Abstract
Rrp12p (Ypl012w) is unusual among characterized ribosome synthesis factors in being associated with late precursors to both the 40S and 60S subunits. Rrp12p is predominately nuclear with nucleolar enrichment at steady state, but shuttled between the nucleus and cytoplasm in a heterokaryon assay. Strains depleted of Rrp12p are impaired in the nuclear export of both ribosomal subunits. Sequence analysis combined with fold recognition and modeling showed that Rrp12p is a member of a family of pre-ribosome-associated HEAT-repeat proteins. Like other HEAT-repeat transport factors, Rrp12p binds in vitro to nucleoporin FG-repeats of both the GLFG and FXFG families and to the GTPase Gsp1p (yeast RAN). Rrp12p also showed robust in vitro binding to a pre-rRNA transcript, in addition to poly(A) and poly(U). We propose that Rrp12p binds to the RNA components of the pre-ribosomes and promotes export of both subunits via its interactions with the nucleoporins and Gsp1p.
Keywords: Ribosome export
Ribosome biogenesis in eukaryotes mainly occurs in a specialized nuclear compartment, the nucleolus, and the newly synthesized subunits must therefore be transported through the nucleoplasm and nuclear pore complexes (NPCs) in order to reach their site of function in the cytoplasm. In actively growing cells, ribosomal subunit export is a major transport activity, but the large, rigid, hydrophilic structures of the pre-ribosomal particles appear to make them difficult transport substrates. The assembly of the pre-ribosomal particles and nuclear export is integrated with the processing of the pre-rRNAs to the mature rRNAs. In the yeast Saccharomyces cerevisiae, the pre-40S particles that are exported from the nucleus contain the 20S pre-rRNA, which is reported to undergo base-dimethylation and final cleavage to mature 18S rRNA in the cytoplasm (see Fig. 1). In contrast, the 25S and 5.8S rRNAs are believed to be fully matured within the nucleus prior to their export in pre-60S particles.
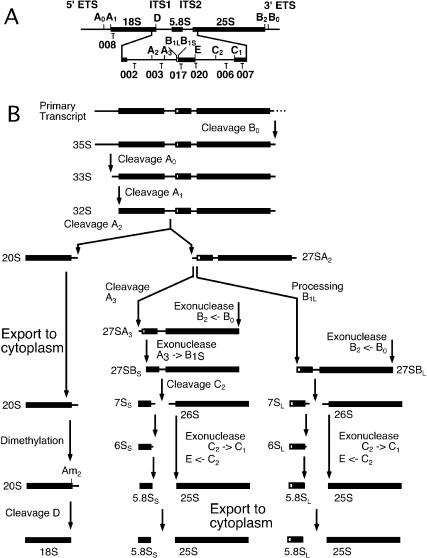
Figure 1.
Pre-rRNA processing in S. cerevisiae. (A) Structure and processing sites of the 35S pre-rRNA. This precursor contains the sequences for the mature 18S, 5.8S, and 25S, which are separated by the two internal transcribed spacers ITS1 and ITS2 and flanked by the two external transcribed spacers 5′ETS and 3′ETS. The positions of the oligonucleotide probes utilized in Northern hybridization and primer extension analyses are indicated. (B) Pre-rRNA processing pathway.
Free diffusion through the lumen of the NPC is likely to be prevented by the FG-repeat regions of the GLFG-repeat and FXFG-repeat families of nucleoporins. Recent studies have proposed that these form a hydrophobic mesh that fills the lumen of the NPC (Ribbeck and Gorlich 2002; Gorlich et al. 2003), although a fringe of FG repeats that masks the entrance to the NPC is favored by others in the field. In either case, diffusion of large molecules and complexes through the NPC will be greatly impeded unless they can interact with the FG repeats. This property can be conferred in trans to transport substrates by binding to nucleocytoplasmic transport factors, including members of the importin-β/karyopherin-β (Kap-β/Imp-β) family. These proteins consist of repeated, predominantly α-helical, motifs termed HEAT repeats (Huntington-elongation-A subunit-TOR) that are related to Armadillo-like repeats (Andrade et al. 2001). In the structures of the PR65/A subunit of protein phosphatase 2A (PP2A) and Kap-β/Imp-β, the HEAT repeats align to form an extended structure with superhelical twist. PR65/A forms an open structure with a regular, left-handed superhelical twist, whereas Imp-β coils up into a tighter, but more irregular and flexible right-handed superhelical structure (Chook and Blobel 1999; Cingolani et al. 1999; Groves et al. 1999; Vetter et al. 1999; Bayliss et al. 2000; Lee et al. 2000).
Analyses in yeast and vertebrates have shown that a member of the Kap-β/Imp-β family, the export receptor Crm1p/Xpo1p, is required for the export of both the pre-40S and pre-60S ribosomal complexes. Subunit export additionally requires a subset of the nucleoporins, and like all active nuclear transport, also requires the small GTPase Ran (Gsp1p in yeast; Hurt et al. 1999; Moy and Silver 1999, 2002; Ho et al. 2000; Stage-Zimmermann et al. 2000; Gadal et al. 2001; Gleizes et al. 2001; Thomas and Kutay 2003; Trotta et al. 2003). Crm1p binds pre-60S particles via the pre-ribosome-associated protein Nmd3p, which may, in turn, bind to the ribosomal protein Rpl10p (Gadal et al. 2001). The mode of interaction of Crm1p with pre-40S particles has not yet been determined. Ribosome subunit export clearly requires Crm1p, but it seems unlikely that a single extrinsic protein factor is able to mediate the transit of the much larger subunits through the lumen of NPC.
A proteomic analysis of purified human nucleoli identified 271 proteins (Andersen et al. 2002), including KIAA0690. A database search identified the yeast homolog as the essential 138-kD protein Ypl012w/Rrp12p. Proteomic analyses identified interactions between Rrp12p and several other proteins involved in ribosome synthesis. These included early binding proteins of the 90S preribosome or U3 processome, and later pre-40S particles (Grandi et al. 2002; Schafer et al. 2003), strongly suggesting its participation in late steps of 40S synthesis. However, interactions were also seen with characterized protein components of pre-60S particles (Gavin et al. 2002), suggesting that it participated in the maturation of both ribosomal subunits. This is an unusual feature, as very few proteins have been shown to participate in the late maturation of both ribosomal subunits, and analyses of pre-ribosomal particles had concluded that the maturation of the two subunits was largely separated (Dragon et al. 2002; Grandi et al. 2002). Interestingly, two other very large proteins, Upt10p (200 kD) and Utp20p (287 kD), showed similar behavior and are postulated to associate with the both 90S ribosome or U3 processome and with pre-60 particles (Dragon et al. 2002; Gavin et al. 2002; Schafer et al. 2003).
We report here that Rrp12p, Upt10p, and Utp20p are each predicted to consist of HEAT-repeat motifs, as are five other ribosome synthesis factors including Noc2p, Noc3p, and Noc4p, which are known to be required for subunit export (Milkereit et al. 2001, 2003). We propose that these large, extended proteins play key roles in ribosome synthesis by providing a framework for pre-ribosome assembly and participate in subunit export to the cytoplasm.
Results
Rrp12p is associated with both pre-ribosomal subunits
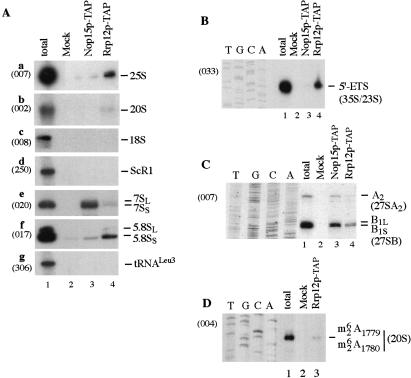
Proteomic analyses suggested that Rrp12p is associated with both the pre-40S and pre-60S ribosomal subunits. To confirm this prediction, Rrp12p was epitope tagged with a tandem-affinity purification (TAP) construct (Rigaut et al. 1999; see Materials and Methods). Following immunoprecipitation, bound RNA was analyzed by Northern hybridization (Fig. 2A) and primer extension (Figs. 2B–D) compared with RNA recovered in parallel from a nontagged control strain (wild-type lanes). Coprecipitation was also performed from a strain expressing Nop15p–TAP, which is required for 60S subunit synthesis, but does not participate in 40S subunit synthesis or subunit export (Oeffinger and Tollervey 2003). The 35S pre-rRNA component of the 90S pre-ribosomes and the 20S pre-rRNA component of the 40S pre-ribosome were coprecipitated with Rrp12p, but not with Nop15p. Primer extension showed that Rrp12p–TAP-precipitated 20S pre-rRNA that had undergone dimethylation at residues m26A1779m26A1780 (Fig. 2D). The 20S pre-rRNA is exported to the cytoplasm (see Fig. 1), where this modification is reported to occur prior to the cleavage that generates the mature 18S (Udem and Warner 1973; Brand et al. 1977). The mature 18S rRNA was not coprecipitated with Rrp12p–TAP, indicating that Rrp12p accompanies the 40S pre-ribosomes to the cytoplasm but dissociates prior to final rRNA maturation. The major pre-60S RNA components, the 27SA2, 27SB, and 7S pre-rRNAs, were coprecipitated with both Rrp12p–TAP and Nop15p–TAP, whereas the mature 25S and 5.8S rRNAs were precipitated only with Rrp12p. Notably, both the 25S and 5.8S rRNAs are fully matured within the nucleus, unlike the 18S rRNA. No coprecipitation was seen for control RNAs, scR1, or tRNALeu 3.
Figure 2.
Analysis of RNAs coprecipitated with TAP-tagged Rrp12p and Nop15p. Rrp12p–TAP and Nop15p–TAP were immunoprecipitated from cell lysates using IgG agarose, with release of bound RNA–protein complexes by cleavage of the protein A linker using TEV protease. RNA was recovered from the released material and from a mock-treated, isogenic wild-type control strain. (A) Northern hybridization. (B) Primer extension with probe 033 complementary to the 5′ETS, 278 nt from the transcription initiation site. The primer extension stop shows the abundance of the 35S pre-rRNA, the excised 5′ ETS fragment, and the aberrant 23S RNA, which is presented at low levels in wild-type strains. (C) Primer extension with probe 007, complementary to the 25S rRNA. This site lies ∼440 nt from sites B1L and B1S, and the corresponding bands are closely spaced. (D) Primer extension with probe 004, in the 5′ region of ITS1. Extension is arrested at site of cytoplasmic dimethylation within the 20S pre-rRNA. Probe names are indicated in parentheses on the left.
These observations demonstrate that Rrp12p is associated with the precursors to both the 40S and 60S subunits, and are consistent with the association of Rrp12p with the export substrates.
Depletion of Rrp12p inhibits 18S rRNA synthesis and causes accumulation of 3′-5.8S rRNA
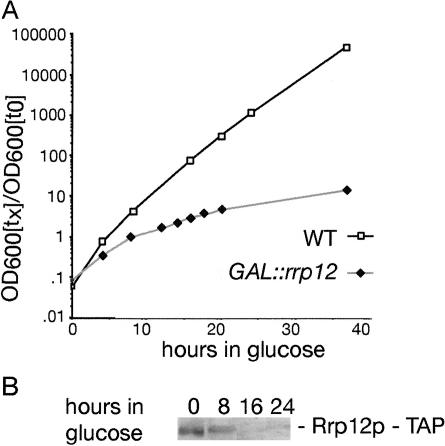
To examine the possible functions of Rrp12p in ribosome synthesis, an Rrp12p–TAP fusion was expressed under the control of the GAL10 promoter (see Materials and Methods), which is repressed in glucose-based medium. The host strain, YDL401, has reduced galactose permease activity leading to reduced GAL-induction. This eliminates the overexpression generally seen with GAL-regulated constructs, and allows faster appearance of phenotypes following transfer to glucose medium. The GAL::rrp12–TAP cells exhibited no detectable growth defect on permissive RGS medium, showing the fusion construct to be functional (data not shown). However, growth declined progressively following transfer to glucose medium commencing around 10 h after transfer (Fig. 3A). Western blotting (Fig. 3B) confirmed the depletion of the fusion protein.
Figure 3.
Depletion of Rrp12p–TAP under GAL-repression inhibits ribosome synthesis. (A) Growth curve of GAL::rrp12-TAP strains following transfer to glucose medium. Strains were pregrown in RGS medium and transferred to glucose medium for the times indicated. Strains were maintained in exponential growth by dilution with prewarmed medium. Cell densities measured by OD600 are shown corrected for dilution. (▪) Wild-type; (♦) GAL::rrp12–TAP. (B) Western analysis of the depletion of Rrp12p–TAP.
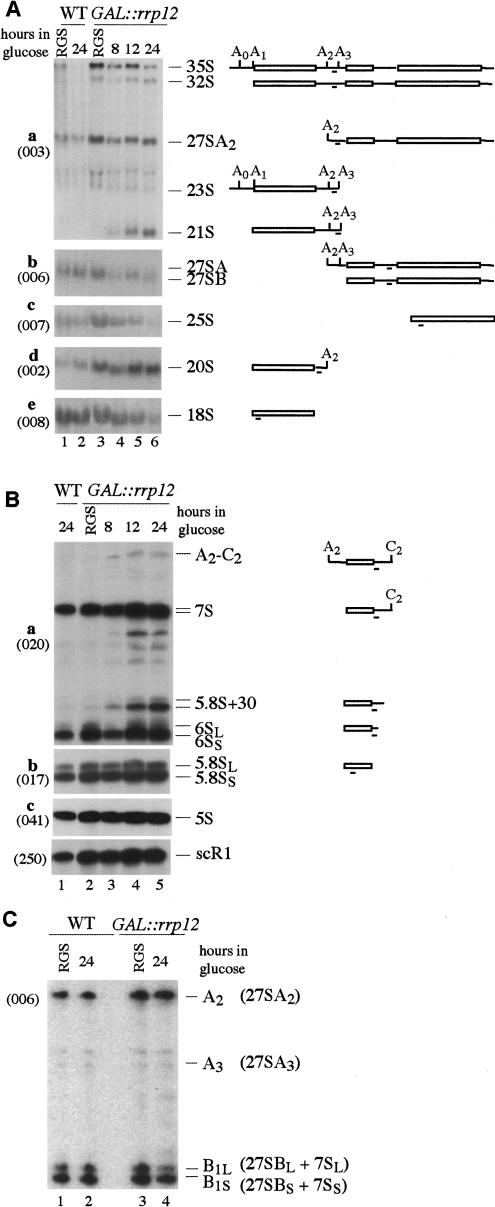
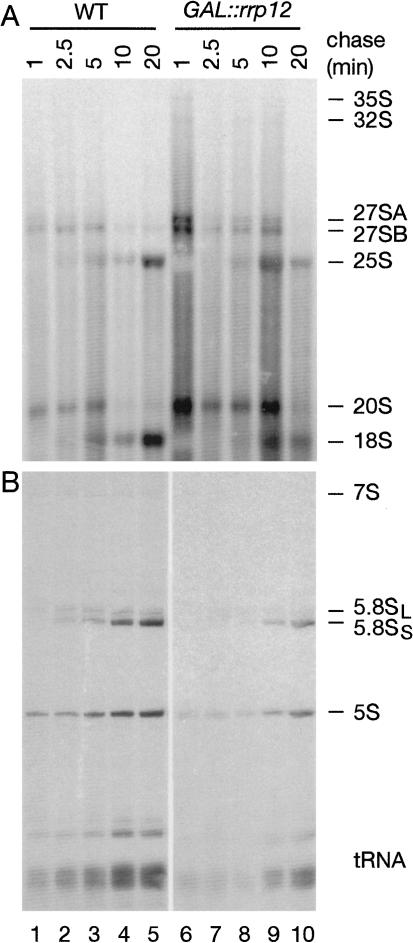
Northern hybridization (Fig. 4A,B) and primer extension (Fig. 4C) showed only mild pre-rRNA processing defects. Some accumulation of the 35S and 32S pre-rRNAs was seen in the GAL::rrp12 strain even in permissive RSG medium (Fig. 4A), indicating a kinetic delay in early processing, which is probably due to reduced Rrp12p expression levels. The aberrant 21S RNA, which results from direct cleavage of the 32S pre-rRNAs at site A3, without prior cleavage at site A2, was accumulated after transfer to glucose. This was not, however, associated with reduced levels of the products of A2 cleavage, the 27SA2, or 20S pre-rRNAs. The level of 20S pre-rRNA was elevated following Rrp12p depletion. The mature 18S rRNA was clearly depleted 24 h after transfer of the GAL::rrp12 strain to glucose medium (Fig. 4A,e), consistent with the inhibition of its synthesis from 20S pre-rRNA.
Figure 4.
Effects of Rrp12p depletion on pre-rRNA processing. (A) Northern analysis of high molecular weight RNA. RNA was extracted from wild-type and GAL::rrp12 strains during growth on permissive RGS medium and after transfer to glucose medium for the times indicated. RNA was separated on a 1.2% agarose/formaldehyde gel. Pre-rRNAs are indicated schematically on the right. Rectangles represent the mature rRNA and thin lines the transcribed spacers, with the position of the probe underlined. Probe names are indicated in parentheses on the left. (B) Northern analysis of low molecular weight RNA separated on 8% acrylamide/urea gels. (C) Primer extension analysis using a primer located in the 5′ region of ITS 2. The pre-rRNAs corresponding to the identified stops are indicated in parentheses.
Little alteration was seen in the major precursors on the pathway of 25S and 5.8S rRNA synthesis, the 27SA2, 27SA3, 27SB, and 7S pre-rRNAs, and no clear alteration was seen in the alternative processing pathways that generate the B1L and B1S intermediates (Fig. 4C) and mature 5.8SL and 5.8SS (Fig. 4B,b). However, the Rrp12p-depleted strain accumulated aberrant 3′-extended forms of 5.8S rRNA (Fig. 4B,a). These comigrated with the 3′-extended 5.8S species previously seen for mutations in components of the exosome complex (data not shown; Mitchell et al. 1997; Briggs et al. 1998). Notably, the same 3′-extended pre-5.8S species were seen in strains with mutations in Gsp1p (Ran; Suzuki et al. 2001), consistent with the functional interaction of Rrp12p and Gsp1p. A low level of aberrant intermediates that extended from sites A2 to C2 (Fig. 4B,a) was also observed. Depletion of the 25S rRNA was seen in the GAL::rrp12 strain (Fig. 4A,c) but analysis of the same RNA preparations did not show any clear change in the level of mature 5.8S rRNA (Fig. 4B,b). We do not know the fate of nuclear-restricted pre-60S particles (see below) but degradation, or at least cleavage of the large 25S rRNA may be more rapid than that of the 5.8S rRNA.
Pre-rRNA processing was also analyzed by pulse-chase labeling with [H3] uracil following depletion of Rrp12p for 16 h (Fig. 5). Processing of both the 27SA and 27SB pre-rRNAs was retarded, as was the accumulation of the mature 25S and 5.8S rRNAs. Notably, the 25S rRNA signal fell between the 10- and 20-min chase points, possibly due to degradation of rRNA that fails to be exported (see below). Consistent with the Northern data, substantial inhibition of processing of the 20S pre-rRNA to 18S rRNA was seen, with marked accumulation of 20S pre-rRNA and reduced 18S synthesis. Some reduction was also seen in labeling of the 5S rRNA and tRNAs, presumably due to the reduced growth rate.
Figure 5.
Pulse-chase labeling of rRNA synthesis in strains depleted of Rrp12p. Cells were transferred to glucose medium for 16 h, labeled with [H3] uracil for 1 min, and then chased with a large excess of unlabeled uracil for the times indicated. (A) RNA separated on a 1.2% agarose/formaldehyde gel. (B) RNA separated on an 8% polyacrylamide/urea gel.
We conclude that, consistent with the coprecipitation data, Rrp12p is required for normal synthesis of both 40S and 60S ribosomal subunits.
Rrp12p is a nucleolar-cytoplasmic shuttling protein required for export of both 40S and 60S subunits
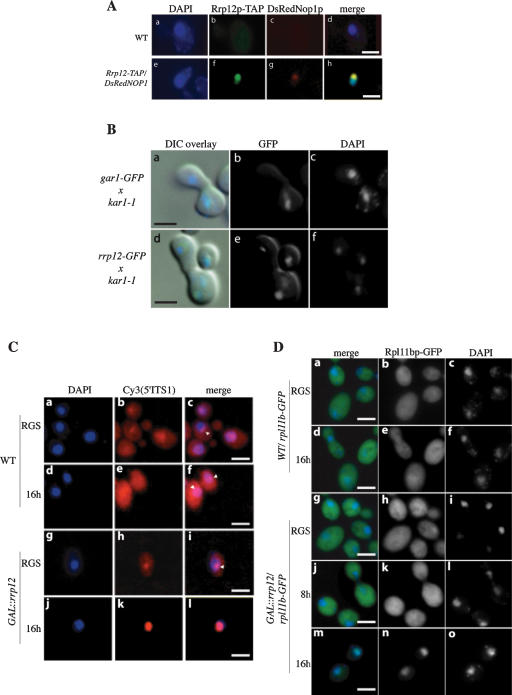
To determine the steady-state location of Rrp12p–TAP, cells were examined by indirect immunofluorescence (Fig. 6A) using a rabbit anti-protein A and a secondary FITC-conjugated goat anti-rabbit antibody to detect the protein A region of the TAP tag (Rigaut et al. 1999). As a marker for the nucleolus, a DsRed fusion with the nucleolar protein Nop1p was coexpressed (Gadal et al. 2001), and the nucleoplasm was identified by DAPI staining. Anti-protein A decorated the nucleolus, with a weaker signal over the nucleoplasm.
Figure 6.
Rrp12p is a nucleolar-cytoplasmic shuttling protein and required for export of both 40S and 60S subunits. (A) Rrp12p is predominately nucleolar. The GAL::rrp12–TAP strain also expressing the nucleolar marker DsRedNop1p (g) was examined by immunofluorescence using an anti-protein A antibody coupled to FITC (f). Also shown is the position of the nucleus visualized by DAPI staining (e), and a wild-type control strain (a–d). (B) Rrp12p can shuttle to an introduced nucleus in a heterokaryon. GAR1–GFP and RRP12–GFP cells were pregrown in YPD, treated with cycloheximide, and mated to kar1-1 cells (MS740). After zygotes were formed (∼1 h), the localization of GFP as well as DAPI staining of the nuclei was determined for Gar1p–GFP (b,c) and Rrp12p–GFP (e,f). Images have been oriented with the receptor nucleus at the top. Signals were merged with differential interference contrast (DIC) microscopy images (a and d, respectively). (C) 40S export assay. FISH localization of the Cy3-labled 5′ITS1 probe, which detects the 35S and 20S pre-rRNAs. DAPI staining is shown in a, d, g, and j. The 5′ITS1 localization is shown in b, e, h, and k. Strains were grown in permissive RGS medium and transferred to glucose medium for 16 h. Nucleoli are indicated with arrows. (D) 60S export assay. Wild-type or GAL::rrp12 cells expressing Rpl11b–GFP from a CEN plasmid were grown in permissive RGS-containing medium and transferred to glucose-containing medium for 8 or 16 h. (c,f,i,l,o) DAPI staining of the nucleoplasm. (b,e,h,k,n) Rpl11b–GFP visualized by fluorescence microscopy.
To assess whether Rrp12p shuttles between the nucleus and cytoplasm, we performed a heterokaryon assay (Fig. 6B; Flach et al. 1994; Peng and Hopper 2000). A strain expressing a Rrp12–GFP fusion under the control of the RRP12 promoter was constructed (see Materials and Methods) and mated with a kar1-1 strain, which is unable to undergo nuclear fusion following cell conjugation (Flach et al. 1994). In the heterokaryon thus formed, Rrp12–GFP was readily detected in both nuclei 1 h after cell fusion in either the presence or the absence of the translation inhibitor cycloheximide, showing that it can be transferred through the cytoplasm. In contrast, a fusion of GFP with another nucleolar protein, Gar1p, remains restricted to one nucleus in the heterokaryon.
We conclude that Rrp12p is largely localized to the nucleus with nucleolar enrichment at steady state, but is able to shuttle between the nucleus and cytoplasm.
Export of the 40S pre-ribosomes was analyzed by fluorescent in situ hybridization (FISH) as previously described (Moy and Silver 1999) using a Cy3-labeled probe specific for the 5′ region of the pre-rRNA ITS1 (Fig. 6C; Cy3[5′ITS1]). This region is present in the early 35S to 32S pre-rRNAs, and is retained in the 20S pre-rRNA, but absent from mature 18S rRNA (see Fig. 1). In the wild-type and in the GAL::rrp12 strain grown in permissive RGS medium, the pre-rRNA signal is concentrated in the nucleolus (arrows in Fig. 6C) and is also detected in the cytoplasm, where some perinuclear enrichment is visible. The cytoplasmic signal is due to the export of the 20S pre-rRNA prior to final processing to 18S rRNA. No cytoplasmic 20S pre-rRNA is seen in the GAL::rrp12 strain following growth in glucose medium for 16 h, whereas the nucleoplasmic 20S signal is strongly elevated, as shown by its colocalization with the DAPI-stained region.
Export of 60S pre-ribosomes was analyzed in wild-type and GAL::rrp12 strains that expressed a 60S subunit ribosomal protein fused to GFP (Rpl11b–GFP) from a CEN plasmid, as previously described (Stage-Zimmermann et al. 2000). In the wild-type strains and in the GAL::rrp12 grown in permissive RGS medium, Rpl1b–GFP was predominately cytoplasmic (Fig. 6D). Depletion of Rrp12p by transfer of the GAL::rrp12strain to glucose medium for 16 h led to a strong reduction in cytoplasmic Rpl11b::GFP signal and its accumulation in the nucleoplasm (Fig. 6D,n).
We conclude that Rrp12p is required for export of both the pre-40S and pre-60S ribosomal subunits from the nucleoplasm to the cytoplasm.
A family of HEAT-repeat proteins associated with pre-ribosomes
In systematic database analyses, Rrp12p was not reported to contain any known protein domains or motifs. However, this seemed unlikely to be the case for a protein of 138 kD, and we therefore undertook independent database searches. Using PSI-BLAST (Altschul et al. 1997), we found that Rrp12p has similarity to yeast proteins Gcn1p (3rd iteration; E = 1 × 10-16) and Tor1p (3rd iteration; E = 6 × 10-6). More specifically, the region of similarity was in parts of Gcn1p and Tor1p containing arrays of anti-parallel α-helices known as HEAT repeats (Andrade et al. 2001). According to secondary structure prediction (Jones 1999), Rrp12p has >60% of its residues in α-helical conformation and only 2% in β-strands, providing further support for its similarity to HEAT repeats. Two independent fold recognition methods, THREADER (Jones et al. 1992) and 3D-PSSM (Kelley et al. 2000), also found HEAT-repeat proteins as the most significant matches, indicating that Rrp12p is composed almost entirely of HEAT repeats or closely related modules.
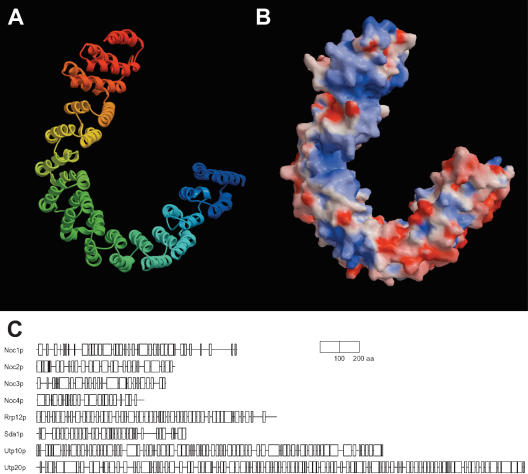
The 3D-PSSM alignment of Rrp12p with the PR65/A subunit of PP2A (Groves et al. 1999) was improved manually and used to build a three-dimensional (3D) model (Sali and Blundell 1993). Figure 7A shows ribbon representation of the Rrp12p model (residues 452–1018). According to evaluation criteria established for protein homology modeling (Sanchez and Sali 1998), the model has a probability of 0.98 to contain a correct fold. Figure 7B shows electrostatic surface representation as assigned by GRASP (Nicholls et al. 1991). Blue color represents regions of positive potential that cluster in larger areas on the inner side of the curved structure. These patches of positive charges are potential candidates for RNA binding.
Figure 7.
Three-dimensional model of Rrp12p and schematic representations of the pre-ribosome-associated HEAT repeat proteins. (A) Ribbon diagram of the predicted structure of Rrp12p. The model comprises residues 34–93 of Rrp12p. Blue color corresponds to the amino terminus. The figure was rendered using MOLSCRIPT (Kraulis 1991) and Raster3D (Merritt and Bacon 1997). (B) Electrostatic surface representation of the Rrp12p model was generated using GRASP (Nicholls et al. 1991). Positive (blue) and negative (red) potentials were rendered at the 10 kT/e level. The concave surface of the molecule has extensive regions of positive charge and is proposed to interact with RNA. (C) Schematic representations of the predicted secondary structure (Jones 1999) of pre-ribosome-associated HEAT repeat proteins. Predicted α-helices are shown as boxes; regions predicted to adopt β-strands comprise only 1%–4% of residues and were omitted for clarity.
Further sequence analysis and fold recognition of ribosome synthesis factors identified seven other proteins that are strongly predicted to consist of HEAT repeats or closely related repeat structures; Utp20p, Utp10p, Sda1p, Noc1p, Noc2p, Noc3p, and Noc4p (Fig. 7C shows regions of predicted α-helical content). Each of these proteins has <4% predicted β-strand content (most have 1%–2%) and 60%–70% α-helical content. All of these proteins are essential for viability and conserved from yeast to humans, indicating that they play important functions in ribosome synthesis. Noc proteins have already been implicated in ribosomal export (Milkereit et al. 2001, 2003), and we have successfully built 3D models that confirm their similarity to HEAT-repeat proteins (M. Dlakić and D. Tollervey, in prep.).
Rrp12p binds to Gsp1p (RAN), FG-repeat nucleoporins and RNA
HEAT-repeat proteins of the Kap-β/Imp-β family interact with the FG-repeat regions of both the GLFG-repeat and FXFG-repeat families of nucleoporins, and with the small GTPase Ran (see Chook and Blobel 2001; Kuersten et al. 2001; Kunzler and Hurt 2001; Weis 2002, 2003). These interactions are of key importance for their transport functions. The potential association of Rrp12p with these proteins was assessed by in vitro-binding assays. Ran (Gsp1p in yeast) and the GLFG-repeat regions of Nup100p and Nup116p (Iovine et al. 1995) and the FXFG-repeat region of Nsp1p were each expressed as GST fusions and purified from Escherichia coli. As controls, we also purified GST itself and GST fusions with three other proteins Prp20p (RCC1), Rrp41p, and Rrp44p (Dis3p). Rrp12p was apparently toxic when expressed in E. coli, and was therefore synthesized by in vitro transcription and translation in the presence of [35S]methionine. The GST-fusion proteins were bound to glutathione Sepharose and then incubated with [35S]-labeled Rrp12p. Following extensive washing, bound proteins were eluted and analyzed by autoradiography to assess the association with Rrp12p.
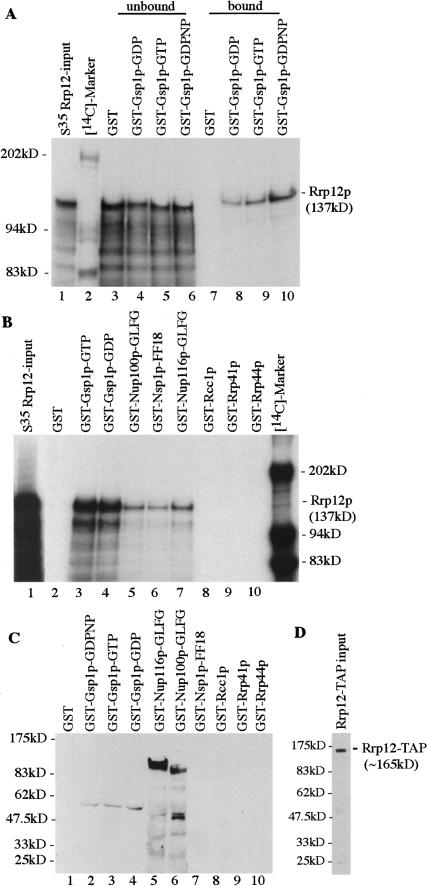
The association of Rrp12p with Gsp1p was assessed for the GTP and GDP-bound forms (Fig. 8A, lanes 2,3). Immediately prior to incubation with Rrp12p, the nucleotide bound to Gsp1p was exchanged for either GDP or GTP, as previously described (Weis et al. 1996). A high efficiency of exchange was confirmed using radiolabeled nucleotides (data not shown). Strong binding was seen between Rrp12p and both Gsp1p–GTP and Gsp1p–GDP. To confirm that apparent binding of Rrp12p to Ran–GTP was not a consequence of hydrolysis of GTP to GDP during the course of the incubation, the binding assay was also performed using the nonhydrolyzable GTP analog GDPNP. Rrp12p associated with Gsp1p–GDPNP with efficiency similar to Gsp1p–GTP and GDP (Fig. 8A). In the input (Fig. 8A, lane 1) and unbound material that was recovered in the supernatant following the incubation (Fig. 8A, lanes 3–6), truncated species are visible, which are likely to arise from premature translation termination in vitro. Notably, these were much less efficiently recovered in the bound fraction than was the full-length Rrp12p, confirming the specificity of the interaction.
Figure 8.
Rrp12p binds Ran and the GLFG/FXFG regions of nucleoporins in vitro. (A) Rrp12p was translated in vitro in the presence of [35S]methionine and incubated with GST, or fusions between GST and Gsp1p preloaded with GDP, GTP or the nonhydrolyzable analog GDPNP. The same relative amounts of the unbound and bound fractions were used and twofold more of these fractions were loaded compared with the input. (B) Rrp12p was translated in vitro with [35S] and incubated with GST, or fusions between GST and Gsp1p–GTP, Gsp1p–GDP, the GLFG region of Nup100p, the GLFG region of Nup116p, Nsp1p–FF18, Rcc1p, Rrp41p, and Rrp44p, which had been immobilized on glutathione-Sepharose. (C) Rrp12p–TAP isolated from a cell lysate by binding to IgG beads was incubated with GST, or fusions between GST and Gsp1p–GTP, Gsp1p–GDP, the GLFG region of Nup100p, the GLFG region of Nup116p, Nsp1p–FF18, Rcc1p, Rrp41p, and Rrp44p. Eluted proteins were separated by SDS-PAGE, immunoblotted, and decorated with anti GST-antibodies. (D) The amount of IgG-bound Rrp12p–TAP used in each binding reaction in C was determined by Western analysis using PAP antibodies.
Strong association of full-length Rrp12p was also seen for the GLFG and FXFG-repeat constructs (Fig. 8B, lanes 4–6). In contrast, no association was detected between Rrp12p and the fusions with Rcc1p, Rrp41p, or Rrp44p (Fig. 8A, lanes 7–9). Western blotting confirmed that the bait proteins were bound and recovered with comparable efficiency (Supplemental Fig. S1).
To further assess binding of Gsp1p and the nucleoporins, Rrp12p–TAP was recovered from a yeast cell lysate by precipitation on IgG-Sepharose beads, followed by washing with 1.6 M MgCl2. The protein A-IgG interact ion is resistant to this stringent wash, but proteins bound to the column indirectly via Rrp12p are expected to be removed (Görlich et al. 1996). Mass-spectroscopic analysis of proteins eluted by SDS treatment following the MgCl2 wash identified only Rrp12p–TAP and the IgG heavy and light chains, whereas only Rrp12p and TEV were observed following elution by TEV cleavage (data not shown). We cannot, however, exclude the possibility that additional proteins were present in substoichiometric amounts, despite the stringent wash conditions. The Rrp12p–TAP column was incubated with GST and the GST-fusion proteins. Following washing and elution, these were identified by Western blotting with anti-GST (Fig. 8B). GST–Gsp1p–GTP, GST–Gsp1p–GDP, GST–Gsp1–GDPNP, GST–Nup100p, and GST–Nup116p were each bound and was recovered from the Rrp12p–TAP column. However, no binding was seen for the FXFG repeat region of Nsp1p, in contrast to the reverse-binding interaction shown in Figure 8B. The reason for this is unclear.
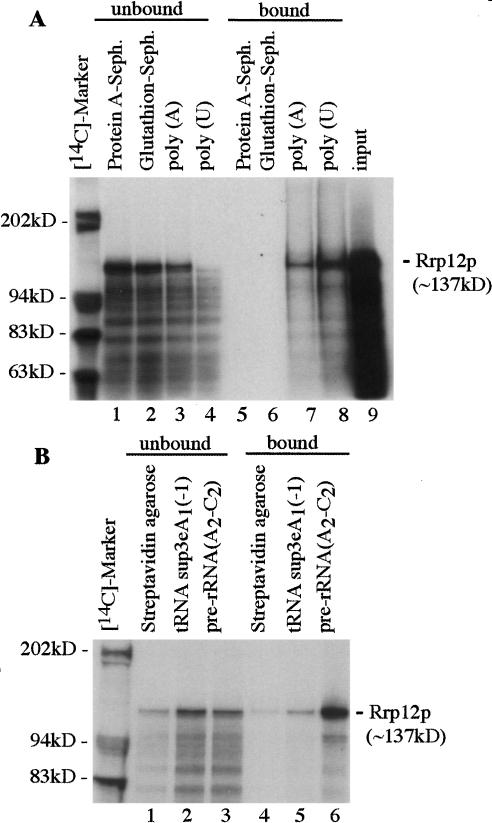
The ability of Rrp12p to associate with RNA was assessed by in vitro binding to the homopolymeric ribonucleotides, poly(A) and poly(U) sepharose (Fig. 9A). Clear binding of [35S]-labeled Rrp12p was seen to poly(A) Sepharose and almost all of the Rrp12p present bound to poly(U) Sepharose, resulting in its substantial depletion from the unbound sample. In contrast, no binding of Rrp12p was detected for the control glutathione Sepharose and protein A Sepharose samples. In vitro binding was further assessed using tRNASup3e (kindly provided by E. Betrand, University of Montpellier) and a transcript corresponding to the pre-rRNA region extending from ITS1 to ITS2 and including processing sites A2, A3, B1, E, and C2 (see Fig. 1A). Transcripts were synthesized in vitro, incorporating biotinylated-UTP immobilized on streptavidin-agarose beads and incubated with [35S]-labeled Rrp12p (Fig. 9B). Comparison of the bound and unbound fractions (twofold more of the bound fraction was loaded) showed substantial binding of Rrp12p (∼50% of the input) to the pre-rRNA transcript and its corresponding depletion from the unbound fraction. In contrast, binding of Rrp12p to the tRNA transcript was not clearly above the background level with streptavidin agarose alone.
Figure 9.
Rrp12p binds to RNA in vitro. (A) [35S]-labeled Rrp12p was incubated with protein A-Sepharose, glutathione-Sepharose, poly(A)-Sepharose, and poly(U)-Sepharose. Proteins recovered from the unbound and bound fractions were separated by SDS-PAGE visualized by autoradiography. (Lane 9) The input of [35S]-labeled Rrp12p used in A and B. (B) [35S]-labeled Rrp12p was incubated with Streptavidin agarose alone or with Streptavidin agarose loaded with biotinylated tRNASup3e or biotinylated pre-rRNA (A2-C2) transcripts. Following incubation, proteins were recovered from the unbound supernatant and bound and eluted fractions and separated by SDS-PAGE visualized by autoradiography. Twofold more of the bound material was loaded compared with the unbound or input.
We conclude that Rrp12p shows substantial in vitro RNA-binding activity. This is unlikely to represent simple electrostatic binding, as it clearly distinguished between the highly structured tRNA and a less-structured pre-rRNA transcript, and full-length Rrp12p showed much more efficient RNA binding than did truncated forms.
Discussion
The translocation of the very large, highly structured, and largely hydrophilic pre-ribosomal particles through the hydrophobic environment of the lumen of the NPC appears to present a difficult problem for the nucleocytoplasmic transport machinery, particularly in view of the high flux of subunit export. Around 30 subunits per minute must be exported through each NPC in a rapidly growing yeast cell. It seems likely that many factors will participate in this task, including Xpo1p and the preribosomal HEAT-repeat proteins described here.
Around 140 proteins have been identified that function in the post-transcriptional steps in ribosome synthesis, but few have been shown to participate in the formation of both the 40S and 60S subunits and only one, Rrp5p, has been shown to function directly in formation of both the 18S and the 25S/5.8S rRNAs (Venema and Tollervey 1996; Torchet et al. 1998; Eppens et al. 1999). Here, we report that Rrp12p is required for the late maturation of the 18S and 5.8S rRNA. Rrp12p is a component of the 90S pre-ribosomes, and remains associated with the 20S pre-rRNA component of the pre-40S ribosomes until its methylation, which probably occurs in the cytoplasm. Rrp12p is also present in the pre-60S ribosomes throughout the maturation of the 25S and 5.8S rRNAs. These data are consistent with the association of Rrp12p with the exported forms of both the pre-40S ribosomes, which contains the 20S pre-rRNA, and pre-60S ribosomes, which contains the mature 25S and 5.8S rRNAs. The 3′-extended pre-5.8S rRNA species seen in strains lacking Rrp12p are the same as those described for mutations in Gsp1p (Ran; Suzuki et al. 2001), suggesting that efficient maturation may be linked to the acquisition of export competence. Recent analyses in Xenopus oocytes also indicate a link between 5.8S rRNA 3′ maturation and 60S subunit export (E. Lund, pers. comm.), so this may be a conserved feature.
Recent models have proposed that gating during nuclear import and export largely depends on the hydrophobic FG repeat regions of the nucleoporins preventing free diffusion (Ribbeck and Gorlich 2002; Gorlich et al. 2003). Hydrophilic RNA and proteins that are unable to interact with the FG repeats are effectively excluded from the lumen of the NPC. Export of both 40S and 60S subunits requires Ran and Crm1p (Xpo1p) in yeast (Ho et al. 2000; Gadal et al. 2001; Moy and Silver 2002) and vertebrates (Thomas and Kutay 2003; Trotta et al. 2003). Crm1p interacts with the 60S subunit via the adaptor protein Nmd3p, which in turn binds to Rpl10p. However, it is unclear how a single export factor, bound indirectly to the much larger pre-60S subunit, could mediate its passage through the lumen of NPC. Transport might be initiated by interaction of Crm1p with the NPC, but we propose that the surfaces of both pre-ribosomal subunits carry Rrp12p, and perhaps other extended HEAT-repeat proteins, which can interact directly with the FG repeats of the nucleoporins and with Ran–GTP. These are envisaged to help cover the hydrophobic RNA surfaces of the ribosomal subunits, allowing them to dissolve into the hydrophobic mesh of the nucleoporin FG repeats.
Of the other seven HEAT-repeat proteins we identified, Sda1p, Noc1p, Noc2p, and Noc3p are each required for pre-rRNA processing on the pathway of 60S subunit synthesis (Milkereit et al. 2001; Ihmels et al. 2002), whereas Noc4p participates in 40S synthesis (Milkereit et al. 2003). Previous analyses indicated that a Noc1/2p complex associates with an early, nucleolar pre-60S particle, whereas a Noc2/3p complex associates with a later nucleoplasmic particle (Milkereit et al. 2001). Mutation of Noc1p leads to nucleolar accumulation of a 60S export reporter Rpl25p–eGFP. In contrast, mutation of Noc3p leads to nucleoplasmic accumulation, whereas mutation of Noc2p gave stronger accumulation in the nucleolus than nucleoplasm (Milkereit et al. 2001). Mutation of Noc4p leads to the nuclear accumulation of a 40S export reporter Rps2p–GFP, consistent with a role in pre-40S subunit export, but did not affect pre-60S export (Milkereit et al. 2003). Subunit export defects have not yet been assessed for Sda1p, Utp10p, or Utp20p.
Previous proteomic analyses clearly implicated Rrp12p, Utp10p, and Utp20p in 40S synthesis, but the evidence for association with pre-60S rested on their co-purification with other known 60S synthesis factors and Bud20p–TAP. The association of Rrp12p with late pre-60S particles is established by the data reported here, and we speculate that this will also be the case for Utp10p and Utp20p. It is notable that many other pre-60S synthesis factors also failed to be detected in previous proteomic analyses. These include all of the known processing enzymes; the endonucleases Rnt1p and the RNase MRP complex, the 5′→3′ exonucleases Rat1p and Xrn1p, and the 3′→5′ exonucleases of the exosome complex as well as Rex1p, Rex3, and Ngl2p. Also absent are several RNA helicases known to be required for ribosome synthesis and almost all of the snoRNAs that have been tested, with the exception of U3. It follows that many of the most important components associated with the pre-ribosomes in vivo dissociate during cell harvesting and/or the purification procedures. In the case of export factors, an obvious possibility is that breakdown of the nuclear envelope during cell lysis disrupts the Ran–GTP/GDP gradient, leading to their rapid dissociation from export-competent pre-ribosomes.
Gsp1p–GTP and Gsp1p–GDP bound to recombinant Rrp12p in vitro with similar efficiency, as did Ran loaded with the nonhydrolyzable analog GDPNP. Yeast Kap-β/Imp-β binds in vitro to both Gsp1p–GTP and Gsp1p–GDP, but binding to the GDP form is much weaker (Floer and Blobel 1996), and the exosome component hDis3p is reported to bind to either GTP– or GDP–Ran (Shiomi et al. 1998). In vivo, Rrp12p may bind to Ran–GTP as components of the export-competent pre-ribosomes, and bind Ran–GDP, while recycling into the nucleus. It is, however, also possible that the in vivo Ran-binding properties of Rrp12p are rather different from those of the recombinant protein in vitro. The structure of Kap-β/Imp-β changes substantially on binding to other proteins (Chook and Blobel 1999; Cingolani et al. 1999; Groves et al. 1999; Vetter et al. 1999; Bayliss et al. 2000; Lee et al. 2000), and the structure of Rrp12p may well be altered on binding to the pre-ribosomes. Binding of Crm1p/Xpo1p to Gsp1p–GTP is stable only in the presence of Yrb1p (yeast Ran–BP1; Maurer et al. 2001). Kap-β/Imp-β makes extensive contacts with Ran–GTP that extend over several HEAT repeats (Vetter et al. 1999). An acidic loop region accounts for part of this interaction, but a similar acidic region is not apparent in the sequence of Rrp12p.
Another, apparently major, problem in eukaryotic ribosome synthesis arises from the use of modification guide snoRNAs to direct the covalent modification of the rRNA sequences. This requires that the core of the rRNA initially remains unfolded and accessible to the many snoRNAs with which it must base pair. Subsequent formation of the mature ribosomal subunits will clearly require large-scale refolding of the rRNA and reorganization of the pre-ribosomal particles. We speculate that the extended HEAT-repeat proteins form a scaffold for pre-ribosome assembly and pre-rRNA unfolding/refolding. A function as a scaffold has been proposed for PR65/A, which assembles the catalytic subunit together with a range of different regulatory B subunits to form the PP2A complex (Groves et al. 1999).
Materials and methods
Strains
Growth and handling of S. cerevisiae were by standard techniques. GAL-regulated strains were pregrown in RGS medium containing 2% raffinose, 2% galactose, and 2% sucrose, and harvested at intervals following a shift to medium containing 2% glucose. Strains for pulse-chase analysis were pregrown in minimal RGS medium lacking uracil and shifted to minimal glucose medium lacking uracil. Strains for immunofluorescence studies were grown in minimal glucose medium lacking tryptophan. Yeast strains used and constructed in this study are listed in Supplemental Table S1. Conditional mutants under the control of the repressible GAL10 promoter were generated by one-step PCR strategy in the strains YDL401 and BMA64. TAP-tagged strains were constructed by a one-step PCR strategy in the GAL-mediated strain GAL::rrp12 (Rigaut et al. 1999). GFP-tagged strains were constructed by one-step PCR strategy in the wild-type strain BMA38 (MATa). Transformants were screened by immunoblotting and PCR. The RRP12–TAP strain was transformed with pUN100DsRedNOP1 (kindly provided by E. Hurt, University of Heidelberg) to allow ready identification of the nucleolus, and the GAL::rrp12 strain was transformed with pRS315-rpl11b–GFP (kindly provided by P. Silver, Dana Faber Institute) to study 60S ribosomal subunit export. Plasmids used and constructed in this study are listed in supplementary Table S2.
RNA extraction, Northern hybridization, and primer extension
For high molecular weight RNA analysis, 7 μg of total RNA was separated on a 1.2% agarose gel containing formaldehyde. Standard 6%, 8%, or 12% acrylamide-8 M urea gels were used to analyze low-molecular weight RNA species and primer extension reactions. Primer extensions were performed on 5 μg of total RNA. For pre-rRNA, rRNA analysis, and FISH, the following oligonucleotides were used: 002; 5′-GCTCTTTGCTCTTGCC; 004; 5′-CGGTTTTAATTGTCCTA; 007; 5′-CTCCGCTTATTGATATGC; 008; 5′-CATGGCTTAATCTTTGAGAC; 017; 5′-GCGTTGTTCATCGATGC; 020; 5′-TGAGAAGGAAATGACGCT; 026; 5′-CCAGATAACTATCTTAAAAG; 033; 5′-CGCTGCTCACCAATGG; 250, 5′-ATCCCGGCCGCCTCCATCAC; 306, 5′-GCATCTTACGATACCTG
Immunoprecipitation of Rrp12p–TAP
Immunoprecipitation of Rrp12p–TAP and subsequent TEV cleavage was performed as described by (Rigaut et al. 1999), except for the addition of 2.5 mM vanadyl-ribonucleoside complexes (VRC) to buffer A. RNA was extracted with buffer AE/phenolchloroform, ethanol precipitated, and analyzed by Northern hybridization and primer extension, respectively. For Solution-binding assays, IgG bead were washed with 1.6M MgCl2 after overnight incubation with Rrp12p–TAP lysate. Washed IgG beads were used for binding assay with GST-proteins.
Immunofluorescence
For localization of yeast Rrp12p–TAP, the protein A region was detected with a rabbit anti-Protein A antibody (Sigma) and a secondary goat anti-rabbit antibody coupled to FITC (Sigma) at a 1:1000 and a 1:200 dilution, respectively. To stain nuclear DNA, DAPI was included in the mounting medium (Vecta-shield, Vector Laboratories).
Fluorescent in situ hybridization
Fluorescent in situ hybridization was performed in the GAL::rrp12 strain as previously described (Amberg et al. 1992) with some modifications. The template for the ITS1-5′ RNA, 5′-TGGACTC5CCATCTCTTGAC5TCTTGCCCAG5AAAAGCTC5CATGCTC5T-3′, was a 50-bp oligonucleotide fluorescently labeled with Cy3. Cells were grown to an optical density of 0.5–1.0 at 600 nm and fixed with 2% (v/v) formaldehyde/5% (v/v) acetic acid for 10 min at 25°C. Samples were hybridized with 100 ng of Cy3-labeled RNA oligonucleotide for 16 h at 37°C.
Heterokaryon analysis
The heterokaryon assay was performed in the Rrp12p–GFP strain as described previously (Flach et al. 1994; Peng and Hopper 2000) with the following modifications. Cells were incubated with cycloheximide at a concentration of 100 μg/μL for 5 min on ice. Mating was initiated by mixing 3 × 106 cells with an equal number of cells from the kar1-1 strain MS740 (kindly provided by M. Rose, Princeton University). Cells were concentrated on a 25-mm diameter, 0.45-μm pore nitrocellulose filter, and by placing the filter on a YPD plate. After 1, 1.5, and 2 h of incubation, cells were washed off the membrane using glucose-containing medium fixed with 4% (v/v) formaldehyde for 30 min at 25°C and spotted onto poly-L-lysine coated coverslips for observation.
Recombinant protein expression
GST-fused constructs were transformed into E. coli BL21 [LysS]. Recombinant GST-fusion proteins were purified as described by previously (Iovine et al. 1995).
Solution-binding assay
[35S]-L-methionine-Rrp12p was expressed in vitro from a PCR-generated template (5′primer-ATTAATACGACTCACTATAGGGAGAGCCACCATGGATCAAGACAAGTTGCT;3′primer–[T]40TCTCCAGGTGTGTAATTAGCCATATTGCTCAGT) using a Quick PCR T7 TNT kit (Promega).
A total of 2 μg of GST–Gsp1p and 1 μg each of GST–Nup116–GLFG, GST–Nup100–GLFG, GST–Nsp1p–FF18, GST–Rcc1p, GST–Rrp41p, GST–Rrp44p, and GST per 10 μL beads were bound to glutathione-agarose beads in 0.5-mL-binding buffer (20 mM Hepes at pH6.8, 150 mM potassium acetate, 2 mM MgCl2, 2 mM DTT, 0.1% Tween-20, 0.1% casamino acids) for 45 min at 4°C. GTP/GDP/GDPNP loading of GST–Gsp1p was performed as previously described (Weis et al. 1996). The reaction was terminated by addition of 30 mM MgCl2, and the beads washed with binding buffer to remove excess nucleotide. Efficiency of Gsp1p–GTP/GDP binding was determined using [3H]GTP or [3H]GDP by filtration and liquid scintillation counting as described (Weis et al. 1996). The binding of labeled Rrp12p to GST proteins and GST proteins to Rrp12p–TAP was carried out as previously described (Rexach and Blobel 1995). Binding of labeled Rrp12p to polyribonucleotides, tRNA, and pre-rRNA,was carried out under similar conditions, but in the presence of 25 mM MgCl2. tRNAsup3e and pre-rRNA were transcribed from a SP6 and T7 promoter, respectively, using biotin-16-UTP (Roche). A total of 100 ng of each biotinylated RNA was bound to Streptavidin-agarose and washed with low-salt buffer. After binding of labeled Rrp12p, the reactions were separated on a 3%–8% Tris-Acetate or 4%–12% Bis-Tris Novex gel (Invitrogen). The 3%–8% Tris-Acetate gel was dried and exposed to a BMS film overnight at -80°C with an intensifying screen. The 4%–12% Bis-Tris gel was immunoblotted and probed with anti-GST antibody conjugated to horseradish peroxidase. Bands were detected by ECL.
Sequence analysis and model building
The nonredundant protein database clustered at 70% identity was searched with PSI-BLAST (Altschul et al. 1997) for 10 iterations or until convergence (E = 0.005). The database was searched separately for three iterations and the checkpoints were saved for secondary structure predictions (Jones 1999). These predictions were also used for fold recognition by THREADER (Jones et al. 1992). We also used 3D-PSSM (Kelley et al. 2000; http://www.sbg.bio.ic.ac.uk/~3dpssm/) as an independent fold-recognition method. In both cases, fold recognition confirmed the results of sequence analysis with high significance. The best match in the 3D-PSSM analysis was the PR65/A subunit of PP2A (Groves et al. 1999). Although Kap-β/Imp-β structure (Cingolani et al. 1999) had only marginally less significant match than the PR65/A subunit of PP2A, it was less suitable as a template because its alignment with Rrp12 contained more insertions and deletions.
Automatic alignments produced by 3D-PSSM were manually corrected where needed and used to build an ensemble of 3D models (Sali and Blundell 1993). The models were evaluated (Sanchez and Sali 1998), and all had 0.97 or better probability of being correct without any further optimizations.
Acknowledgments
We thank Pam Silver for the Rp11b–GFP construct, Mark Rose for the kar1-1 strain, Edouard Bertrand for the Gar1–eGFP and tRNASup3 constructs, Murray Stewart for the Nsp1p–FF18 construct, and Elsebet Lund for communicating results prior to publication. M.O. was supported by the Darwin Trust of Edinburgh. M.D. was supported in part by a Special Fellowship from the Leukemia & Lymphoma Society. D.T. is a Wellcome Trust principal fellow. This work was supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.285604.
References
- Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein, A.L., and Cole, C.N. 1992. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 6: 1173-1189. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12: 1-11. [DOI] [PubMed] [Google Scholar]
- Andrade M.A., Petosa, C., O'Donoghue, S.I., Muller, C.W., and Bork, P. 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309: 1-18. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood, T., and Stewart, M. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 102: 99-108. [DOI] [PubMed] [Google Scholar]
- Brand R.C., Klootwijk, J., van Steenbergen, T.J.M., de Kok, A.J., and Planta, R.J. 1977. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 75: 311-318. [DOI] [PubMed] [Google Scholar]
- Briggs M.W., Burkard, K.T., and Butler, J.S. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273: 13255-13263. [DOI] [PubMed] [Google Scholar]
- Chook Y.M. and Blobel, G. 1999. Structure of the nuclear transport complex karyopherin-β2-Ran x GppNHp. Nature 399: 230-237. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11: 703-715. [DOI] [PubMed] [Google Scholar]
- Cingolani G., Petosa, C., Weis, K., and Muller, C.W. 1999. Structure of importin-β bound to the IBB domain of importin-α. Nature 399: 221-229. [DOI] [PubMed] [Google Scholar]
- Dragon F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppens N.A., Rensen, S., Granneman, S., Raue, H.A., and Venema, J. 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5: 779-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach J., Bossie, M., Vogel, J., Corbett, A., Jinks, T., Willins, D.A., and Silver, P.A. 1994. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol. Cell. Biol. 14: 8399-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M. and Blobel, G. 1996. The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem. 271: 5313-5316. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss, D., Kessl, J., Trumpower, B., Tollervey, D., and Hurt, E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21: 3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Gleizes P.E., Noaillac-Depeyre, J., Leger-Silvestre, I., Teulieres, F., Dauxois, J.Y., Pommet, D., Azum-Gelade, M.C., and Gas, N. 2001. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol. 155: 923-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Kraft, R., Kostka, S., Vogel, F., Hartmann, E., Laskey, R.A., Mattaj, I.W., and Izaurraide, E. 1996. Importin provides a link between nuclear protein import and U snRNA export. Cell 87: 21-32. [DOI] [PubMed] [Google Scholar]
- Görlich D., Seewald, M.J., and Ribbeck, K. 2003. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22: 1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Rybin, V., Bassler, J., Petfalski, E., Strauss, D., Marzioch, M., Schafer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10: 105-115. [DOI] [PubMed] [Google Scholar]
- Groves M.R., Hanlon, N., Turowski, P., Hemmings, B.A., and Barford, D. 1999. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96: 99-110. [DOI] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom, G., and Johnson, A.W. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell. Biol. 151: 1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Hannus, S., Schmelzl, B., Lau, D., Tollervey, D., and Simos, G. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell. Biol. 144: 389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J., Friedlander, G., Bergmann, S., Sarig, O., Ziv, Y., and Barkai, N. 2002. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31: 370-377. [DOI] [PubMed] [Google Scholar]
- Iovine M.K., Watkins, J.L., and Wente, S.R. 1995. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 131: 1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195-202. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor, W.R., and Thornton, J.M. 1992. A new approach to protein fold recognition. Nature 358: 86-89. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., MacCallum, R.M., and Sternberg, M.J. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299: 499-520. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24: 946-950. [Google Scholar]
- Kuersten S., Ohno, M., and Mattaj, I.W. 2001. Nucleocytoplasmic transport: Ran, β and beyond. Trends Cell Biol. 11: 497-503. [DOI] [PubMed] [Google Scholar]
- Kunzler M. and Hurt, E. 2001. Targeting of Ran: Variation on a common theme? J. Cell Sci. 114: 3233-3241. [DOI] [PubMed] [Google Scholar]
- Lee S.J., Imamoto, N., Sakai, H., Nakagawa, A., Kose, S., Koike, M., Yamamoto, M., Kumasaka, T., Yoneda, Y., and Tsukihara, T. 2000. The adoption of a twisted structure of importin-β is essential for the protein-protein interaction required for nuclear transport. J. Mol. Biol. 302: 251-264. [DOI] [PubMed] [Google Scholar]
- Maurer P., Redd, M., Solsbacher, J., Bischoff, F.R., Greiner, M., Podtelejnikov, A.V., Mann, M., Stade, K., Weis, K., and Schlenstedt, G. 2001. The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p). Mol. Biol. Cell 12: 539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon, D.J. 1997. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277: 505-524. [DOI] [PubMed] [Google Scholar]
- Milkereit P., Gadal, O., Podtelejnikov, A., Trumtel, S., Gas, N., Petfalski, E., Tollervey, D., Mann, M., Hurt, E., and Tschochner, H. 2001. Maturation and intranuclear transport of preribosomes requires Noc proteins. Cell 105: 499-509. [DOI] [PubMed] [Google Scholar]
- Milkereit P., Strauss, D., Bassler, J., Gadal, O., Kuhn, H., Schutz, S., Gas, N., Lechner, J., Hurt, E., and Tschochner, H. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278: 4072-4081. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome; a conserved eukaryotic RNA processing complex containing multiple 3′→5′exoribonuclease activities. Cell 91: 457-466. [DOI] [PubMed] [Google Scholar]
- Moy T.I. and Silver, P.A. 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes & Dev. 13: 2118-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 115: 2985-2995. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281-296. [DOI] [PubMed] [Google Scholar]
- Oeffinger M. and Tollervey, D. 2003. Yeast Nop15p is an RNA binding protein required for pre-ribosomal RNA processing and cytokinesis. EMBO J. 22: 6573-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G. and Hopper, J.E. 2000. Evidence for Gal3p's cytoplasmic location and Gal18p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 5140-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M. and Blobel, G. 1995. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83: 683-692. [DOI] [PubMed] [Google Scholar]
- Ribbeck K. and Gorlich, D. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21: 2664-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030-1032. [DOI] [PubMed] [Google Scholar]
- Sali A. and Blundell, T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779-815. [DOI] [PubMed] [Google Scholar]
- Sanchez R. and Sali, A. 1998. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. 95: 13597-13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer T., Strauss, D., Petfalski, E., Tollervey, D., and Hurt, E. 2003. The path from nucleolar 90S to cytoplasmic 40S preribosomes. EMBO J. 22: 1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi T., Fukushima, K., Suzuki, N., Nakashima, N., Noguchi, E., and Nishimoto, T. 1998. Human Dis3p, which binds to either GTP- or GDP-Ran, complements Saccharomyces cerevisiae dis3. J. Biochem. 123: 883-890. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T., Schmidt, U., and Silver, P.A. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11: 3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Noguchi, E., Nakashima, N., Oki, M., Ohba, T., Tartakoff, A., Ohishi, M., and Nishimoto, T. 2001. The Saccharomyces cerevisiae small GTPase, Gsp1p/Ran, is involved in 3′ processing of 7S-to-5.8S rRNA and in degradation of the excised 5′-A0 fragment of 35S pre-rRNA, both of which are carried out by the exosome. Genetics 158: 613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F. and Kutay, U. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116: 2409-2419. [DOI] [PubMed] [Google Scholar]
- Torchet C., Jacq, C., and Hermann-Le Denmat, S. 1998. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA 4: 1636-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta C.R., Lund, E., Kahan, L., Johnson, A.W., and Dahlberg, J.E. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 22: 2841-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem S.A. and Warner, J.R. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248: 1412-1416. [PubMed] [Google Scholar]
- Venema J. and Tollervey, D. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15: 5701-5714. [PMC free article] [PubMed] [Google Scholar]
- Vetter I.R., Arndt, A., Kutay, U., Gorlich, D., and Wittinghofer, A. 1999. Structural view of the Ran-Importin β interaction at 2.3 A resolution. Cell 97: 635-646. [DOI] [PubMed] [Google Scholar]
- Weis K. 2002. Nucleocytoplasmic transport: Cargo trafficking across the border. Curr. Opin. Cell Biol. 14: 328-335. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell 112: 441-451. [DOI] [PubMed] [Google Scholar]
- Weis K., Dingwall, C., and Lamond, A. 1996. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 15: 7120-7128. [PMC free article] [PubMed] [Google Scholar]