Abstract
Background
It has been demonstrated that PALB2 acts as a bridging molecule between the BRCA1 and BRCA2 proteins and is responsible for facilitating BRCA2-mediated DNA repair. Truncating mutations in the PALB2 gene have been reported to be enriched in Fanconi anemia and breast cancer patients in various populations.
Methods
We evaluated the contribution of PALB2 germline mutations in 279 African-American breast cancer patients including 29 patients with a strong family history, 29 patients with a moderate family history, 75 patients with a weak family history, and 146 non-familial or sporadic breast cancer cases.
Results
After direct sequencing of all the coding exons, exon/intron boundaries, 5′UTR and 3′UTR of PALB2, three (1.08%; 3 in 279) novel monoallelic truncating mutations were identified: c.758dupT (exon4), c.1479delC (exon4) and c.3048delT (exon 10); together with 50 sequence variants, 27 of which are novel. None of the truncating mutations were found in 262 controls from the same population.
Conclusions
PALB2 mutations are present in both familial and non-familial breast cancer among African-Americans. Rare PALB2 mutations account for a small but substantial proportion of breast cancer patients.
Keywords: Breast cancer, PALB2, Mutation, African-American, Sequencing
INTRODUCTION
Breast cancer is a global health problem as about 1 million women are diagnosed with it annually and it is the second leading cause of cancer death among women in the United States. In 2010, more than 207,090 new cases of invasive breast cancer were diagnosed.1 A deadly combination of genetic, as well as endogenous and exogenous factors, lead to an increased risk for breast cancer. Study of germline mutations in BRCA1 and BRCA2 over the past two decades has established their role in inherited breast and/or ovarian cancers. Mutation screening of genes functionally related to BRCA1 and/or BRCA2 has revealed mutations in genes such as CHEK2, ATM, BRIP1, and PALB2. As mutations in BRCA1 and BRCA2 have been suggested to be rare high-penetrance mutations, mutations in ATM, CHEK2, BRIP1 and PALB2 were classified as rare moderate-penetrance mutations, while single nucleotide polymorphisms (SNPs) identified by genome-wide association studies were characterized as common low-penetrance genetic variants. Although mutations/variants in the above-mentioned breast cancer susceptibility genes as well as TP53 and PTEN account for a significant fraction of breast cancer risk, more than 70% of the genetic component of breast cancer risk remains uncharacterized.2, 3
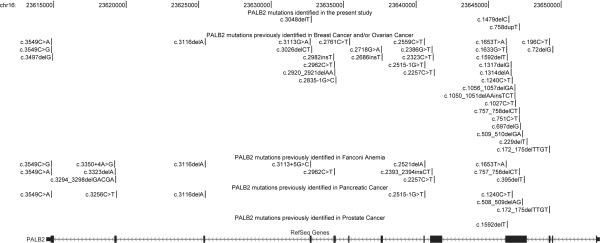
As an essential component of the BRCA1-PALB2-BRCA2-RAD51 pathway, the protein PALB2 (partner and localizer of BRCA2) acts as a bridging molecule that directly connects BRCA1 and BRCA2 to form a “BRCA complex” and subsequently facilitates BRCA2-mediated DNA repair.4 It has been demonstrated that PALB2 plays a crucial role in maintenance of genomic integrity and stability by executing double-strand break repair via homologous recombination, and tumor suppression of Fanconi anemia and cancers. Monoallelic/heterozygous mutations in Fanconi anemia genes have been implicated in breast cancer.4, 5 The similarity of clinical phenotypes caused by biallelic/homozygous mutations in BRCA2/FANCD1 and PALB2/FANCN pointed out that Fanconi anemia and breast cancer might share some genetic risk factors. To date, deleterious mutations in PALB2 have been identified in Fanconi anemia,6, 7 breast cancer,8-24 ovarian cancer,17, 25 pancreatic cancer,24, 26-28 and prostate cancer9, 29 patients in various populations (Figure 1). The spectrum, penetrance and clinical significance of PALB2 mutations in diverse populations remain to be further characterized.
Figure 1.
Deleterious mutations in the PALB2 gene
Chromosomal positions were based on Human Feb. 2009 assembly (GRCh37/hg19).
In this study, we have conducted mutational analysis and elucidated the mutation spectrum of the PALB2 gene in a cohort of 279 female African-American breast cancer patients unselected by family history, early age of onset, or BRCA1/2 mutation status.
MATERIAL AND METHODS
Study subjects
Two hundred and seventy-nine African-American women diagnosed with breast cancer were recruited at the Cancer Risk Clinic at the University of Chicago, Chicago, from 1993 to 2008. The physicians who participated in this project were asked to inquire about the family history of cancer in as much detail as possible and to obtain accurate patient information. In order to have a comprehensive understanding of the mutation rate in both familial and non-familial breast cancer patients, we did not select the participants based on positive family history, early age of onset, or BRCA1/2 mutation status. Our cohort contains 279 breast cancer patients, including 29 patients with strong family history (three or more first-degree relatives [FDR] and/or second-degree relatives [SDR] affected by breast and/or ovarian cancer, FDR + SDR n ≥ 3); 29 patients with moderate family history (FDR + SDR n = 2); 75 patients with weak family history (FDR + SDR n = 1); and 146 non-familial or sporadic breast cancer cases). 19 patients were previously tested positive for BRCA1/2, 51 patients were tested negative, whereas the mutation screening has not been conducted in the rest 209 patients. The characteristics of the breast cancer cases are listed in Table 1. A total of 262 cancer-free African-American female volunteers were recruited as normal controls to examine whether the deleterious mutations/variants identified occur in the normal population. Informed consents were obtained from all the individuals before the collection of blood samples. This study was approved by the Institutional Review Boards of the University of Chicago.
Table 1.
279 African-American breast cancer patients in the present study
| Subject group | Family history of breast and/or ovarian caner |
Number | Age of onset, mean ± SD (years) |
|---|---|---|---|
|
BRCA1/2 mutation - positive |
high | 8 | 43.63 ± 12.52 |
| moderate | 5 | 43.60 ± 4.39 | |
| weak | 6 | 37.17 ± 10.44 | |
| sporadic | 0 | - | |
| total | 19 | 41.58 ± 10.25 | |
|
BRCA1/2 mutation - negative |
high | 11 | 43.19 ± 12.14 |
| moderate | 9 | 49.00 ± 8.19 | |
| weak | 20 | 44.63 ± 11.75 | |
| sporadic | 11 | 38.27 ± 7.24 | |
| total | 51 | 43.68 ± 10.71 | |
|
BRCA1/2 mutation - unknown |
high | 10 | 45.50 ± 11.30 |
| moderate | 15 | 42.27 ± 6.08 | |
| weak | 49 | 44.73 ± 8.77 | |
| sporadic | 135 | 44.04 ± 10.06 | |
| total | 209 | 44.14 ± 9.55 | |
| All | high | 29 | 44.07 ± 11.58 |
| moderate | 29 | 44.59 ± 7.05 | |
| weak | 75 | 44.08 ± 9.83 | |
| sporadic | 146 | 43.59 ± 9.97 | |
| total | 279 | 43.88 ± 9.81 |
Mutation identification by sequencing
Genomic DNA was isolated from peripheral blood leukocytes and stored at −20°C. Oligonucleotide primers were designed using Primer3 v0.4.030 to cover all the thirteen coding exons, exon/intron boundaries (at least 50 bp), 5′UTR and 3′UTR of the PALB2 gene (Table 2). Polymerase chain reaction (PCR) reactions were performed in a final volume of 10 μl containing 10 ng DNA, 5 pmol of each primer, 25 nmol MgCl2, 2 nmol of each dNTP, 1 μl of 10× PCR Buffer II, and 0.5U of AmpliTaq Gold® DNA Polymerase (Applied Biosystems, USA). The cycling profile included: an initial denaturation at 95°C for 5 min, followed by 20 touchdown cycles consisting of a denaturation step of 95°C for 30 s, primer annealing started from 64°C to 54°C (0.5°C decrease per cycle) for 30 s and an elongation step at 72°C for 1 min; then followed by 20 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 1 min; and a final extension step at 72°C for 10 min. PCR products were subsequently checked by agarose gel electrophoresis and cleaned by ExoSap-IT (USB, USA) prior to the cycle-sequencing reactions using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Capillary electrophoretic sequencing was performed on an automated Applied Biosystems 3730xl DNA Analyzer according to the manufacturer’s instructions. SeqScape analysis software v2.6.0 (Applied Biosystems, USA) was used to conduct base calling, quality assessment, and assembly. The average sequencing success rate was 99.08% for all amplicons. The amplicons harboring truncating mutations and one predicted deleterious variant of the PALB2 gene (see below) were also analyzed in the normal control subjects with an average call rate of 99.50%. In addition, we confirmed those three truncating mutations by bi-direction sequencing of the same DNA samples as well as other batches of DNA extracted from bloods of the corresponding patients.
Table 2.
Primers for PALB2 sequencing
| Primer | Primer sequence (5′->3′) | Amplicon size (bp) |
|---|---|---|
| PALB2_Ex1-F | ACAGCGCGGCTCTCCTTTAG | 410 |
| PALB2_Ex1-R | ATACTGCTGCCCTCGGACTG | |
| PALB2_Ex2&3-F | GTAGATTGTTATGGACCAGTGCTACT | 511 |
| PALB2_Ex2&3-R | GTCTATTGCTAGTCATTATCTTCACAC | |
| PALB2_Ex4A-F | TCTGCCTGAATGAAATGTCACTGA | 645 |
| PALB2_Ex4A-R | GGTCTTCTTAGGAATGTATCAACACC | |
| PALB2_Ex4B-F | TGAAATAAGAACTCACCTTTTAAGTCT | 600 |
| PALB2_Ex4B-R | TTTTCTGCAGAAAGAGGAGAGGTT | |
| PALB2_Ex4C-F | TGACACTCTTGATGGCAGGA | 683 |
| PALB2_Ex4C-R | AAGGAAGTGCCAGGCAAATA | |
| PALB2_Ex5A-F | TTAAATCTAGGAGATCCTATTCTCTTTG | 531 |
| PALB2_Ex5A-R | GTATAAAGTAATATGGATGAAGAAAGGC | |
| PALB2_Ex5B-F | GAGGGAAGCTGTATTTTTCCA | 636 |
| PALB2_Ex5B-R | CATGCTGTTTACATTCACTAAGGC | |
| PALB2_Ex6-F | GCTGCTGTTATAAGAGGAAATAAAGACA | 344 |
| PALB2_Ex6-R | GGGAAAAATAACCAATCCAAATCTG | |
| PALB2_Ex7-F | GCTCTTTCTTTTCACCTGCATAAGA | 492 |
| PALB2_Ex7-R | TGGGTATATGGGTGCTCACTATACA | |
| PALB2_Ex8-F | CCTTGTACAGTGAGAATACAAAAGAATGTGA | 271 |
| PALB2_Ex8-R | TAGGTTATTACCTGCACTTAAAACCA | |
| PALB2_Ex9-F | AAAAAAGTGAACCTAGTCCTTTAATATT | 581 |
| PALB2_Ex9-R | GCTCAAACTTCTGCCTTGGC | |
| PALB2_Ex10-F | ATATTATGCAGTTCAACAATGCGG | 386 |
| PALB2_Ex10-R | ACCTGGGTGATAGGAGGAGACTC | |
| PALB2_Ex11-F | ACCTCCTAAGACATGCTATGATGAATAA | 379 |
| PALB2_Ex11-R | GCCAGAAAATTTACCAAGCAATCA | |
| PALB2_Ex12-F | CAGAGCCTATCGGTCATTGCTT | 438 |
| PALB2_Ex12-R | TCTGGGGTTTGACTCAAGTCCA | |
| PALB2_Ex13-F | TTTAATTGTTTTTTGGATATGTAATCTGAA | 678 |
| PALB2_Ex13-R | TGAAATTTACACTTACTAGGCAAAGA |
Assessment of pathogenicity
All variants detected were classified based on their variant type as frameshift/truncating, nonsynonymous, synonymous, untranslated-5′UTR, untranslated-3′UTR, intronic, and 3′ downstream. Five different computational algorithms, SIFT,31 PolyPhen,32 PMUT,33 PANTHER,34 and Align GVGD35 were used to predict the functional impact of missense variants detected in this study. We constructed protein multiple sequence alignments (PMSAs) for use with Align GVGD by EXPRESSO(3DCoffee)::Regular36 which takes protein structure into account when performing multiple sequence alignment. Sequences of thirty-six sequences species were extracted from ENSEMBL37 for creating PMSAs file: Anolis carolinensis, Bos taurus, Callithrix jacchus, Cavia porcellus, Choloepus hoffmanni, Dasypus novemcinctus, Dipodomys ordii, Echinops telfairi, Equus caballus, Erinaceus europaeus, Felis catus, Gallus gallus, Gorilla gorilla, Homo sapiens, Loxodonta africana, Macaca mulatta, Macropus eugenii, Microcebus murinus, Monodelphis domestica, Mus musculus, Ochotona princeps, Ornithorhynchus anatinus, Oryctolagus cuniculus, Otolemur garnettii, Pan troglodytes, Pongo pygmaeus, Procavia capensis, Pteropus vampyrus, Rattus norvegicus, Sorex araneus, Spermophilus tridecemlineatus, Sus scrofa, Tarsius syrichta, Tupaia belangeri, Tursiops truncatus, Vicugna pacos.
Classification scores
Above prediction methods rely on PMSAs, and/or protein structural considerations, and/or evolutionary evaluation, additionally, their specificity and sensitivity vary. Hence, concordance between these in silico assessment programs is suggested to be an effective predictor to differentiate pathological missense variants from benign ones. We established a classification system based on the combined computational analysis for each nonsynonymous variant detected in this study. Firstly, a score of 0 (benign), 0.5 (possibly deleterious) and 1 (deleterious) was given to results of each of the five predictions: SIFT − tolerated = 0 and affect = 1; PolyPhen − benign = 0, possible damaging = 0.5 and probably damaging = 1; PMUT − neutral = 0 and pathologic =1; PANTHER − neutral = 0 and pathologic = 1; Align GVGD − C0 = 0, C15, C25, or C35 = 0.5, and, C45, C55, or C65 = 1. Secondly, for each variant, the scores for all five algorithms were summed and subsequently classified as follows: benign = 0, possibly benign = 0.5-2, possibly deleterious = 2.5-4, and deleterious = 4.5-5.
Nomenclature
The variants found in this study were named according to the GenBank PALB2 reference sequence NM_024675.3 and NP_078951, following the instructions provided by the Human Genome Variation Society.38 In addition, we checked the nomenclature by Mutalyzer v2.0.39
RESULTS
Variants identified
After direct sequencing of all the coding exons, exon/intron boundaries, 5′UTR and 3′UTR of the PALB2 gene, we identified a total of fifty-three sequence variants. Although neither nonsense mutations nor splicing variants which occur in canonical AG or GT splice acceptor or donor sites were detected, the nucleotide alterations found belong to frameshift-duplication, frameshift-deletion, nonsynonymous, synonymous, untranslated-5′UTR, untranslated-3′UTR, intronic, and 3′ downstream. Basically, all the sequence variants appear to be located throughout the PALB2 gene. Table 3 lists the truncating mutations and nonsynonymous variants identified in this study, and Table 4 lists all the novel synonymous and non-coding variants identified in the present study.
Table 3.
Truncating mutations and nonsynonymous variants of PALB2 discovered in the present study
| Location | Nucleotide change |
Protein change | rs# (SNP132) | Variant type | Computational prediction | Minor allele frequency (MAF) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | PolyPhen | PMUT | PANTHER | Align GVGD |
Re-score |
BRCA1/2 mutation positive cases (n = 19) |
BRCA1/2 mutation negative cases (n = 51) |
BRCA1/2 mutation unknown cases (n = 209) |
All cases (n = 279) |
Controls (n = 262) |
|||||
| exon2 | c.53A>G | p.Lys18Arg | - | nonsynonymous | tolerated (0.21) |
benign (1.252) |
neutral (0.046) |
neutral (−1.770) | C0 | benign (0) | 0.000 | 0.000 | 0.005 | 0.004 | - |
| exon4 | c.400G>A | p.Asp134Asn | - | nonsynonymous | tolerated (0.41) |
benign (0.979) |
neutral (0.154) |
neutral (−1.352) | C0 | benign (0) | 0.000 | 0.000 | 0.002 | 0.002 | - |
| exon4 | c.629C>T | p.Pro210Leu | rs57605939 | nonsynonymous | tolerated (0.44) |
benign (1.139) |
pathological (0.708) |
neutral (−0.666) | C0 | possibly benign (1) |
0.079 | 0.029 | 0.060 | 0.056 | - |
| exon4 | c.721A>G | p.Asn241Asp | rs113217267 | nonsynonymous | tolerated (0.62) |
benign (0.114) |
neutral (0.032) |
neutral (−0.597) | C0 | benign (0) | 0.000 | 0.030 | 0.014 | 0.016 | 0.004 |
| exon4 | c.758dupT | p.Ser254Ilefs*3 | - | frameshift- duplication |
truncating | truncating | truncating | truncating | truncating | truncating | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 |
| exon4 | c.925A>G | p.Ile309Val | rs3809683 | nonsynonymous | tolerated (0.30) |
benign (0.097) |
neutral (0.045) |
neutral (−0.266) | C0 | benign (0) | 0.056 | 0.010 | 0.012 | 0.015 | 0.035 |
| exon4 | c.1010T>C | p.Leu337Ser | rs45494092 | nonsynonymous | tolerated (0.73) |
possibly damaging (1.691) |
pathological (0.734) |
neutral (−2.282) | C0 | possibly benign (1.5) |
0.000 | 0.010 | 0.005 | 0.005 | 0.004 |
| exon4 | c.1479delC | p.Thr494Leufs*67 | - | frameshift- deletion |
truncating | truncating | truncating | truncating | truncating | truncating | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 |
| exon4 | c.1676A>G | p.Gln559Arg | rs152451 | nonsynonymous | tolerated (0.56) |
benign (0.175) |
neutral (0.070) |
neutral (−0.476) | C0 | benign (0) | 0.316 | 0.163 | 0.189 | 0.193 | 0.215 |
| exon5 | c.2014G>C | p.Glu672Gln | rs45532440 | nonsynonymous | tolerated (0.44) |
benign (1.348) |
neutral (0.107) |
neutral (−2.154) | C0 | benign (0) | 0.000 | 0.000 | 0.007 | 0.005 | - |
| exon7 | c.2590C>T | p.Pro864Ser | rs45568339 | nonsynonymous | tolerated (0.46) |
probably damaging (2.274) |
pathological (0.819) |
neutral (−1.5704) | C0 | possibly benign (2) |
0.000 | 0.020 | 0.002 | 0.005 | - |
| exon9 | c.2851T>C | p.Ser951Pro | - | nonsynonymous | tolerated (0.37) |
benign (0.044) |
pathological (0.652) |
neutral (−1.955) | C0 | possibly benign (1) |
0.000 | 0.010 | 0.000 | 0.002 | 0.002 |
| exon9 | c.2993G>A | p.Gly998Glu | rs45551636 | nonsynonymous | affect (0.01) | probably damaging (2.172) |
pathological (0.840) |
deleterious (− 4.581) |
C65 | deleterious (5) |
0.000 | 0.000 | 0.007 | 0.005 | 0.004 |
| exon10 | c.3048delT | p.Phe1016Leufs*17 | - | frameshift- deletion |
truncating | truncating | truncating | truncating | truncating | truncating | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 |
| exon10 | c.3056T>C | p.Val1019Ala | - | nonsynonymous | affect (0.01) | possibly damaging (1.583) |
pathological (0.577) |
deleterious (− 3.190) |
C25 | possibly deleterious (4) |
0.000 | 0.000 | 0.002 | 0.002 | 0.000 |
Three truncating mutations, p.Ser254Ilefs*3, p.Thr494Leufs*67, and p.Phe1016Leufs*17 identified in this study are shown in bold.
Table 4.
Novel synonymous and non-coding variants of PALB2 identified in the present study
| Location | Nucleotide change |
Variant type |
BRCA1/2 mutation positive cases (n = 19) |
BRCA1/2 mutation negative cases (n = 51) |
BRCA1/2 mutation unknown cases (n = 209) |
All cases (n = 279) |
Controls (n = 260) |
|---|---|---|---|---|---|---|---|
| 5′UTR | c.−194C>G | untranslated-5′UTR | 0.000 | 0.000 | 0.002 | 0.002 | - |
| exon4 | c.909C>T | synonymous | 0.000 | 0.000 | 0.005 | 0.004 | 0.002 |
| exon4 | c.1038A>G | synonymous | 0.000 | 0.010 | 0.000 | 0.002 | 0.000 |
| exon4 | c.1606C>T | synonymous | 0.000 | 0.031 | 0.007 | 0.011 | 0.002 |
| exon5 | c.1810C>T | synonymous | 0.000 | 0.000 | 0.005 | 0.004 | - |
| exon5 | c.2256A>G | synonymous | 0.000 | 0.020 | 0.007 | 0.009 | - |
| exon5 | c.2301C>A | synonymous | 0.000 | 0.000 | 0.002 | 0.002 | - |
| exon5 | c.2365C>T | synonymous | 0.000 | 0.000 | 0.002 | 0.002 | - |
| intron5 | c.2515−24A>G | intronic | 0.000 | 0.010 | 0.000 | 0.002 | - |
| intron7 | c.2748+121T>C | intronic | 0.000 | 0.010 | 0.005 | 0.005 | - |
| intron8 | c.2835−27C>T | intronic | 0.000 | 0.010 | 0.000 | 0.002 | 0.002 |
| intron9 | c.2996+124C>T | intronic | 0.000 | 0.010 | 0.000 | 0.002 | 0.002 |
| intron9 | c.2996+183delA | intronic | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 |
| intron10 | c.3113+131G>T | intronic | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 |
| intron10 | c.3114−40T>G | intronic | 0.000 | 0.000 | 0.002 | 0.002 | - |
| intron11 | c.3201+96G>A | intronic | 0.000 | 0.010 | 0.000 | 0.002 | - |
| intron11 | c.3201+112A>G | intronic | 0.000 | 0.000 | 0.002 | 0.002 | - |
| intron11 | c.3201+115T>C | intronic | 0.000 | 0.000 | 0.005 | 0.004 | - |
| intron11 | c.3202−45A>G | intronic | 0.000 | 0.000 | 0.005 | 0.004 | - |
| exon12 | c.3252G>A | synonymous | 0.000 | 0.000 | 0.002 | 0.002 | - |
| intron12 | c.3350+16T>G | intronic | 0.000 | 0.010 | 0.000 | 0.002 | - |
| 3′UTR | c.*232G>T | untranslated-3′UTR | 0.000 | 0.010 | 0.000 | 0.002 | - |
| 3′ downstream | c.*347A>C | 3′ downstream | 0.000 | 0.010 | 0.000 | 0.002 | - |
In 279 African-American women diagnosed with breast cancer, we found one deletion and two duplication mutations (totally 1.08%, 3 in 279) (Figure 1). They are all frameshift truncating mutations due to their possibility of altering the encoded protein, and turn out to be singleton mutations, namely, each of them was found only once in the present study. These three novel truncating mutations have not been deposited in The Single Nucleotide Polymorphism database (dbSNP) Build 13240 and have not been reported in the literature. They were not detected in any of the 262 normal controls in our cohort. We did not detect any nonsense, frameshift truncating, or deleterious missense mutations in 19 BRCA1/2 mutation carriers.
We also uncovered twelve nonsynonymous/missense variants (four novel and eight known), thirteen synonymous/silent variants (eight novel and five known), two untranslated-5′UTR variants (one novel and one known), one novel untranslated-3′UTR variant, twenty-one intronic variants (twelve novel and nine known), and one novel 3′ downstream variant. Forty (75.5%) variants are very rare (minor allele frequency, MAF < 0.01), six (11.3%) variants are rare (0.01 ≤ MAF ≤ 0.05), and the remaining seven (13.2%) variants are common (MAF > 0.05).
Patients with truncating mutations
The c.758dupT (p.Ser254Ilefs*3) mutation was identified in a patient diagnosed at the age of 45 years with infiltrating ductal carcinoma of the breast (IDC) not otherwise specified (NOS). Immunohistochemistry of her grade II tumor showed the expression of estrogen receptor (ER) and progesterone receptor (PR), as well as borderline expression of human epidermal growth factor receptor 2 (HER2/neu). Her family history of breast cancer is weak as only a half-sister was reported as having breast cancer at the age of 50 years and her maternal aunt had colorectal cancer at the age of 65 years, with the remaining family members reported as cancer-free. No additional family members were available for testing.
The patient who carried c.1479delC (p.Thr494Leufs*67) has a strong family history. She was diagnosed with grade III IDC NOS (ER+, PR+, but HER2/neu status unknown) at the age of 36 years. Her daughter, siblings, and parents never had cancer. However, she had maternal and paternal history of breast cancer and ovarian cancer. Her maternal grandmother had four sisters: one was healthy; two of them had breast cancer at the age of 45 and 68 years, respectively; while another one was diagnosed with breast cancer at the age of 58 years and subsequent ovarian cancer fourteen years later. In addition, three of four sisters of the patient’s father were diagnosed with breast cancer at the age of 37, 45 and >50 years, respectively. Because of the lack of DNA from the family members of the index patient, we were not able to determine from which side the mutation was inherited.
The patient with c.3048delT (p.Phe1016Leufs*17) was diagnosed with grade II IDC NOS (ER-, PR-, but HER2/neu status unknown) at the age of 60 years (died four years later). A family history of breast and/or ovarian cancer was not reported, but, she had a sister who died of colorectal cancer at the age of 55 years, a mother who died from leukemia at the age of 33 years, and her maternal grandfather who had pancreatic cancer at the age of 52 years. It is very likely that the deleterious mutation was inherited from her maternal side; however, we could not confirm this due to the absence of DNA samples.
In silico analyses of nonsynonymous variants
Of the fifty-three variants identified in the breast cancer cases, twelve were nonsynonymous/missense and analyzed by the above-mentioned five computational algorithms: SIFT, PolyPhen, PMUT, PANTHER, and Align GVGD (Table 3). In order to obtain a better classification of the variants, a classification/re-score system was established based on the summed scores of these individual analyses to help assess the possible pathological effects of the nonsynonymous variants. Of the twelve missense variants, six were consistently (all the five programs) evaluated as benign, four as possibly benign (three of five, or four of five), one as possibly deleterious (four of five), and another as deleterious (all the five predictions).
Occurrences of variants in controls
We sequenced four amplicons containing all the three frameshift/truncating mutations, one predicted deleterious (c.2993G>A/p.Gly998Glu) and one possibly deleterious missense variant (c.3056T>C/p.Val1019Ala), in 262 cancer-free African-American women. None of the three truncating mutations were found in control subjects from the same population. The MAF of c.2993G>A was 0.005 in caseswhich is comparable to the MAF of 0.004 in 262 controls. The c.3056T>C was found with a MAF of 0.002 in cases but not in controls.
DISCUSSION
PALB2 as a breast cancer susceptibility gene
Efforts have been made to investigate the mutation spectrum of PALB2 in breast cancer patients, and deleterious mutations have been reported in various populations such as the British,8, 23 Irish and Scot,23 Finnish,9, 15 Scottish-Canadian,10 French-Canadian,11 Spanish,12 Chinese,13 South African,14 Italian,16, 19, 22 Polish,17 Netherlander,18 French,23 German,20, 23 Russian,20 African-American,23 European-American,24 as well as Australian and New Zealander.21, 41 All the deleterious mutations associated with breast cancer found in PALB2 appeared to be singletons, except a few were recurrent mutations (c.3116delA, c.3113G>A, and c.3549C>G in British,8 c.751C>T in Chinese,13 c.509-510delGA in Polish,17 and c.3113G>A in Australian and New Zealander21), and founder mutations (c.1592delT in Finnish9, 15 and c.2323C>T in French-Canadian11). Five PALB2 mutations found in the UK were estimated to be associated with a 2.3 fold increased risk.8 The recurrent mutation c.3113G>A detected in Australia and New Zealand was speculated to originate from the UK and conferred an age-specific cumulative risk of 49% (95% CI: 15-93) to age 50 years and 91% (95% CI: 44-100) to age 70 years.21 The founder mutation c.1592delT was estimated to contribute a 6-fold increased breast cancer risk (95% CI: 2-17, equivalent to a risk of 40% to age 70 years) in Finland.9, 15
In spite of different study designs of the above mutational analyses, namely, full gene sequencing versus individual mutation screening, familial cases versus non-familial cases, selected versus unselected early age onset, female versus male patients, and the various sample size, the majority of mutations identified turned out to be rare with frequencies from 0.0011 (1/923, c.2982insT and c.2386G>T in British)8 to 0.0208 (1/48, c.697delG in South Africans).14 Mutation prevalence of PALB2 per study varied, from 0.56 to 2.65% in familial breast cancer patients, and < 1% in unselected cases. We found three truncating PALB2 mutations in our cohort of 279 female African-American breast cancer patients which consists of 133 familial and 146 non-familial or sporadic breast cancer cases. Of these three mutations, two (c.758dupT and c.1479delC) were found in individuals with family history of breast and/or ovarian cancer, and one (c.3048delT) was found in a non-familial patient. Therefore, the mutation prevalence in the present study are 1.08, 1.50 and 0.68% for overall, familial and non-familial, respectively. There is no significant discrepancy of mutation prevalence between our study and the literature. However, it is worth noting that, Ding et al. recently sequenced PALB2 in 139 African-American breast cancer cases (134 familial and 5 non-familial) and failed to detect any truncating mutation.42 Recently, Casadei et al. screened 1,144 familial breast cancer patients (172 Ashkenazi Jewish and 972 cases of multiple ancestries) and found two (out of 18) African-American patients carried PALB2 mutations.23 Taking their results into account, mutation prevalence of PALB2 in African-American women with breast cancer turn out to be 1.15 (5/436), 1.40 (4/285) and 0.66% (1/151) for overall, familial and non-familial, respectively. Given the low or undetectable frequencies of PALB2 mutations in different racial/ethnic groups, study sample size would have to be large to capture rare mutations. Moreover, the majority of the studies employed criteria that would enrich for PALB2 mutation carriers, for instance, positive family history of breast and/or ovarian cancer and early age of onset. It is therefore difficult to apply these estimates to the general population. Future studies evaluating mutation spectrum and prevalence in non-familial patients are warranted. To date, there is paucity of data on PALB2 germline mutation screenings in breast cancer patients of African ancestry, additional studies are needed to fill in this gap.
Genotype-phenotype correlation
To date, all the PALB2 deleterious mutations found in breast cancer are frameshift deletion/insertion, nonsense mutations, and mutations affecting splicing (Figure 1). It seems that mutations causing breast and/or ovarian cancer tend to cluster at the 5′ of the PALB2 gene; mutations in Fanconi anemia spread throughout the gene, mutations in pancreatic cancer might be enriched at the 5′ of PALB2 (although few studies have been carried out), and only one mutation, c.1592delT, has been reported in prostate cancer.
PALB2 has a coiled-coil domain (p.9-p.43, corresponding to c.25-c.129) at the N terminus which binds to the coiled-coil pre-BRCT motif of BRCA1; WD40 motifs (p.848-p.1186, corresponding to c.2542-c.3558) enriched in the C-terminal of PALB2 interacts with the N-terminal of BRCA2; and a region (p.611-p.738, corresponding to c.1831-c.2214) approximately in the middle has been identified as a binding site of MRG15 which is a MRG domain-containing protein involved in chromatin remodeling complexes.43 It appears that PALB2 mutations identified in Fanconi anemia and cancers do not affect the binding sites of BRCA1 and MRG15 directly (except c.72delG), but some are predicted to disrupt the BRCA2 binding site. Most of the mutations were mapped to exons 4 and 5 of the PALB2 gene, where the biological functions of the corresponding protein structure remain unknown. One possible explanation for the enrichment in exons 4 and 5 is that they are the largest two exons in PALB2. Thus, full-gene sequencing of PALB2 gene may be necessary, especially in populations in which recurrent and founder mutations have not yet been identified.
In addition, it has been proposed that, similar to BRCA1- but not BRCA2-related breast cancers, around 40% of PALB2-related breast cancer patients might be triple-negative (ER-, PR-, and HER2/neu-) tumors.4 In our African-American cohort, none of three mutation carriers had triple-negative tumor, which does not support the previous observation. A better understanding of PALB2-related breast cancer subtypes awaits future investigation in the same population. It is also worth noting that PALB2 mutations have been detected in <5% of familial pancreatic cancer patients, however, it remains to be determined whether screening for deleterious mutations of the PALB2 gene is warranted in families enriched for both breast cancer and pancreatic cancer cases.24, 26-28
CONCLUSIONS
Accumulated evidence suggests that PALB2 is a breast cancer susceptibility gene. We performed direct full gene sequencing of PALB2 in 133 familial and 146 non-familial female African-American breast cancer cases. Besides twelve nonsynonymous, thirteen synonymous, two untranslated-5′UTR, one untranslated-3′UTR, one 3′ downstream, and twenty-one intronic variants, we were able to identify three novel truncating mutations. Interestingly, these deleterious mutations were detectable in both familial and non-familial groups. Our data suggest that PALB2 mutations may be linked to both familial and non-familial breast cancer among African-Americans. Taking into account the fact that approximately 1-2% of familial breast cancer and 3-5% of familial pancreatic cancer patients have been identified to carry germline PALB2 mutations, better characterizing the mutation spectrum in diverse populations will definitely benefit the evaluation and consideration of the routine clinical testing of the PALB2 gene for patients with breast and/or other cancers.
Acknowledgments
We would like to thank all the families for providing samples voluntarily for this study.
Funding sources: This work was supported by grants from the National Cancer Institute at the National Institutes of Health (R01 CA141712, R01 CA142996-01, P50 CA125183), Entertainment Industry Foundation and Falk Medical Research Trust.
Footnotes
Conflict of interest statement: None of the authors has any potential financial conflict of interest related to this manuscript.
Conference presentation: Presented at 60th Annual Meeting of The American Society of Human Genetics (ASHG), Washington, DC, November 2-6 2010.
Contributor Information
Yonglan Zheng, Center for Clinical Cancer Genetics and Global Health, Department of Medicine, The University of Chicago, Chicago, IL 60637, USA
Jing Zhang, Center for Clinical Cancer Genetics and Global Health, Department of Medicine, The University of Chicago, Chicago, IL 60637, USA
Qun Niu, Center for Clinical Cancer Genetics and Global Health, Department of Medicine, The University of Chicago, Chicago, IL 60637, USA
Dezheng Huo, Department of Health Studies, The University of Chicago, Chicago, IL 60637, USA
Olufunmilayo I. Olopade, Center for Clinical Cancer Genetics and Global Health, Department of Medicine, The University of Chicago, Chicago, IL 60637, USA
REFERENCES
- 1.Society AC. [accessed May 25, 2011];Breast Cancer Overview. Available from URL: http://www.cancer.org/Cancer/BreastCancer/OverviewGuide/breast-cancer-overview-key-statistics.
- 2.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7(12):937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 3.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321–345. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 4.Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70(19):7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med. 2010;362(20):1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 7.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 8.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erkko H, Xia B, Nikkila J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446(7133):316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 10.Tischkowitz M, Xia B, Sabbaghian N, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104(16):6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foulkes WD, Ghadirian P, Akbari MR, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9(6):R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia MJ, Fernandez V, Osorio A, et al. Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Res Treat. 2009;113(3):545–551. doi: 10.1007/s10549-008-9945-0. [DOI] [PubMed] [Google Scholar]
- 13.Cao AY, Huang J, Hu Z, et al. The prevalence of PALB2 germline mutations in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res Treat. 2009;114(3):457–462. doi: 10.1007/s10549-008-0036-z. [DOI] [PubMed] [Google Scholar]
- 14.Sluiter M, Mew S, van Rensburg EJ. PALB2 sequence variants in young South African breast cancer patients. Fam Cancer. 2009;8(4):347–353. doi: 10.1007/s10689-009-9241-0. [DOI] [PubMed] [Google Scholar]
- 15.Heikkinen T, Karkkainen H, Aaltonen K, et al. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res. 2009;15(9):3214–3222. doi: 10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]
- 16.Papi L, Putignano AL, Congregati C, et al. A PALB2 germline mutation associated with hereditary breast cancer in Italy. Fam Cancer. 2010;9(2):181–185. doi: 10.1007/s10689-009-9295-z. [DOI] [PubMed] [Google Scholar]
- 17.Dansonka-Mieszkowska A, Kluska A, Moes J, et al. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med Genet. 2010;11:20. doi: 10.1186/1471-2350-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adank MA, van Mil SE, Gille JJ, Waisfisz Q, Meijers-Heijboer H. PALB2 analysis in BRCA2-like families. Breast Cancer Res Treat. 2011;127(2):357–362. doi: 10.1007/s10549-010-1001-1. [DOI] [PubMed] [Google Scholar]
- 19.Balia C, Sensi E, Lombardi G, Roncella M, Bevilacqua G, Caligo MA. PALB2: a novel inactivating mutation in a Italian breast cancer family. Fam Cancer. 2010;9(4):531–536. doi: 10.1007/s10689-010-9382-1. [DOI] [PubMed] [Google Scholar]
- 20.Bogdanova N, Sokolenko AP, Iyevleva AG, et al. PALB2 mutations in German and Russian patients with bilateral breast cancer. Breast Cancer Res Treat. 2011;126(2):545–550. doi: 10.1007/s10549-010-1290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Southey MC, Teo ZL, Dowty JG, et al. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 2010;12(6):R109. doi: 10.1186/bcr2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterlongo P, Catucci I, Pasquini G, et al. PALB2 germline mutations in familial breast cancer cases with personal and family history of pancreatic cancer. Breast Cancer Res Treat. 2011;126(3):825–828. doi: 10.1007/s10549-010-1305-1. [DOI] [PubMed] [Google Scholar]
- 23.Casadei S, Norquist BM, Walsh T, et al. Contribution to Familial Breast Cancer of Inherited Mutations in the BRCA2-interacting Protein PALB2. Cancer Res. 2011;71(6):2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofstatter EW, Domchek SM, Miron A, et al. PALB2 mutations in familial breast and pancreatic cancer. Fam Cancer. 2011;10(2):225–231. doi: 10.1007/s10689-011-9426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erkko H, Nikkilä J, Bützow R, et al. Occurrence of germline PALB2 mutations in ovarian cancer. Proceedings of the 57th Annual Meeting of the American Society of Human Genetics; 2007. Abstract A404. [Google Scholar]
- 26.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324(5924):217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tischkowitz MD, Sabbaghian N, Hamel N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137(3):1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slater EP, Langer P, Niemczyk E, et al. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78(5):490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 29.Pakkanen S, Wahlfors T, Siltanen S, et al. PALB2 variants in hereditary and unselected Finnish prostate cancer cases. J Negat Results Biomed. 2009;8:12. doi: 10.1186/1477-5751-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 31.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21(14):3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 34.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavtigian SV, Deffenbaugh AM, Yin L, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43(4):295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 37.Hubbard T, Barker D, Birney E, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30(1):38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29(1):6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 40.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong MW, Nordfors C, Mossman D, et al. BRIP1, PALB2, and RAD51C mutation analysis reveals their relative importance as genetic susceptibility factors for breast cancer. Breast Cancer Res Treat. 2011;127(3):853–859. doi: 10.1007/s10549-011-1443-0. [DOI] [PubMed] [Google Scholar]
- 42.Ding YC, Steele L, Chu LH, et al. Germline mutations in PALB2 in African-American breast cancer cases. Breast Cancer Res Treat. 2011;126(1):227–230. doi: 10.1007/s10549-010-1271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem. 2009;284(32):21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]