Abstract
The NCBI Taxonomy database (http://www.ncbi.nlm.nih.gov/taxonomy) is the standard nomenclature and classification repository for the International Nucleotide Sequence Database Collaboration (INSDC), comprising the GenBank, ENA (EMBL) and DDBJ databases. It includes organism names and taxonomic lineages for each of the sequences represented in the INSDC’s nucleotide and protein sequence databases. The taxonomy database is manually curated by a small group of scientists at the NCBI who use the current taxonomic literature to maintain a phylogenetic taxonomy for the source organisms represented in the sequence databases. The taxonomy database is a central organizing hub for many of the resources at the NCBI, and provides a means for clustering elements within other domains of NCBI web site, for internal linking between domains of the Entrez system and for linking out to taxon-specific external resources on the web. Our primary purpose is to index the domain of sequences as conveniently as possible for our user community.
A BRIEF HISTORY
The NCBI Taxonomy project began in 1991, when we designed the first version of the Entrez information retrieval system. At that time, each of the partners of what was to become the International Nucleotide Sequence Database Collaboration (INSDC)—GenBank, EMBL and the DDBJ—maintained the taxonomic nomenclature and classification in their own sequence entries independently. The classifications used by the three partners were clearly derived from a common source, but had drifted apart over the years. Sequence entries were regularly exchanged within the collaboration, but the source organism nomenclature and taxonomic classifications were inconsistent and were updated irregularly. Protein sequences were maintained separately from the nucleotide sequences, in two different databases—Swiss-Prot (1) and PIR (2). Each of these databases maintained their own taxonomies, each very different from the other and from the more closely related taxonomies in use by the INSDC partners.
Entrez (3) was the first system to link nucleotide sequences and protein sequences (from all of these sources) together with relevant abstracts from the scientific literature in a single unified resource. It was obviously important to provide a single taxonomic classification to index the entire set of entries in Entrez. The first step was to shuffle together the taxonomies from each of the contributing databases, each of which covered a somewhat different set of species with often very different internal classifications. The end result of this process was a hideous abomination, but it did provide a single classification that spanned all of the entries in Entrez, which we set out to improve. At this point we hosted series of taxonomy workshops to provide advice and direction for the project. David Hillis, John Taylor and Gary Olsen, in particular, put in a significant amount of time and effort in the initial cleanup of our merged classification.
The next step forward was the 1997 agreement by the INSDC members to resolve taxonomic issues of nomenclature and classification prior to the release of new sequence data. Sequences submitted to GenBank are screened for organism names that are new to the taxonomy database, and result in a taxonomy consult sent to the taxonomy group. Prior to this agreement, we would not see the organism names in entries from the collaborating databases until they had been released to the public—issues involving synonymies, misspellings and alternate classifications had to be resolved and corrected after the fact. To improve this situation, the INSDC partners agreed to send taxonomy consults to the NCBI when they first processed their entries, just as the GenBank indexing group does. As a consequence, the NCBI agreed that our public taxonomy pages would only show taxa that are linked to public sequence entries.
THE NCBI TAXONOMY DATABASE
The NCBI Taxonomy database was developed to fill a practical and very specific need—to provide nomenclature and classification for the source organisms in the sequence databases. In this respect, it differs from most existing taxonomy databases—we do not have the luxury of focusing on a particular area of expertise; we have to deal with names of all sorts that walk in the door on a daily basis with new sequence submissions. By its very nature, the taxonomy database is closely tied to the sequence databases—updates to the nomenclature and taxonomy are automatically reflected in the corresponding sequence entries. We try to maintain a phylogenetic taxonomy—one in which the structure of the classification corresponds with the evolutionary history of the tree of life. A phylogenetic classification aims to include only monophyletic groups—groups in which all of the members are more closely related to each other than any of them are to anything outside of the group. The traditional Reptilia, for example, is not a monophyletic group, since the crocodiles are more closely related to the birds than they are to the lizards and turtles. At the same time, the NCBI taxonomy is not generated automatically from the sequence data—rather, we try to reflect the current consensus in the systematic literature.
There are several large taxonomy database projects that seek to aggregate names from other sources into more or less comprehensive collections—the Catalog of Life, the Encyclopedia of Life, NameBank and WikiSpecies, for example. These are useful resources for the taxonomy group when we research the names that we add to our database, and we maintain reciprocal links with many of them. Even more useful are the curated specialty databases that are devoted to a particular group—IPNI for the plants, Index Fungorum and MycoBank for the fungi, Algaebase for the algae, AmphibiaWeb and Amphibian Species of the World for the amphibians, the Catalog of Fishes and FishBase for the fish, Bergey's Manual for the prokaryotes and so on. More than 150 outside groups are registered to maintain LinkOut (http://www.ncbi.nlm.nih.gov/projects/linkout/) links in the NCBI Taxonomy database. But in every case, the ultimate authoritative source for the nomenclature and classification is the primary taxonomic literature itself.
The NCBI Taxonomy database serves as an important entry point into the Entrez system for users who want to find all available information about a particular taxon, from the species level (and below) on up to genus, family, order and higher (or unranked) levels of the hierarchy. Many of the domains of Entrez (sequence, structure, genes, genomes, literature, etc.) are indexed by taxonomy in the [organism] search field, and these indices support reciprocal links between the taxonomy and the other domains of Entrez that are surfaced in the taxonomy browser.
HOW MANY SPECIES?
Since its inception, the NCBI Taxonomy database has paralleled the growth of the sequence databases themselves. How many species are represented in the database? This requires a little background into the structure of the database. By INSDC collaborative agreement, each entry in the sequence database must map into the taxonomy at or below the species level (an exception is made for patent entries). Each entry in the taxonomy database includes a primary name (the ‘scientific name’) and any number of secondary names, of several different name types. The primary name may either be a formal name (with standing in the relevant code of nomenclature) or an informal name (which represent putative species that have not yet been described in the literature, or specimens that have not been identified to a particular species). Environmental sample sequences constitute a special subset of informal names—these are sequences that have been recovered directly from the environment, with no direct knowledge of the source organism (apart from the sequence itself). The public taxonomy database currently (as of 26 September 2011) includes 234 991 species with formal names and another 405 546 ‘species’ with informal names (33 406 of which represent environmental samples). Counts of ‘species’ with informal names must be interpreted carefully, since many of these represent individual strains or specimens, not real putative species.
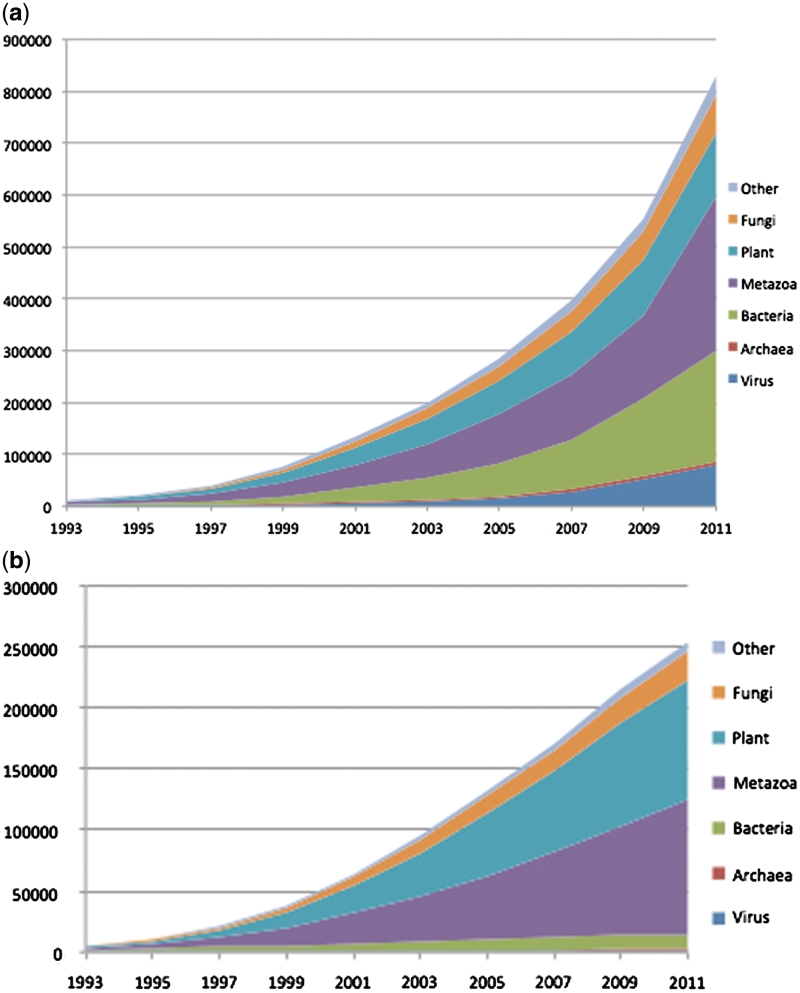
The taxonomy statistics page gives a summary of counts in the taxonomy database that can be customized in several ways—the default settings display counts only for species with formal names, but this page can be configured in many different ways (Figure 1)
Figure 1.
(a) Total growth of the taxonomy database. This includes formal and informal taxa at all levels, from unranked isolate-level taxids added for the influenza genome project to genera, families and higher taxa. (b) Valid species in the taxonomy database. This includes only valid binomial and trinomial species, subspecies, varietas and forma (infraspecific taxa with standing in the nomenclature). The viruses and bacteria are basically flat in this figure, since the rate-limiting step is the description of new species, not the sequencing.
There are three main codes of nomenclature—one for the animals (the ICZN) (4), one for plants, algae and fungi (the ICN, formerly the ICBN) (5,6) and one for the prokaryotes (the ICNB) (7,8). Each of these codes consists of a set of rules for publishing new taxonomic names in the scientific literature. There is also the ICTV for the viruses, which is not so much a code of nomenclature as an approved list of valid species names and classifications, maintained by a large set of committees, each responsible for a particular group of viruses (9). Formal names (except viruses) have ‘authorities’. The authority for a name is a reference to the taxonomic publication where the name was first described—much like a structured literature reference, e.g. Homo sapiens Linnaeus, 1758 and Caenorhabditis elegans (Maupas, 1900). These can take many complicated forms, but most are quite simple—the parenthesis in the second case indicate that this species was originally described under a different name, in this case Rhabditis elegans Maupas, 1900, and was transferred to the genus Caenorhabditis by a later author.
The taxonomy database currently includes 11 110 prokaryotic species with formal scientific names (as of 26 September 2011). This includes virtually all of the formally described species of prokaryotes (Bacteria and Archaea)—most are represented by at least a 16S rRNA sequence, as is every description of a new bacterial species. There are several wrinkles. If you sample the 16S rRNA sequences found in almost any environment, the vast majority of them do not closely resemble any of the formally described species of bacteria that are commonly studied in the laboratory. Furthermore, the bacterial code of nomenclature requires that the description of each new species include the designation of a ‘type strain’, a pure culture that must be deposited in at least two different culture collections. This means that bacteria that can not (or have not) been cultured cannot be formally described in the literature. ‘Candidatus’ nomenclature is an attempt to address this problem—names like Candidatus Liberibacter africanus are semi-formal species that can be cited in the literature, but have not been cultured in the laboratory. As of 26 September 2011, there were only 287 Candidatus species listed in the taxonomy database. The vast majority of prokaryotic diversity lies outside of the currently described taxa, and is likely to number in millions of species.
The taxonomy database currently includes 221 263 eukaryotic species with formal scientific names (as of 26 September 2011). Estimates of the number of eukaryotic species that have already been described in the literature vary widely, typically between 1.25 and 2 million. Given this uncertainty, the sequence databases currently contain at least a snippet of sequence from 10% to 20% of the described species of life on earth. Estimates of the total number of species on earth vary even more widely, typically 10 million or more (10).
The taxonomy database also includes 95 extinct species that are represented in the sequence databases, ranging in time from the woolly mammoth to Tyrannosaurus rex. In this context, it is important to note that GenBank and the taxonomy group do not (and cannot) attempt to verify the taxonomic identification that is provided by the submitter, unless the sequence itself points to an egregious misidentification. We rejected an earlier submission of dinosaur DNA that proved to be 99% identical to E. coli sequence, but the collagen protein fragment sequences submitted as coming from T. rex are not inconsistent with this identification.
MORE ABOUT NAMES
Names can be duplicated in many ways. For example, ‘black darter’ is the common name for both a fish (Sympetrum danae) and a dragonfly (Etheostoma duryi), while geranium is the common name for a species of plant (Pelargonium x hortorum) and the scientific name for a different genus of plants (Geranium). Names that actually mean the same thing can appear in multiple places in the classification. As mentioned above, we list the birds (within the Dinosauria) as sister group to the crocodilians (their closest living relatives). For retrieval purposes, we list the common name ‘reptiles’ and the formal name ‘Reptilia’ at three different nodes in our taxonomy (to pick up the turtles, the crocodiles and the lizards and snakes).
Duplicated scientific names are of particular interest to us. As mentioned above, formal names are regulated by codes of nomenclature. Each of the codes of nomenclature is different—they regulate different classes of names under different sets of rules. There is no real attempt to ensure that names are not duplicated between the domains of the codes of nomenclature—and in some cases, even within them. For example, the zoological code of nomenclature regulates names at the species, genus and family levels, but it does not require that names be unique between these sets. As a consequence, it is perfectly legal to find the damselfly genus Lestoidea in a superfamily of the same name. The zoological code does not regulate names above the family level, so we list the superclass Gnathostomata (the jawed vertebrates) and the superorder Gnathostomata (the sand dollars). Duplications between the codes are a bigger problem—we have come across hundreds of generic names that are valid under more than one code. Bacillus, for example, is a genus of bacteria and a genus of stick insects. Leptonema is a genus of plants, of bacteria, and of insects—and a genus of fossil fungi (fossils have a separate nomenclature of their own). The real problem (for the sequence database application) lies at the species level. With a large number of duplicated genus names, it is to be expected that the commonly used species epithets (americana, robusta, elegans, etc.) will result in duplicated names at the species level. We have come across six examples of duplicated binomials that are represented in the sequence databases (Table 1). In these cases, we use the full binomial name with the authority to disambiguate the entries.
Table 1.
Duplicated binomials in the sequence database
| Agathis montana Shestakov, 1932 | wasp | AJ302786 |
| Agathis montana de Laub., 1969 | conifer | U96478 |
| Rhaphidophora angulata (Miq.) Schott, 1860 | angiosperm | AY398512 |
| Rhaphidophora angulata Ingrisch, 2002 | cricket | |
| Rhaphidophora beccarii Engl., 1881 | angiosperm | AY398526 |
| Rhaphidophora beccarii Griffini, 1908 | cricket | |
| Gaussia princeps Scott, 1894 | copepod | AY015993 and CQ977721 |
| Gaussia princeps H. Wendl., 1865 | angiosperm | DQ227206 |
| Clusia flava Jacq., 1760 | angiosperm | AY145176, etc. |
| Clusia flava Meigen, 1830 | fly | FJ435902 |
| Tayloria grandis (Long) Goffinet and Shaw, 2002 | moss | AY039052 and AY039077 |
| Tayloria grandis Thiele, 1934 | land snail | HQ328315 and HQ328433 |
We use informal names for entries that are not identified to the species level with formal names. We try to avoid names like ‘Bacillus sp.’, and even names like ‘Bacillus sp. 1’ and ‘Bacillus sp. A’, which can easily be used by different researchers to denote different species. We do not distinguish between informal names that represent putative undescribed species (like Danio sp. ‘Hikari’ and Etheostoma cf. bellator A TJN-2011) and names that represent individual specimens which have not been assigned to a species (like Maytenus aff. obtusifolia Lombardi 7213 and Corallium sp. USNM 1075800).
Table 2 shows the various name types that are allowed in the taxonomy database.
Table 2.
TAXON name types
| Scientific name | Exactly one per node |
| Synonym | |
| Acronym | |
| anamorph | Asexual fungal name |
| teleomorph | Sexual fungal name |
| misspelling | Data not shown on public pages |
| misnomer | |
| equivalent name | |
| Includes | |
| in-part | |
| blast name | |
| Common name | |
| genbank common name | At most one per node |
| Genbank synonym | At most one per node |
| Genbank acronym | At most one per node |
| Genbank anamorph | At most one per node |
| unpublished name | Data not shown on public pages |
| Authority |
Unless otherwise specified, each name type may appear any number of times at a given node.
The ‘scientific name’ is the primary name for the node, and may either be a formal or an informal name. Synonyms may also be formal or informal names. The ‘equivalent name’ name type was added to tighten up our usage of synonyms—informal synonyms of formal scientific names should appear here, although this usage is not enforced. Acronyms are primarily used for the viruses, and common names for the higher eukaryotes. Misspellings are for incorrect forms of names that have previously appeared in sequence entries, as well as for misspellings that are found in the literature. These can be used in taxonomy lookups, but they do not appear on our web displays. Misnomers are for incorrect forms of names that aren’t quite misspellings—and for misspellings that we want to appear on our web displays. Other name types (includes and in-part) are for names which are useful as retrieval terms but which do not correspond with unique taxa in our classification (e.g. Reptilia).
The anamorph and teleomorph name types are specifically for use in the Fungi. Current practice allows fungal species to have two completely different scientific names depending on whether they are in the asexual, haploid (anamorph) or sexual, diploid (teleomorph) phase of their growth cycle. The most recent meeting of the botanical nomenclature section has addressed this confusing situation, and the new botanical code of nomenclature will mandate ‘one fungus, one name’. The current multiplicity of names should be resolved over the coming decades.
The ‘unpublished name’ is a particularly important new name type. It is becoming increasingly common to include a little bit of DNA sequence when describing a new species in the literature—the 16S rRNA sequence in prokaryotes, the barcode locus (COI for the animals, rbcL and matK for the plants, ITS for the fungi) and/or one of the other standard phylogenetic loci. This means that authors are coming to GenBank prior to publication to get accession numbers for sequences with ‘manuscript names’—proposed new species names that have not yet appeared in print. Our experience with the bacteria proved that the proposed new name would very often be changed during the editorial review process before the description of the new species was published, but that submitters would rarely get back in touch with us to update the name in their sequence entries. Furthermore, it can be very dangerous to expose these unpublished names—if they make their way into a taxonomic publication before the corresponding description is published they become nomen nudum (literally ‘naked name’) and are subsequently invalid. For these reasons we added the ‘unpublished name’ name type. These nodes are indexed with an informal name—our default formula uses the submitters’ initials and year of submission (rather like an informal authority for an informal name). For example, FN677936–FN677950 were originally submitted with the unpublished name Parapercis lutevittatus. These were indexed and released with the informal name Parapercis sp. TYC-2010. This species was eventually published as Parapercis lutevittata (11), and the name was updated in the taxonomy. At no point did the name Parapercis lutevittatus appear on our public web pages, although it could always be used as a successful search term (first as an unpublished name, and now as a misspelling).
The ‘GenBank’ name types are the way that we identify the ‘first among equals’ for use in display purposes. For common names and acronyms (which are informal name types), the ‘GenBank’ name type identifies the name that should appear in the GenBank flatfile. The ‘GenBank’ formal names (synonym and anamorph) are used in a much more limited manner—these are only assigned when two different names are in common use for the same species. For example, the valid taxonomic name for the torafugu pufferfish is Takifugu rubripes, but the junior synonym Fugu rubripes is common in much of the molecular biology literature (12). The ‘GenBank synonym’ name type ensures that both names will appear prominently in all the GenBank flatfiles from this species. We would do the same thing if a taxonomic revision forces a name change from Drosophila melanogaster to Sophophora melanogaster (13).
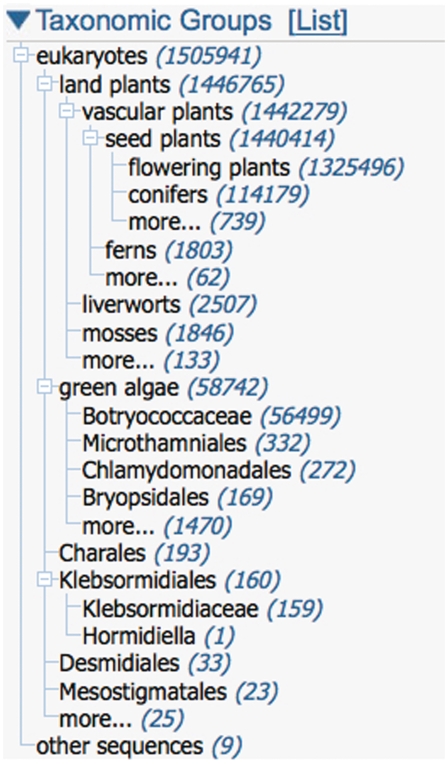
The ‘blast names’ are a special subset of common names for large, well-known taxa like the red algae, the mammals or the beetles. We have assigned 222 ‘blast names’ in the taxonomy. These are used for display purposes (in BLAST, in Taxonomy Entrez etc.) when a species name might not be generally recognizable. For example, many users will not recognize Cibotium barometz. Even when a common name is listed (Scythian lamb, in this case) it may not be informative—but the ‘blast name’ (ferns) is very helpful. Blast names are also used in the interactive taxonomy portlet found in many Entrez domains, since they provide an abbreviated, vernacular view of the classification (Figure 2).
Figure 2.
The taxonomy portlet in Nucleotide Entrez. This particular display summarizes the taxonomic distribution of plant sequences released in 2011, given by the Entrez query viridiplantae[orgn] AND 2011[pdat]. http://www.ncbi.nlm.nih.gov/nuccore?term=viridiplantae[orgn]+AND+2011[pdat] The taxonomy portlet toggles between a list of top taxa by entry count in the Entrez results list, and the taxonomic overview shown above.
ACCESS TO THE TAXONOMY DATABASE
The NCBI taxonomy is stored in an SQL Server relational database, called TAXON. The NCBI taxonomy group maintains the database with taxedit, a customized software tool. The database is taxon-centric; each node represents a taxonomic element (a taxon) and is identified with a numerical unique identifier (the taxid). Taxids are stable and persistent—they may be deleted (when taxa are removed from the database) and they may be merged (when taxa are synonymized), but they will never be reused to identify a different taxon. Names are associated with nodes, and each taxid is linked to its parent taxid. The root node (taxid 1) links to itself.
Public access to the taxonomy database is provided in three different ways—the Taxonomy Browser (which is updated in real time as we edit the database), the Taxonomy domain of Entrez (which is updated daily) and the taxonomy ftp site (which is updated hourly).
http://www.ncbi.nlm.nih.gov/taxonomy
http://www.ncbi.nlm.nih.gov/Taxonomy/Browser.wwwtax.cgi
ftp://ftp.ncbi.nih.gov/pub/taxonomy
Taxonomy was the first database to be added to the Entrez system after the initial triad of Nucleotide, Protein and PubMed. As with other Entrez databases, Taxonomy Entrez supports Boolean queries, a History function and an array of search fields. Some of the search fields are common across all Entrez databases—Date (the date the object first appeared in Entrez), Filter (links to internal and external databases) and Properties (many useful search terms)—others are specific to Taxonomy (e.g. Rank). Taxonomy was the first Entrez database to have an internal hierarchical structure. Taxonomy search fields and search history can be browsed on the Advanced Search page. Because Entrez deals with unordered sets of objects in a given domain, we introduced two new fields to represent the hierarchy—the Lineage field indexes all of the taxa in the hierarchy above a given node in the taxonomy, and Subtree indexes all of the taxa below it. Several useful queries are shown in Table 3.
Table 3.
Some useful Entrez queries
| all [filter] | Retrieves everthing |
| Specified [property] | Formal binomial and trinomial |
| at or below species level [property] | |
| family [rank] | Rank-based query |
| taxonomy genome [filter] | Taxa with a direct link to a genome sequence |
| 2009/10/21:2020 [date] | Date-bounded query |
| mammalia [subtree] | All taxa within the Mammalia |
| extinct [property] | Extinct organisms |
| Terminal [property] | Terminal nodes in the tree |
| loprovencyclife [filter] | Entries with LinkOut links to the Encyclopedia of Life |
These can be combined in Boolean expressions, e.g. mammalia [subtree] AND specified [prop] AND subspecies [rank] AND 2009 [date].
All of the tools developed for Entrez are available for use with Taxonomy Entrez. Taxonomy Entrez search results can be downloaded in several formats using the ‘Send to File’. The E-utilities (14) facility can be used to query and retrieve entries from Taxonomy in Perl scripts. Taxonomy Entrez queries can be saved in MyNCBI (15), and the user can register to receive periodic email updates (What's New) whenever anything new in Entrez satisfies the query. For example, one can register the query ‘specified [property]’, and ask to receive a weekly (or monthly, or daily) email with the list of species that have appeared in the sequence databases for the first time in the last week.
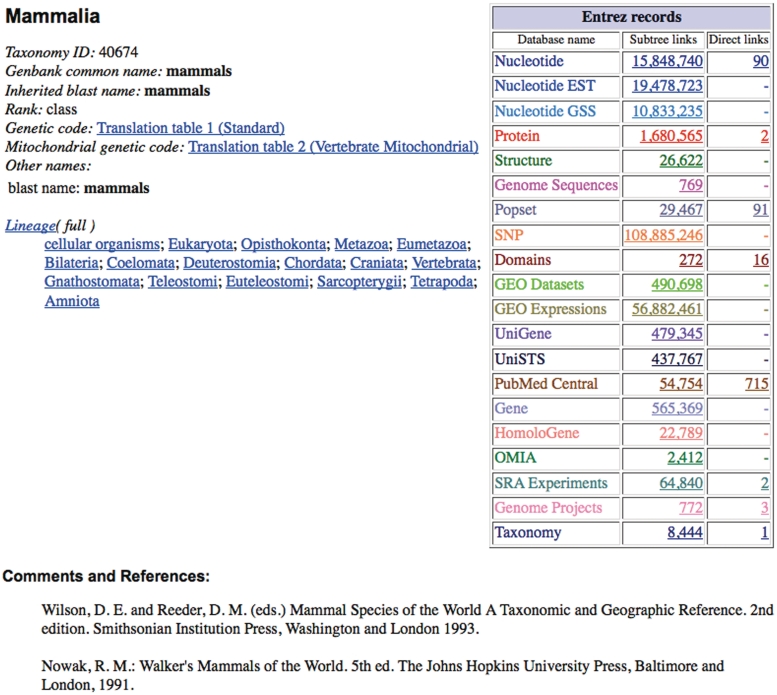
Taxonomy Entrez provides some powerful tools for searching the taxonomy, but it is not a natural way to explore a hierarchical data set. The Taxonomy Browser provides this facility. The browser supports two different kinds of web pages—hierarchy pages, which present the familiar indented view of the taxonomic classification, and taxon-specific pages, which summarize all of the information that we associate with a particular taxonomic entry in the database. By default, the hierarchy displays three levels in the classification, but this can be changed (asking for zero levels displays the taxon-specific page). The hierarchy pages can also be customized to display hotlinked counts of entries in other Entrez databases (Figure 3).
Figure 3.
Taxonomy browser page for the Mammalia. Exploded and unexploded links to other Entrez database are shown in ‘Entrez records’. LinkOut links to external databases are displayed below the Comments and References (data not shown).
The taxon-specific pages display several different kinds of information, starting with all of the names associated with the entry in the taxonomy database (except for misspellings and unpublished names, as discussed above). The lineage line displays toggles between the full and abbreviated taxonomic classification for the entry (the abbreviated lineage appears in the GenBank sequence entries). The taxonomy group may also manually curate comments, and hotlinks to literature references either in PubMed or at arbitrary URL addresses in the Web. The ‘Entrez records’ table shows links to other Entrez databases, in two columns—‘Subtree links’ and ‘Direct links’ (also called ‘exploded’ and ‘unexploded’ links). The direct (unexploded) links retrieve entries that map directly to this taxon; the subtree (exploded) links retrieve all of the entries that map into the taxonomy at or below this taxon. Many databases (Nucleotide, Protein, Structure, etc.) typically map into the taxonomy at or below the sequence level, and entries that break that rule are either annotation errors or exceptions (e.g. the 47 entries with /organism=‘Hominidae’ all patent sequences). The default Entrez links from Taxonomy to these Entrez databases follow the exploded links—from Mammalia in taxonomy, we want to retrieve all of the mammalian sequences in GenBank (not just the ones with /organism=‘Mammalia’). The literature domains are different—following the direct links to PubMed Central will find all of the articles that mention the ‘Mammalia’. These are likely to be the articles of interest, and not every paper that uses Chinese hamster cell lines or inbred mouse strains. Links to the Entrez Popset domain (the database of population studies and phylogenetic sets) are another special case—the direct links will retrieve every phylogenetic set that spans the taxon of interest, while the subtree links will include all of the sets that are completely contained within the taxon.
LinkOut links are also prominently displayed on the browser pages. LinkOut is a facility supported by the NCBI that allow outside users to maintain detailed sets of hotlinks from entries in Entrez back to specific web pages on their own sites. It was first developed to allow publishers to put links on PubMed abstracts back to the full-text articles on their own sites, but it has since been extended to serve all of the domains of Entrez. LinkOut users are given an ftp site at the NCBI where they can upload files that describe how to build the links they would like to support. For example, the Encyclopedia of Life supports links back to species pages at eol.org, and Rod Page supports the links to WikiSpecies.
It is easy to build URLs that link to specific pages in the Taxonomy browser (see Linking to Taxonomy, on the Taxonomy home page) and to build URLs that evaluate specific queries in Taxonomy Entrez (see Linking to Records in the Entrez System, in Entrez Help on the NCBI Bookshelf).
The Taxonomy browser also supports several search capabilities that are not available in the generic Entrez search—in particular, the ‘wild card’ search mode uncovers two entries that match ‘E* coli’, and 79 entries that match ‘C* elegans’. There is only a very limited wild-card search capability within Entrez itself.
We provide two other useful tools relevant to the taxonomy database—the name/id status page and the common tree viewer. Upload a list of names (or a list of taxids) into the status page to see a report of their current status in the NCBI taxonomy database. Save copies of the report and track differences to follow changes in the classification and nomenclature of a set of taxa of particular interest. A command-line version of this function (taxident) is available in the NCBI C++ toolkit. Upload a list of names (or a list of taxids) into the common tree viewer to see the subset of the NCBI taxonomy that spans that set of nodes. The common tree view is also one of the display formats once you have selected a set of nodes in Taxonomy Entrez. The tree can be saved in several standard formats—text file, phylip tree (Newick format) and taxid list.
The taxonomy ftp site includes table dumps from the TAXON database that are sufficient to recreate the taxonomy. There is a terse README, but the two crucial files are nodes.dmp (which maps taxids to their parent taxids) and names.dmp (which maps names to taxids). delnodes.dmp lists nodes that have been deleted from the database, as well as nodes that were once public but are no longer linked to any public sequence entries. merged.dmp maps secondary taxids onto primary taxids for taxa that have been synonymized in the database.
FUTURE DIRECTIONS
There are several initiatives underway, notably the Barcodes of Life (16) initiative, that are actively focused on sequencing reference specimens from every eukaryotic species of life on the planet. These efforts should lead to a rapid expansion of the NCBI taxonomy database over the coming years.
FUNDING
Intramural Research Program of the National Institutes of Health, National Library of Medicine. Funding for open access charge: Intramural Research Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991;19:2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker WC, George DG, Hunt LT, Garavelli JS. The PIR protein sequence database. Nucleic Acids Res. 1991;19:2231–2236. doi: 10.1093/nar/19.suppl.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuler GD, Epstein JA, Ohkawa H, Kans JA. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1966;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 4.Ride WDL, Cogger HG, Dupuis C, Kraus O, Minelli A, Thompson FC, Tubbs PK, editors. International Code of Zoological Nomenclature. 1999. 4th edn. International Trust for Zoological Nomenclature, The Natural History Museum, London http://www.nhm.ac.uk/hosted-sites/iczn/code/ (23 November 2011, date last accessed) [Google Scholar]
- 5.McNeill J, Barrie FR, Burdet HM, Demoulin V, Hawksworth DL, Marhold K, Nicolson DH, Prado J, Silva PC, Skog JE, et al., editors. International Code of Botanical Nomenclature (Vienna Code). Regnum Vegetabile. 2006;Vol. 146 A.R.G. Ruggell, Liechtenstein, Gantner Verlag KG. http://ibot.sav.sk/icbn/main.htm (23 November 2011, date last accessed) [Google Scholar]
- 6.Miller J, Funk V, Wagner W, Barrie FR, Hoch PC, Herendeen P. Outcomes of the 2011 Botanical Nomenclature Section at the XVIII International Botanical Congress. Phytokeys. 2011;5:1–3. doi: 10.3897/phytokeys.5.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaPage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR, Clark WA, editors. International Code of Nomenclature of Bacteria: Bacteriological Code (1990 Revision) 1992. ASM Press, Washington D.C. http://www.ncbi.nlm.nih.gov/books/NBK8817/ (23 November 2011, date last accessed) [PubMed] [Google Scholar]
- 8.Euzéby JP. Altertions to the bacteriological code (1990 Revision) In: Euzéby JP, editor. List of Prokaryotic Names with Standing in Nomenclature. 2011. http://www.bacterio.cict.fr/code.html (23 November 2011, date last accessed) [Google Scholar]
- 9.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2011. [Google Scholar]
- 10.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. How many species are there on earth and in the ocean? PLoS Biol. 2011;9:e100127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao YC, Cheng TY, Shao KT. Parapercis lutevittata, a new cryptic species of Parapercis (Teleostei: Pinguipedidae) from the western Pacific based on morphological evidence and DNA barcoding. Zootaxa. 2011;2867:32–42. [Google Scholar]
- 12.Matsuura K. The pufferfish genus Fugu Abe, 1952, a junior subjective synonym of akifugu Abe, 1949. Bull. Natn. Sci. Mus. Tokyo. 1990 Ser. A, 18, 15–20. [Google Scholar]
- 13.Dalton R. What's in a name? Fly world is abuzz. Nature. 2010;464:825. doi: 10.1038/464825a. [DOI] [PubMed] [Google Scholar]
- 14.NCBI Help Manual. Entrez Programming Utilities Help. 2010. National Center for Biotechnology Information, Bethesda, MD. http://www.ncbi.nlm.nih.gov/books/NBK25501/ (23 November 2011, date last accessed) [Google Scholar]
- 15.NCBI Help Manual. My NCBI Help. 2010. National Center for Biotechnology Information, Bethesda, MD. http://www.ncbi.nlm.nih.gov/books/NBK3843/ (23 November 2011, date last accessed) [Google Scholar]
- 16.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]