Abstract
Purpose
Two previous genome-wide association studies (GWAS) of high myopia in a Japanese population found several single nucleotide polymorphisms (SNPs) associated with the disease. The present study examined whether these markers are associated with myopia in a Chinese population.
Methods
Individuals with or without complex myopia were recruited from Chinese university students, and probands with early onset high myopia were identified in the Pediatric and Genetic Eye Clinic of the Zhongshan Ophthalmic Center. DNA was prepared from venous leukocytes. Three SNPs, rs577948 and rs11218544 at chromosome position 11q24.1 and rs2839471 at chromosome position 21q22.3, were genotyped. The allele and genotype frequencies of these SNPs were compared between the myopia cases and controls using a χ2 test.
Results
A total of 2,870 subjects were examined in this study, including 1,255 individuals with complex myopia (−10.00 diopter (D)<spherical refraction≤-4.00 D), 563 with early onset high myopia (spherical refraction≤-6.00 D), and 1,052 healthy controls (−0.50 D≤spherical equivalent≤ +2.00 D). There were no statistically significant differences found for the genotype or allele frequencies of the three SNPs between the myopia cases and controls in the Chinese population under study.
Conclusions
We did not find evidence for the association of myopia with rs577948, rs11218544, or rs2839471 in the Chinese population studied.
Introduction
Myopia is the most common cause of vision impairment worldwide, with a prevalence of approximately 25.4%–80% [1–3]. The prevalence of myopia reaches 71%–96% in students living in the developed region of East Asia [4,5] and is expected to increase [6–8]. Both environmental and genetic factors are believed to play important roles in the development of myopia, but the exact molecular basis of this condition is still unknown [1,9].
Molecular genetic studies have identified several loci associated with a predisposition to myopia [10–27]. However, the genes at most of these loci remain to be identified [28,29]. In the past three years, several independent genome-wide association studies (GWAS) have focused on the search for genetic factors contributing to myopia [20,30–34]. Two of these studies were based on Japanese populations, and identified several single nucleotide polymorphisms (SNPs) significantly associated with high myopia, including rs577948 and rs11218544 at chromosome 11q24.1 and rs2839471 at chromosome 21q22.3 [20,30]. Replication is an important step to confirm the original findings of association studies; although the original reports suggested replication studies to validate their findings, these have yet to be performed.
Both Chinese and Japanese populations exhibit a very high prevalence of myopia. Additionally, the populations of these two regions are closely related [35]. The LD structures around rs577948, rs11218544, and rs2839471 are very similar between Chinese and Japanese populations according to data from HapMap Phase 3. Therefore, we performed a replication study to examine whether the SNPs rs577948, rs11218544, and rs2839471 are also associated with myopia in Chinese individuals.
Methods
Subjects
A total of 2,870 unrelated individuals were examined in this study, including 1,052 healthy controls and 1,818 myopia cases. Based on degrees of spherical refraction, the 1,818 subjects with myopia were classified into three groups: −6.00 diopter (D)<spherical refraction≤-4.00 D for 423 cases, −9.25 D<spherical refraction≤-6.00 D for 1,124 cases, and spherical refraction≤-9.25 D (−9.25 D was the criterion for myopia used in the original study [20]) for 271 cases. The 1,818 myopia cases were divided into two groups based on genetic contribution: 563 cases with early onset high myopia and 1,255 cases with complex myopia. The 563 individuals with early onset high myopia were identified at the Pediatric and Genetic Clinic of the Zhongshan Ophthalmic Center. The enrollment criteria used were basically the same as previously described [15]: 1) myopia was present before attending primary school; 2) spherical refraction ≤-6.00 D; 3) no other known ocular or systemic diseases. The 1,255 subjects with complex myopia and 1,052 healthy controls were college students recruited from 12 universities in Guangzhou. The 1,255 subjects with myopia met the following criteria: 1) spherical refraction at each meridian ≤-4.00 D; 2) best corrected visual acuity (BCVA) ≤0.1 in logMAR; 3) myopia onset occurred after 7 years of age; 4) no other known eye or related systemic diseases were present; 5) no family history of high myopia existed. The 1,052 subjects in the healthy control group met the following criteria: 1) bilateral refraction measured between −0.50 D and +2.00 D (spherical equivalent); 2) best unaided visual acuity ≤0 in logMAR; 3) no other known eye or related systemic diseases were present; 4) no family history of high myopia was documented.
The results of ophthalmologic examinations were recorded, including visual acuity (unaided, near, and/or best), color vision, slit lamp and direct ophthalmoscope examination. Refractive errors were measured using an autorefractor (Topcon KR-8000, Paramus, NJ) after mydriasis with compound tropicamide (Mydrin®-P, Santen Pharmaceutical Co. Ltd., Osaka, Japan). All of the college students recruited from 12 universities in Guangzhou received an ocular biometry examination using IOL master V5 (Carl Zeiss Meditec AG, Jena, Germany). Additional examinations included an electroretinogram and fundus photograph in selected individuals. The study adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Board of Zhongshan Ophthalmic Center, Guangzhou China. As this study was part of a project with the aim of identifying genetic factors related to myopia, the procedure for obtaining informed consent and collecting subjects was the same as previously described [36]. Genomic DNA was prepared from venous blood samples obtained from the study subjects.
Genotyping
Restriction fragment length polymorphism (RFLP) analysis was used to detect the SNP rs2839471. The primers that were used to amplify the DNA fragment that included rs2839471 [provided by the National Center for Biotechnology Information (NCBI)] are listed in Table 1. The amplicon was digested using the restriction endonuclease Hpy188I according to the manufacturer’s instructions (New England Biolabs [Beijing], Ltd, Beijing, China]. The digested products were analyzed by PAGE.
Table 1. Primers used for genotyping.
| SNP | Direction | Primer sequence (5′-3′) |
|---|---|---|
|
rs2839471 |
F |
TGGGGCTGCAGGTGTTGTG |
| |
R |
AGTGGGCCAGCTAGGAAAAGAAA |
|
rs577948 |
Nest-F |
CACAATGCAAAGGATCAGAGC |
| |
Nest-R |
AGCGTGATCAGAATACAAGATGG |
| |
F-A |
ACTGTCAGAAACTCAACTCTCA |
| |
F-G |
TATAACTGTCAGAAACTCAACTCTCG |
| |
R-Pub+M13 |
CGTTGTAAAACGACGGCCAGTatcagaatacaagatggagcgtagg |
|
rs11218544 |
Nest-F |
GCAAAGGAAACAATCAACAGAG |
| |
Nest-R |
AAAGGTGTCCAAATGATAGCC |
| |
F-T |
CAAATCACTAACCTTTCCAGAACAT |
| |
F-G |
TATACAAATCACTAACCTTTCCAGAACAG |
| |
R-Pub+M13 |
CGTTGTAAAACGACGGCCAGTggaccaaagacagctcaaacc |
| M13 probe | FAM-CGTTGTAAAACGACGGCCAGT |
The SNPs rs577948 and rs11218544 were genotyped by nested polymerase chain reaction (nest-PCR) combined with fluorescence-labeled allele-specific PCR (AS-PCR). Briefly, two pairs of primers were used. The first pair of primers was designed to amplify fragments encompassing the SNPs. The amplicons were then used as templates for a second round of PCR, in which three internal primers and a labeled common primer were used: two SNP allele-specific primers with a length difference of 4 bp, a reverse specific primer with an M13 tail, and a 5′-FAM-labeled M13 probe (Table 1). Finally, the labeled allele-specific amplicons were analyzed using capillary electrophoresis in an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA) and analyzed using GeneMapper software v3.5 (Applied Biosystems).
For each SNP, 48 samples were simultaneously analyzed by cycle sequencing to ensure that either the RFLP or M13-tailed method was reliable. Genotyping results were read independently by two researchers. Two types of additional validation were conducted: 1) direct sequencing of the first round of PCR products from four samples in each plate (96 samples); and 2) any sample that was read differently by the two researchers was confirmed by sequencing.
Statistical analysis
The distribution of sexes was compared between the myopia cases and healthy controls with a χ2 test. The average age was compared between the myopia cases and healthy controls with a t-test. The distribution of all three SNPs was evaluated with respect to Hardy–Weinberg equilibrium (HWE). The frequencies of alleles and genotypes in the early onset high myopia and complex myopia groups, as well as in the classified spherical refraction groups were compared with those in the healthy control group using a χ2 test. A p<0.05 was used as the level of statistical significance (α), and a Bonferroni correction was applied for multiple testing, correcting the threshold for significance from 0.05 to 0.017 (α/3=0.05/3).
Results
The basic clinical information for all 2870 subjects is listed in Table 2. Genotyping of rs2839471, rs577948, and rs11218544 was successfully performed in the 2,870 unrelated subjects (Figure 1, Table 3, and Table 4). The genotyping success rate was 100% for the three SNPs. There was no significant deviation from HWE for any of the three SNPs in the healthy controls. No statistically significant difference in genotype frequencies or allele frequencies was found between the myopia cases and controls, regardless of whether myopia was classified into early onset high myopia and complex myopia or classified into three groups based on spherical refraction (Table 3 and Table 4). Although the frequency of rs11218544 exhibited a slight difference between the complex myopia group and healthy control group (allele p=0.032), the difference was not significant after Bonferroni correction with a corrected significance level of p=0.017.
Table 2. Basic information for the 2,870 subjects.
| Category | Early onset high myopia | Complex myopia | Healthy control | ||||
|---|---|---|---|---|---|---|---|
| |
|
−9.25 D<S≤-6.00 D |
S≤-9.25 D |
−6.00 D<S≤-4.00 D |
−9.25 D<S≤-6.00 D |
S≤-9.25 D |
−0.50 D≤SE≤+2.00 D |
| Number of subjects |
355 |
208 |
615 |
610 |
30 |
1052 |
|
| Age (years) |
Range |
0.3~79 |
2~64 |
16~27 |
15~33 |
19~27 |
16~32 |
| |
Average |
13.44±9.28 |
20.06±14.01 |
20.70±1.59 |
20.85±1.73 |
20.94±1.53 |
20.91±1.78 |
| Gender |
Male |
0.535 |
0.474 |
0.503 |
0.476 |
0.486 |
0.597 |
| |
Female |
0.465 |
0.526 |
0.497 |
0.524 |
0.514 |
0.403 |
| Refraction (D) |
Range |
−6.00~-9.00 |
−30.00~-9.25 |
−4.00D~-5.87D |
−6.00D~-9.00D |
−10.00~-9.25 |
−0.5~+2.00 |
| |
Average |
−7.70±0.95 |
−14.46±4.51 |
−5.06±0.52 |
−7.00±0.88 |
−9.64±0.31 |
0.25±0.52 |
| Axial length (mm) |
Range |
not available |
not available |
23.39~28.36 |
24.00~29.30 |
25.29~29.32 |
20.30~26.53 |
| Average | 25.79±0.81 | 26.48±0.91 | 27.29±0.83 | 23.60±0.79 | |||
Abbreviations: S,spherical refraction; SE,spherical equivalent.
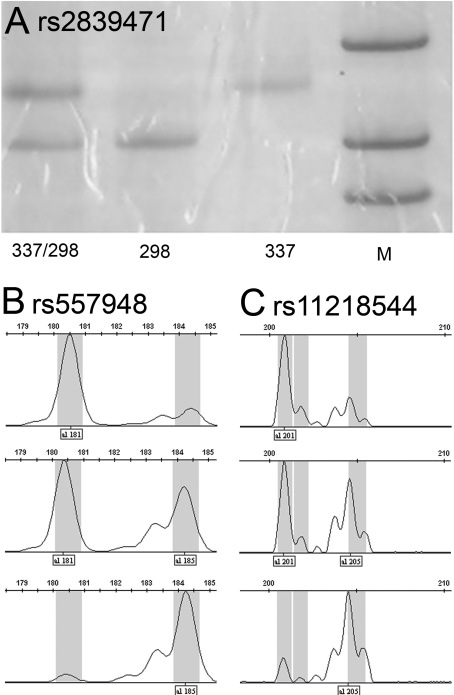
Figure 1.
Genotyping of rs2839471, rs577948, and rs11218544. The three SNPs were successfully genotyped in 2,870 subjects. M: 100 bp DNA ladder. A: RFLP analysis of SNP rs2839471. The double bands at 337 bp and 298 bp represent genotype C/T, the single band at 298 bp represents C/C, and the single band at 337 bp represents T/T. Capillary electrophoresis analysis was used for genotyping rs577948 and rs11218544. The three peak patterns represent three different SNP genotypes. B: SNP rs577948: the single peak at 181 bp is genotype A/A, the double peaks at 181 bp and 185 bp are A/G, and the single peak at 185 bp is G/G. C: SNP rs11218544: the single peak at 201 bp is T/T, the double peaks at 201 bp and 205 bp are T/G, and the single peak at 205 bp is G/G.
Table 3. Allele frequency and genotypes of the three SNPs in myopia cases and healthy controls. Myopia was classified as early onset high myopia and complex myopia.
| SNPs | Allele (A/B) | Classification of subjects | Number of subjects | MAF | Genotype | p value* | OR | 95%CI | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
A/A |
A/B |
B/B |
Allele |
Genotype |
|
|
|
rs2839471 |
C/T |
myopia |
complex (−10.00D≤S≤-4.00D) |
1255 |
0.489 |
293 |
641 |
321 |
0.393 |
0.266 |
1.052 |
0.937–1.181 |
| |
|
|
early onset (S≤-6.00D) |
563 |
0.472 |
116 |
300 |
147 |
0.838 |
0.131 |
0.985 |
0.852–1.139 |
| |
|
|
complex+early onset |
1818 |
0.484 |
409 |
941 |
468 |
0.582 |
0.151 |
1.031 |
0.926–1.148 |
| |
|
healthy control |
−0.50D≤SE≤+2.00D |
1052 |
0.476 |
248 |
506 |
298 |
|
|
|
|
|
rs577948 |
A/G |
myopia |
complex (−10.00D≤S≤-4.00D) |
1255 |
0.494 |
302 |
665 |
288 |
0.307 |
0.269 |
1.062 |
0.946–1.193 |
| |
|
|
early onset (S≤-6.00D) |
563 |
0.491 |
123 |
307 |
133 |
0.973 |
0.282 |
1.003 |
0.867–1.159 |
| |
|
|
complex+early onset |
1818 |
0.499 |
425 |
972 |
421 |
0.439 |
0.207 |
1.043 |
0.937–1.162 |
| |
|
healthy control |
−0.50D≤SE≤+2.00D |
1052 |
0.49 |
251 |
530 |
271 |
|
|
|
|
|
rs11218544 |
G/T |
myopia |
complex (−10.00D≤S≤-4.00D) |
1255 |
0.401 |
200 |
607 |
448 |
0.032 |
0.098 |
1.14 |
1.012–1.284 |
| |
|
|
early onset (S≤-6.00D) |
563 |
0.382 |
83 |
264 |
216 |
0.515 |
0.771 |
1.051 |
0.905–1.220 |
| |
|
|
complex+early onset |
1818 |
0.395 |
283 |
871 |
664 |
0.061 |
0.17 |
1.112 |
0.995–1.242 |
| healthy control | −0.50D≤SE≤+2.00D | 1052 | 0.37 | 142 | 495 | 415 | ||||||
Note: MAF=minor allele frequency; S=spherical refraction; SE=spherical equivalent. *P value was calculated by chi-square test between myopia and normal control.
Table 4. Allele frequency and genotypes of the three SNPs in myopia cases and healthy controls. Myopia was classified based on the degrees of refractive errors.
| SNPs | Allele (A/B) | Classification of subjects | Number of subjects | MAF | Genotype | p value* | OR | 95%CI | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
A/A |
A/B |
B/B |
Allele |
Genotype |
|
|
|
rs2839471 |
C/T |
myopia |
−6.00D<S≤-4.00D |
615 |
0.497 |
153 |
313 |
149 |
0.132 |
0.19 |
1.114 |
0.968~1.283 |
| |
|
|
−9.25D<S≤-6.00D |
965 |
0.478 |
212 |
499 |
254 |
0.899 |
0.269 |
1.008 |
0.891~1.141 |
| |
|
|
S≤-9.25D |
238 |
0.456 |
44 |
129 |
65 |
0.422 |
0.153 |
0.921 |
0.755~1.125 |
| |
|
Healthy control |
−0.50D≤SE≤+2.00D |
1052 |
0.476 |
248 |
506 |
298 |
|
|
|
|
|
rs577948 |
A/G |
myopia |
−6.00D<S≤-4.00D |
615 |
0.497 |
152 |
315 |
148 |
0.477 |
0.735 |
1.052 |
0.914–1.211 |
| |
|
|
−9.25D<S≤-6.00D |
965 |
0.499 |
216 |
531 |
218 |
0.591 |
0.099 |
1.034 |
0.914–1.171 |
| |
|
|
S≤-9.25D |
238 |
0.496 |
57 |
126 |
55 |
0.589 |
0.673 |
1.056 |
0.866–1.289 |
| |
|
Healthy control |
−0.50D≤SE≤+2.00D |
1052 |
0.49 |
251 |
530 |
271 |
|
|
|
|
|
rs11218544 |
G/T |
myopia |
−6.00D<S≤-4.00D |
615 |
0.402 |
103 |
289 |
223 |
0.065 |
0.148 |
1.146 |
0.992–1.323 |
| |
|
|
−9.25D<S≤-6.00D |
965 |
0.397 |
153 |
460 |
352 |
0.082 |
0.21 |
1.119 |
0.986–1.271 |
| |
|
|
S≤-9.25D |
238 |
0.37 |
27 |
122 |
89 |
0.984 |
0.446 |
0.998 |
0.812–1.226 |
| Healthy control | −0.50D≤SE≤+2.00D | 1052 | 0.37 | 142 | 495 | 415 | ||||||
Note: MAF=minor allele frequency; S=spherical refraction; SE=spherical equivalent. *P value was calculated by χ2 test between myopia and normal control.
Discussion
In this study, we genotyped three SNPs that were previously reported to be significantly associated with myopia. The SNP rs2839471 is located in the region of the uromodulin-like 1 (UMODL1) gene, which may be associated with extracellular matrix and involved in the formation of sclera [37,38]. The SNPs rs577948 and rs11218544 are located in a region that includes two genes, BH3-like motif containing protein (BLID) and LOC39959. BLID is expressed in mitochondria, and may function in apoptosis in pathological myopia [39–41]. LOC399959 is a hypothetical noncoding RNA [30]. Further study may elaborate the function of these genes. In the previous Japanese studies, axial length or spherical equivalent were used as criteria for recruiting the myopia patients. The proportion of early onset myopia or complex myopia was not reported [20,30]. In this study, no significant association was found between myopia and the three SNPs examined, regardless of whether myopia was classified based on genetic contribution (early onset high myopia and complex myopia) or based on degrees of spherical refraction. Therefore, our results suggest that these three SNPs may not be associated with myopia in the Chinese subjects we studied. Currently, multiple association studies are inconsistent and the causes of these contradictions have been widely analyzed [42,43]. The reasons why our results in the Chinese population were not identical to the Japanese results are likely related to the recruitment criteria, environmental influences, and genetic differences between the Chinese population and Japanese population because an earlier Chinese replication of a Japanese study on chromosome 11q24.1 (rs577948) also failed to confirm the reported association [44]. Furthermore, another explanation may be that the genetic variants interact with one another, and the interaction between genes and environmental factors vary in different human populations, although this has not been systematically assessed [45]. Although our results do not support the previous findings related to these SNPs [20,30], the results obtained with a Japanese population cannot be considered false-positives at this stage. However, this result must be analyzed with great caution until additional replication studies confirm a reported association, especially for myopia.
The distribution of sexes was not completely matched in the myopia and healthy control groups. However, this does not affect our conclusions because the results were unaffected when male subjects were randomly excluded to reach a balanced proportion of sexes. In addition, on average our study subjects were younger than those in the Japanese study for both cases and controls. This should not influence our conclusion because myopia is an early onset disease, and the degree of refractive error in myopia usually increases with age [46–48]. In our study females are more myopic (odds ratio>1, p<0.05) than males, which is consistent with previous reports [49,50]. On the other hand, males with a higher prevalence of myopia or no difference among genders were also reported [45,51,52]. Lower age as a predictor of myopia (odds ratio<1,p<0.001) was also consistent with results reported in other populations [53,54].
Myopia is a genetically heterogeneous disease, and both early onset and complex traits have been suggested to be associated with myopia. Traditionally, myopia has generally been classified based on degrees of spherical refraction or their closely related axial length [20,30]. In this study, we attempted to classify myopia based on either the type of genetic contribution, or based on degrees of spherical refraction. We did not find a significant association between the SNPs analyzed and myopia in either case. Additional studies are expected to validate the original findings [20,30].
Acknowledgments
We would like to thank all subjects for their participation. This study was supported in part by the National Science Foundation for Distinguished Young Scholars (30725044 to Q.Z.) and Fundamental Research Funds of the State Key Lab of Ophthalmology, Sun Yat-sen University.
References
- 1.Ip JM, Huynh SC, Robaei D, Rose KA, Morgan IG, Smith W, Kifley A, Mitchell P. Ethnic differences in the impact of parental myopia: findings from a population-based study of 12-year-old Australian children. Invest Ophthalmol Vis Sci. 2007;48:2520–8. doi: 10.1167/iovs.06-0716. [DOI] [PubMed] [Google Scholar]
- 2.Fan DS, Lam DS, Lam RF, Lau JT, Chong KS, Cheung EY, Lai RY, Chew SJ. Prevalence, incidence, and progression of myopia of school children in Hong Kong. Invest Ophthalmol Vis Sci. 2004;45:1071–5. doi: 10.1167/iovs.03-1151. [DOI] [PubMed] [Google Scholar]
- 3.Choo V. A look at slowing progression of myopia. Lancet. 2003;361:1622–3. doi: 10.1016/S0140-6736(03)13315-6. [DOI] [PubMed] [Google Scholar]
- 4.Lam CS, Goldschmidt E, Edwards MH. Prevalence of myopia in local and international schools in Hong Kong. Optom Vis Sci. 2004;81:317–22. doi: 10.1097/01.opx.0000134905.98403.18. [DOI] [PubMed] [Google Scholar]
- 5.Goh WS, Lam CS. Changes in refractive trends and optical components of Hong Kong Chinese aged 19–39 years. Ophthalmic Physiol Opt. 1994;14:378–82. [PubMed] [Google Scholar]
- 6.Bloom RI, Friedman IB, Chuck RS. Increasing rates of myopia: the long view. Curr Opin Ophthalmol. 2010;21:247–8. doi: 10.1097/ICU.0b013e328339f1dd. [DOI] [PubMed] [Google Scholar]
- 7.Bar Dayan Y, Levin A, Morad Y, Grotto I, Ben-David R, Goldberg A, Onn E, Avni I, Levi Y, Benyamini OG. The changing prevalence of myopia in young adults: a 13-year series of population-based prevalence surveys. Invest Ophthalmol Vis Sci. 2005;46:2760–5. doi: 10.1167/iovs.04-0260. [DOI] [PubMed] [Google Scholar]
- 8.He M, Zheng Y, Xiang F. Prevalence of myopia in urban and rural children in mainland China. Optom Vis Sci. 2009;86:40–4. doi: 10.1097/OPX.0b013e3181940719. [DOI] [PubMed] [Google Scholar]
- 9.Dirani M, Shekar SN, Baird PN. Adult-onset myopia: the Genes in Myopia (GEM) twin study. Invest Ophthalmol Vis Sci. 2008;49:3324–7. doi: 10.1167/iovs.07-1498. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–6. [PubMed] [Google Scholar]
- 11.Young TL, Ronan SM, Drahozal LA, Wildenberg SC, Alvear AB, Oetting WS, Atwood LD, Wilkin DJ, King RA. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998;63:109–19. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–24. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naiglin L, Gazagne C, Dallongeville F, Thalamas C, Idder A, Rascol O, Malecaze F, Calvas P. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002;39:118–24. doi: 10.1136/jmg.39.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–6. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–60. [PubMed] [Google Scholar]
- 16.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–7. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23-q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007;13:229–36. [PMC free article] [PubMed] [Google Scholar]
- 19.Lam CY, Tam PO, Fan DS, Fan BJ, Wang DY, Lee CW, Pang CP, Lam DS. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 20.Nishizaki R, Ota M, Inoko H, Meguro A, Shiota T, Okada E, Mok J, Oka A, Ohno S, Mizuki N. New susceptibility locus for high myopia is linked to the uromodulin-like 1 (UMODL1) gene region on chromosome 21q22.3. Eye (Lond) 2009;23:222–9. doi: 10.1038/eye.2008.152. [DOI] [PubMed] [Google Scholar]
- 21.Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes T, Ciner E, Bailey-Wilson JE. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet. 2004;75:448–59. doi: 10.1086/423789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojciechowski R, Moy C, Ciner E, Ibay G, Reider L, Bailey-Wilson JE, Stambolian D. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–99. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008;32:454–63. doi: 10.1002/gepi.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nürnberg G, Jacobi FK, Broghammer M, Becker C, Blin N, Nurnberg P, Stephani U, Pusch CM. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008;21:429–38. [PubMed] [Google Scholar]
- 26.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Confirmation of linkage to ocular refraction on chromosome 22q and identification of a novel linkage region on 1q. Arch Ophthalmol. 2007;125:80–5. doi: 10.1001/archopht.125.1.80. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Li S, Xiao X, Jia X, Guo X. Confirmation of a genetic locus for X-linked recessive high myopia outside MYP1. J Hum Genet. 2007;52:469–72. doi: 10.1007/s10038-007-0130-9. [DOI] [PubMed] [Google Scholar]
- 28.Andrew T, Maniatis N, Carbonaro F, Liew SH, Lau W, Spector TD, Hammond CJ. Identification and replication of three novel myopia common susceptibility gene loci on chromosome 3q26 using linkage and linkage disequilibrium mapping. PLoS Genet. 2008;4:e1000220. doi: 10.1371/journal.pgen.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metlapally R, Li YJ, Tran-Viet KN, Abbott D, Czaja GR, Malecaze F, Calvas P, Mackey D, Rosenberg T, Paget S, Zayats T, Owen MJ, Guggenheim JA, Young TL. COL1A1 and COL2A1 genes and myopia susceptibility: evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009;50:4080–6. doi: 10.1167/iovs.08-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Iida T, Matsuo K, Tajima K, Yoshimura N, Matsuda F. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, Hysi PG, Despriet DD, van Koolwijk LM, Ho L, Ramdas WD, Czudowska M, Kuijpers RW, Amin N, Struchalin M, Aulchenko YS, van Rij G, Riemslag FC, Young TL, Mackey DA, Spector TD, Gorgels TG, Willemse-Assink JJ, Isaacs A, Kramer R, Swagemakers SM, Bergen AA, van Oosterhout AA, Oostra BA, Rivadeneira F, Uitterlinden AG, Hofman A, de Jong PT, Hammond CJ, Vingerling JR, Klaver CC. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hysi PG, Young TL, Mackey DA, Andrew T, Fernandez-Medarde A, Solouki AM, Hewitt AW, Macgregor S, Vingerling JR, Li YJ, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CC, Hammond CJ. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y, Zhang D, Zhao P, Zhang Q, Tam PO, Sun L, Zuo X, Zhou X, Xiao X, Hu J, Li Y, Cai L, Liu X, Lu F, Liao S, Chen B, He F, Gong B, Lin H, Ma S, Cheng J, Zhang J, Chen Y, Zhao F, Yang X, Chen Y, Yang C, Lam DS, Li X, Shi F, Wu Z, Lin Y, Yang J, Li S, Ren Y, Xue A, Fan Y, Li D, Pang CP, Zhang X, Yang Z. Genetic variants at 13q12.12 are associated with high myopia in the han chinese population. Am J Hum Genet. 2011;88:805–13. doi: 10.1016/j.ajhg.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Qu J, Xu X, Zhou X, Zou H, Wang N, Li T, Hu X, Zhao Q, Chen P, Li W, Huang K, Yang J, He Z, Ji J, Wang T, Li J, Li Y, Liu J, Zeng Z, Feng G, He L, Shi Y. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011;20:2861–8. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 35.Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC, Chaurasia A, Chen CH, Chen J, Chen YT, Chu J, Cutiongco-de la Paz EM, De Ungria MC, Delfin FC, Edo J, Fuchareon S, Ghang H, Gojobori T, Han J, Ho SF, Hoh BP, Huang W, Inoko H, Jha P, Jinam TA, Jin L, Jung J, Kangwanpong D, Kampuansai J, Kennedy GC, Khurana P, Kim HL, Kim K, Kim S, Kim WY, Kimm K, Kimura R, Koike T, Kulawonganunchai S, Kumar V, Lai PS, Lee JY, Lee S, Liu ET, Majumder PP, Mandapati KK, Marzuki S, Mitchell W, Mukerji M, Naritomi K, Ngamphiw C, Niikawa N, Nishida N, Oh B, Oh S, Ohashi J, Oka A, Ong R, Padilla CD, Palittapongarnpim P, Perdigon HB, Phipps ME, Png E, Sakaki Y, Salvador JM, Sandraling Y, Scaria V, Seielstad M, Sidek MR, Sinha A, Srikummool M, Sudoyo H, Sugano S, Suryadi H, Suzuki Y, Tabbada KA, Tan A, Tokunaga K, Tongsima S, Villamor LP, Wang E, Wang Y, Wang H, Wu JY, Xiao H, Xu S, Yang JO, Shugart YY, Yoo HS, Yuan W, Zhao G, Zilfalil BA. Mapping human genetic diversity in Asia. Science. 2009;326:1541–5. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Wang P, Li S, Xiao X, Jia X, Guo X, Kong QP, Yao YG, Zhang Q. Mitochondrial DNA haplogroup distribution in Chaoshanese with and without myopia. Mol Vis. 2010;16:303–9. [PMC free article] [PubMed] [Google Scholar]
- 37.Shibuya K, Nagamine K, Okui M, Ohsawa Y, Asakawa S, Minoshima S, Hase T, Kudoh J, Shimizu N. Initial characterization of an uromodulin-like 1 gene on human chromosome 21q22.3. Biochem Biophys Res Commun. 2004;319:1181–9. doi: 10.1016/j.bbrc.2004.05.094. [DOI] [PubMed] [Google Scholar]
- 38.Di Schiavi E, Riano E, Heye B, Bazzicalupo P, Rugarli EI. UMODL1/Olfactorin is an extracellular membrane-bound molecule with a restricted spatial expression in olfactory and vomeronasal neurons. Eur J Neurosci. 2005;21:3291–300. doi: 10.1111/j.1460-9568.2005.04164.x. [DOI] [PubMed] [Google Scholar]
- 39.Xu GZ, Li WW, Tso MO. Apoptosis in human retinal degenerations. Trans Am Ophthalmol Soc. 1996;94:411–30. [PMC free article] [PubMed] [Google Scholar]
- 40.Mao J, Liu S, Wen D, Tan X, Fu C. Basic fibroblast growth factor suppresses retinal neuronal apoptosis in form-deprivation myopia in chicks. Curr Eye Res. 2006;31:983–7. doi: 10.1080/02713680600910510. [DOI] [PubMed] [Google Scholar]
- 41.Broustas CG, Gokhale PC, Rahman A, Dritschilo A, Ahmad I, Kasid U. BRCC2, a novel BH3-like domain-containing protein, induces apoptosis in a caspase-dependent manner. J Biol Chem. 2004;279:26780–8. doi: 10.1074/jbc.M400159200. [DOI] [PubMed] [Google Scholar]
- 42.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Murillo L, Greenberg DA. Genetic association analysis: a primer on how it works, its strengths and its weaknesses. Int J Androl. 2008;31:546–56. doi: 10.1111/j.1365-2605.2008.00896.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhao F, Bai J, Chen W, Xue A, Li C, Yan Z, Chen H, Lu F, Hu Y, Qu J, Zeng C, Zhou X. Evaluation of BLID and LOC399959 as candidate genes for high myopia in the Chinese Han population. Mol Vis. 2010;16:1920–7. [PMC free article] [PubMed] [Google Scholar]
- 45.Wojciechowski R. Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet. 2011;79:301–20. doi: 10.1111/j.1399-0004.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin LL, Shih YF, Hsiao CK, Chen CJ. Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singapore. 2004;33:27–33. [PubMed] [Google Scholar]
- 47.Saka N, Ohno-Matsui K, Shimada N, Sueyoshi S, Nagaoka N, Hayashi W, Hayashi K, Moriyama M, Kojima A, Yasuzumi K, Yoshida T, Tokoro T, Mochizuki M. Long-term changes in axial length in adult eyes with pathologic myopia. Am J Ophthalmol. 2010;150:562–8. doi: 10.1016/j.ajo.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Low W, Dirani M, Gazzard G, Chan YH, Zhou HJ, Selvaraj P, Au Eong KG, Young TL, Mitchell P, Wong TY, Saw SM. Family history, near work, outdoor activity, and myopia in Singapore Chinese preschool children. Br J Ophthalmol. 2010;94:1012–6. doi: 10.1136/bjo.2009.173187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai MY, Lin LL, Lee V, Chen CJ, Shih YF. Estimation of heritability in myopic twin studies. Jpn J Ophthalmol. 2009;53:615–22. doi: 10.1007/s10384-009-0724-1. [DOI] [PubMed] [Google Scholar]
- 50.Wong TY, Foster PJ, Hee J, Ng TP, Tielsch JM, Chew SJ, Johnson GJ, Seah SK. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Invest Ophthalmol Vis Sci. 2000;41:2486–94. [PubMed] [Google Scholar]
- 51.Zylbermann R, Landau D, Berson D. The influence of study habits on myopia in Jewish teenagers. J Pediatr Ophthalmol Strabismus. 1993;30:319–22. doi: 10.3928/0191-3913-19930901-12. [DOI] [PubMed] [Google Scholar]
- 52.Tan CS, Chan YH, Wong TY, Gazzard G, Niti M, Ng TP, Saw SM. Prevalence and risk factors for refractive errors and ocular biometry parameters in an elderly Asian population: the Singapore Longitudinal Aging Study (SLAS). Eye (Lond) 2011;25:1294–301. doi: 10.1038/eye.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu N, Nomura H, Ando F, Niino N, Miyake Y, Shimokata H. Refractive errors and factors associated with myopia in an adult Japanese population. Jpn J Ophthalmol. 2003;47:6–12. doi: 10.1016/s0021-5155(02)00620-2. [DOI] [PubMed] [Google Scholar]
- 54.Sherwin JC, Kelly J, Hewitt AW, Kearns LS, Griffiths LR, Mackey DA. Prevalence and predictors of refractive error in a genetically isolated population: the Norfolk Island Eye Study. Clin Experiment Ophthalmol. 2011;39:734–42. doi: 10.1111/j.1442-9071.2011.02579.x. [DOI] [PubMed] [Google Scholar]