publishers, New York.